NUCLEOTIDES NUCLEIC ACID STRUCTURE AND FUNCTION Prof Dr







































- Slides: 113

NUCLEOTIDES, NUCLEIC ACID STRUCTURE AND FUNCTION Prof. Dr. Yıldız DİNÇER Cerrahpaşa Medical Faculty Department of Biochemistry

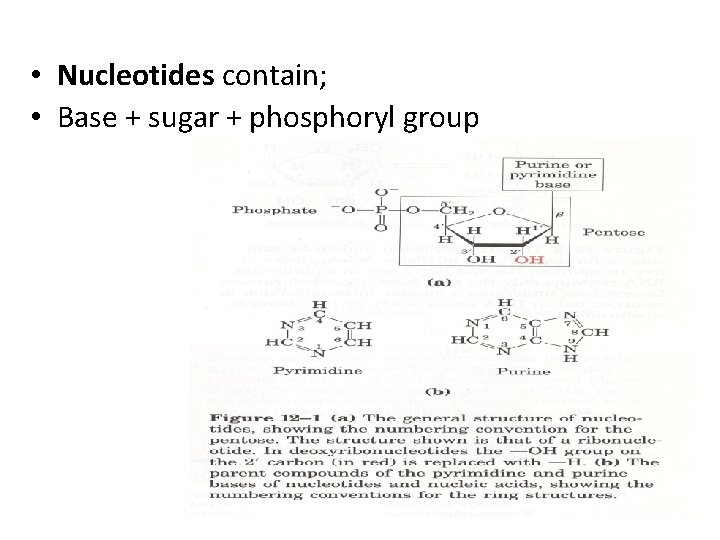
• Nucleotides contain; • Base + sugar + phosphoryl group

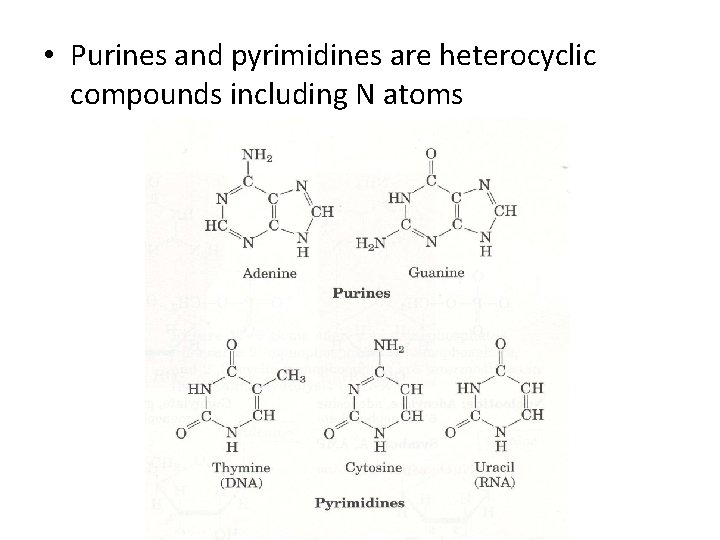
• Purines and pyrimidines are heterocyclic compounds including N atoms

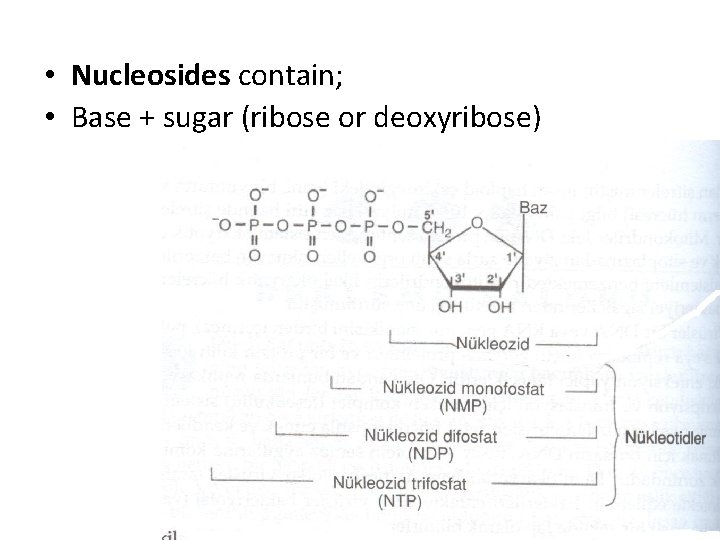
• Nucleosides contain; • Base + sugar (ribose or deoxyribose)

• Sugar, D-ribose or 2’-deoxy D-ribose, is linked to base via a covalent β-N-glycosidic bond to N -9 of a purine or to N-1 of a pyrimidine • Nucleotides are termed ribonucleotides or deoxyribonucleotides based on whether the sugar is ribose or 2’-deoxyribose

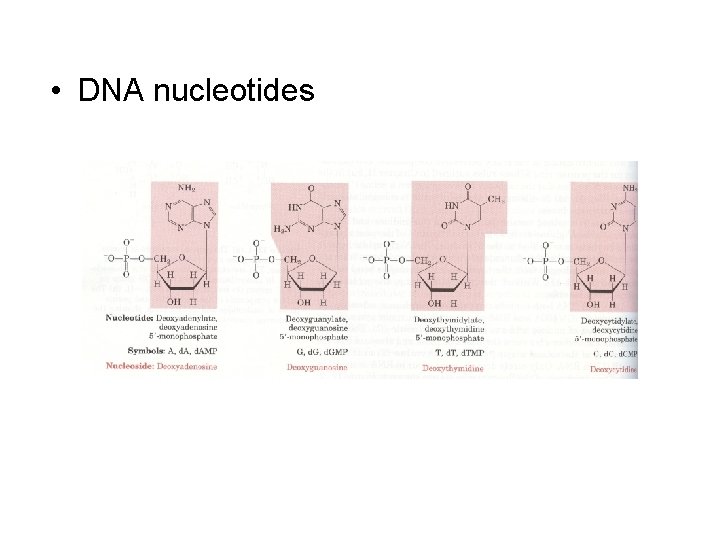
• DNA nucleotides

• RNA nucleotides

Nucleotides are phosphorolated nucleosides • Mononucleotides are nucleotides singly phosphorolated on hydroxyl group of the sugar • AMP → Adenosine monophosphate • Adenine + ribose +phosphate • Additional phosphates linked by acid anhydride bonds to the existing phosphate of a mononucleotide form nucleoside di- and triphosphates

• ADP → Adenosine diphosphate • Adenine + ribose +phosphate • ATP → Adenosine triphosphate • Adenine + ribose +phosphate + phosphate

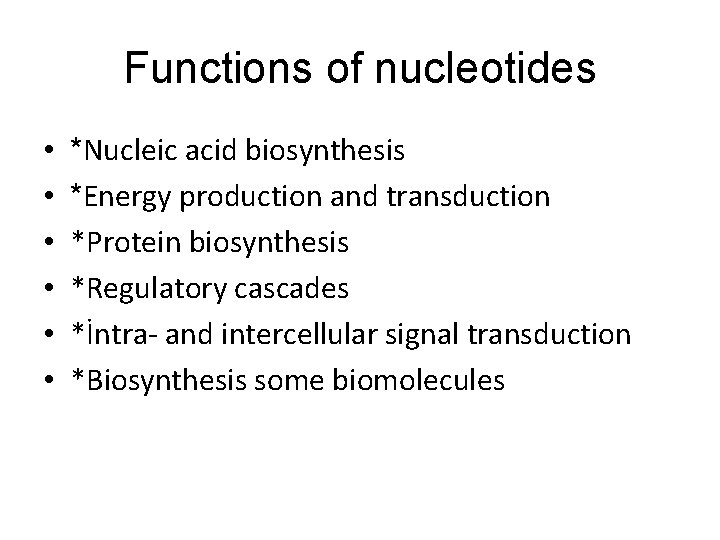
Functions of nucleotides • • • *Nucleic acid biosynthesis *Energy production and transduction *Protein biosynthesis *Regulatory cascades *İntra- and intercellular signal transduction *Biosynthesis some biomolecules

Some properties of nucleotides • 1. Mononucleotides have a negative charge at physiological p. H • 2. Nucleotides absorb UV light • 3. Many coenzymes are nucleotide derivatives • 4. Synthetic nucleotide analogs are used in chemotherapy • 5. Nucleoside triphosphates have high group transfer potential • 6. Some nucleotides are involved in signal transduction

• 1. Mononucleotides have a negative charge at physiological p. H. The p. Ks of the primary and secondary phosphoryl groups are about 1. 0 and 6. 2, respectively • Nucleosides and or free purine or pyrimidine bases are uncharged at physiological p. H

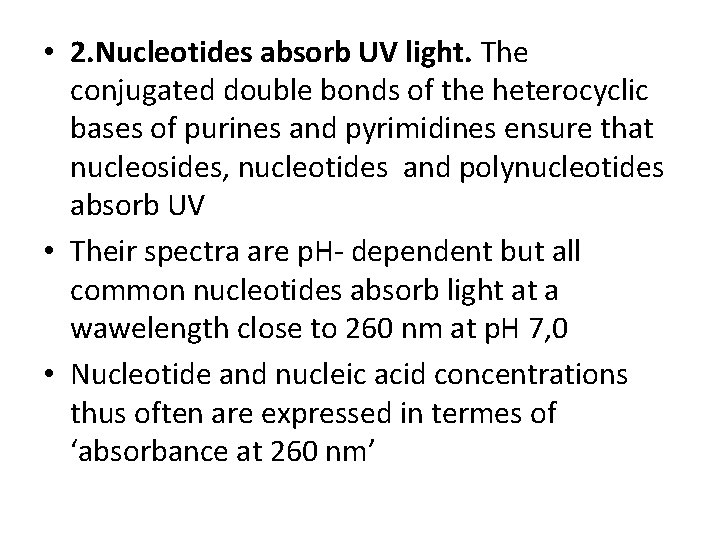
• 2. Nucleotides absorb UV light. The conjugated double bonds of the heterocyclic bases of purines and pyrimidines ensure that nucleosides, nucleotides and polynucleotides absorb UV • Their spectra are p. H- dependent but all common nucleotides absorb light at a wawelength close to 260 nm at p. H 7, 0 • Nucleotide and nucleic acid concentrations thus often are expressed in termes of ‘absorbance at 260 nm’

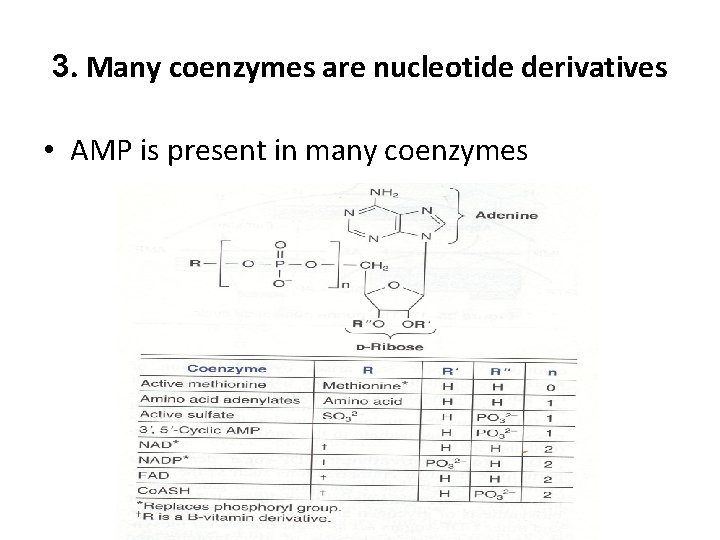
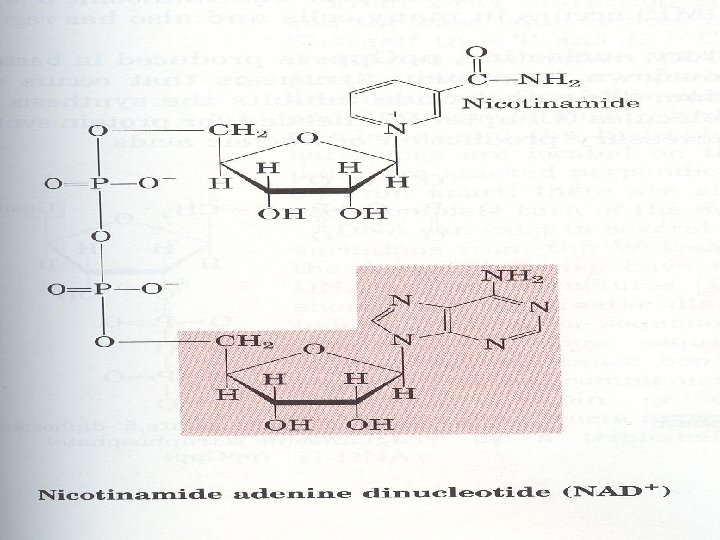
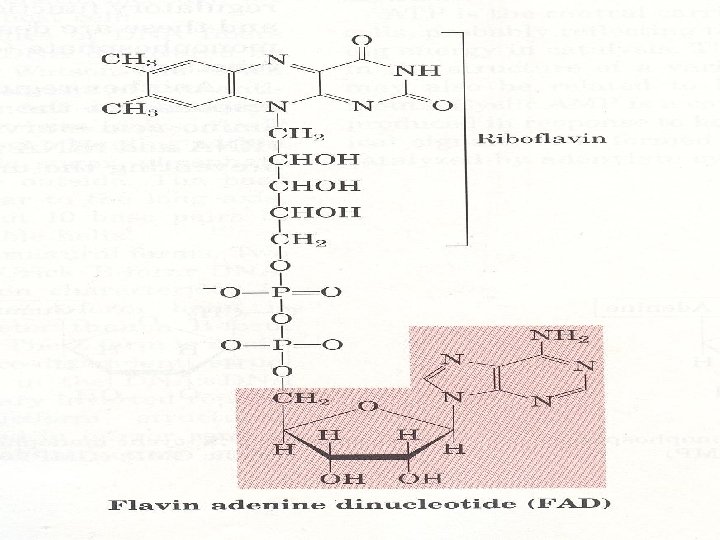
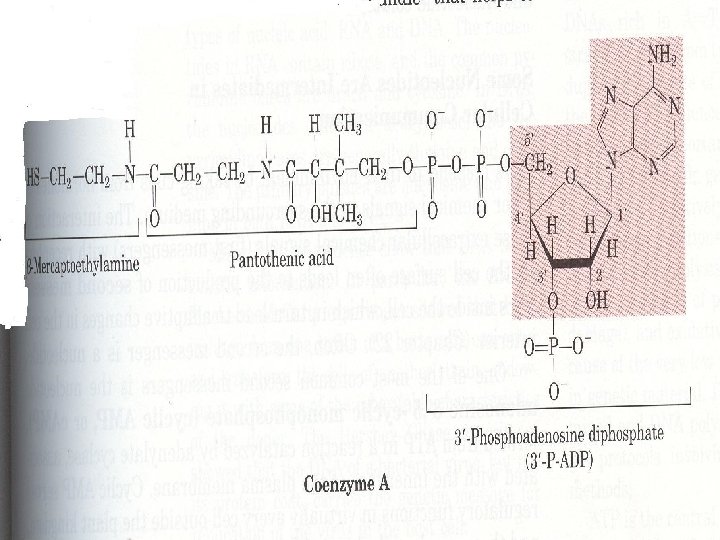
3. Many coenzymes are nucleotide derivatives • AMP is present in many coenzymes




• 4. Synthetic nucleotide analogs are used in chemotherapy • Chemically synthesized analogs of purines and pyrimidines, their nucleosides and their nucleotides find numerious applications in clinical medicine • Administration of an analog in which either the heterocyclic ring or the sugar moiety has been altered induces toxic effect when the analog is incorporated into specific cellular constituents

• Their effects reflect one of two processes: • a)Inhibition by the drug of specific enzymes essential for nucleic acid synthesis • b)Incorporation of metabolites of the drug into nucleic acids, thus they effect the base pairing required for accurate replication

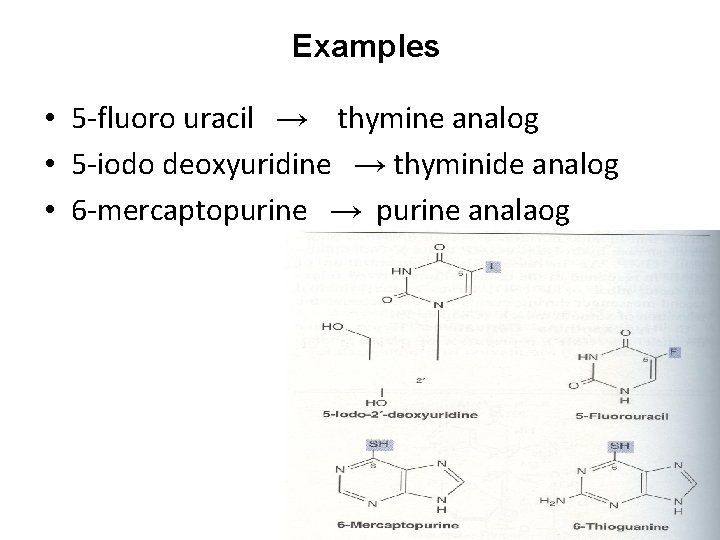
Examples • 5 -fluoro uracil → thymine analog • 5 -iodo deoxyuridine → thyminide analog • 6 -mercaptopurine → purine analaog

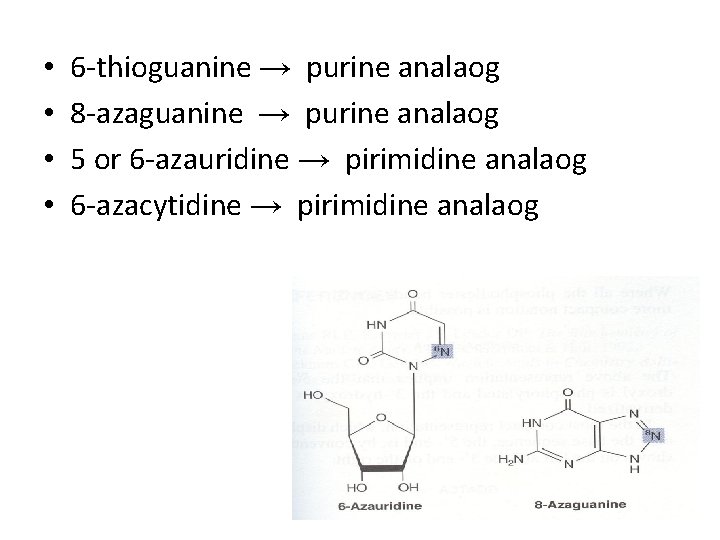
• • 6 -thioguanine → purine analaog 8 -azaguanine → purine analaog 5 or 6 -azauridine → pirimidine analaog 6 -azacytidine → pirimidine analaog

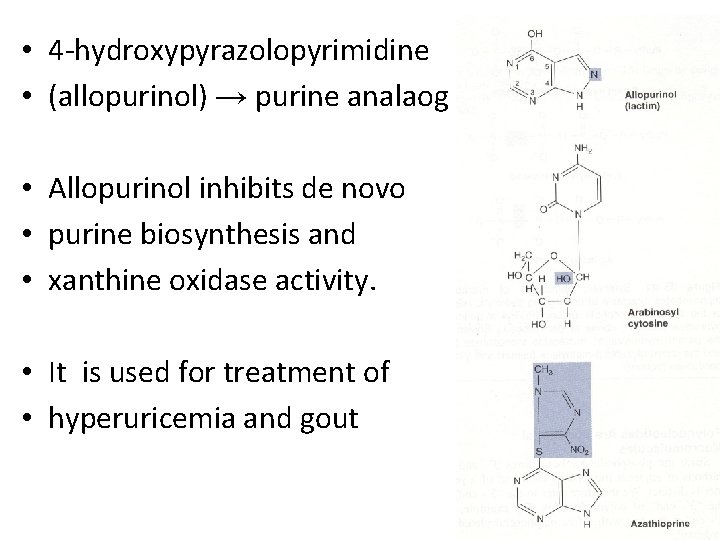
• 4 -hydroxypyrazolopyrimidine • (allopurinol) → purine analaog • Allopurinol inhibits de novo • purine biosynthesis and • xanthine oxidase activity. • It is used for treatment of • hyperuricemia and gout

• The nucleoside, cytarabine (arabinosyl cytosine), in which arabinose replaces ribose, is used in chemotherapy and in treatment of viral infections • Azathioprine is catabolized to 6 mercaptopurine and is used during organ transplantation to suppress immunological rejection

• 5. Nucleoside triphosphates have high group transfer potential because of acid anhydride bonds • High group transfer potential of nucleoside triphosphates allows them to participate as group transfer reagents in various reactions. • In these reactions, cleavage of an acid anhydride bond is coupled with a highly endergonic reaction such as protein synthesis or nucleic acid synthesis.

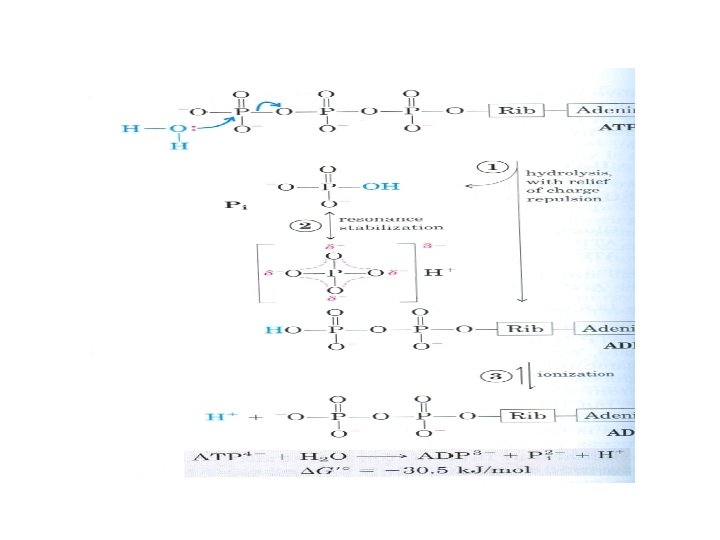
• ADP and ATP are substrates and products, respectively, for oxidative phosphorylation • ATP serves as the major biologic transducer of free energy • It is the most abundant free nucleotide in mammalian cells (1 m. M) • ATP donates some of its chemical energy by hydrolysis of the terminal phosphoanhydride bond

• The hydrolytic cleavage of the terminal phosphoanhydride bond in ATP separates one of the three negatively charged phosphates and thus relieves some of the electrostatic repulsion in ATP • Released Pi is stabilized by the formation of several resonance forms not possible in ATP • ADP 2 - is the other product of hydrolysis and it immediately ionizes, releasing H+ into a medium of very low [H+]


• For hydrolysis reactions with large, negative standard free energy changes, the products are more stable than the reactants

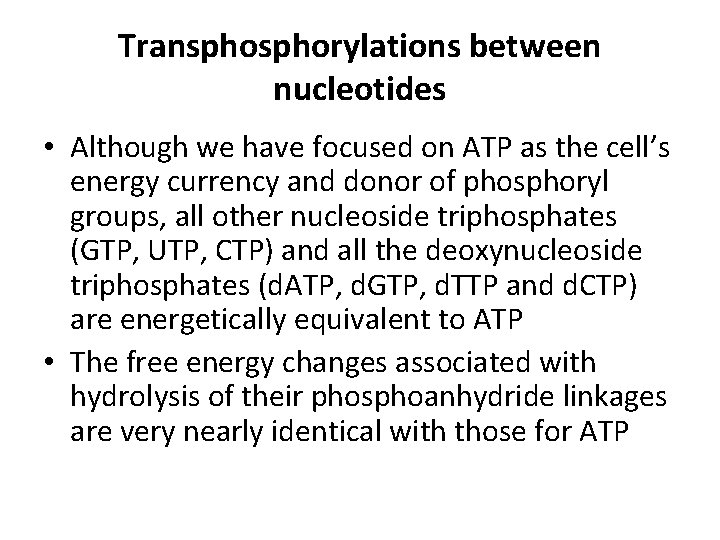
Transphorylations between nucleotides • Although we have focused on ATP as the cell’s energy currency and donor of phosphoryl groups, all other nucleoside triphosphates (GTP, UTP, CTP) and all the deoxynucleoside triphosphates (d. ATP, d. GTP, d. TTP and d. CTP) are energetically equivalent to ATP • The free energy changes associated with hydrolysis of their phosphoanhydride linkages are very nearly identical with those for ATP

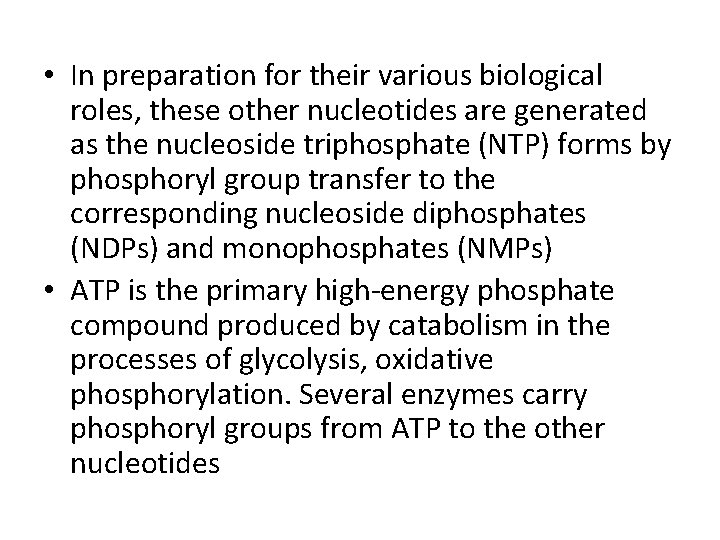
• In preparation for their various biological roles, these other nucleotides are generated as the nucleoside triphosphate (NTP) forms by phosphoryl group transfer to the corresponding nucleoside diphosphates (NDPs) and monophosphates (NMPs) • ATP is the primary high-energy phosphate compound produced by catabolism in the processes of glycolysis, oxidative phosphorylation. Several enzymes carry phosphoryl groups from ATP to the other nucleotides

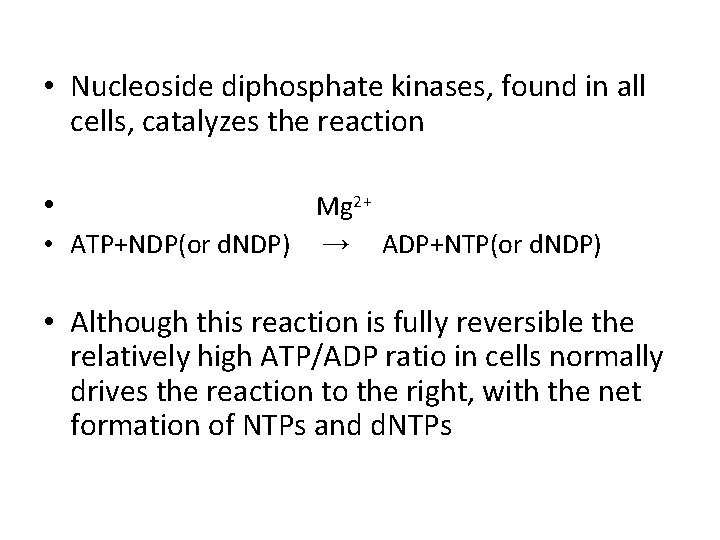
• Nucleoside diphosphate kinases, found in all cells, catalyzes the reaction • Mg 2+ • ATP+NDP(or d. NDP) → ADP+NTP(or d. NDP) • Although this reaction is fully reversible the relatively high ATP/ADP ratio in cells normally drives the reaction to the right, with the net formation of NTPs and d. NTPs

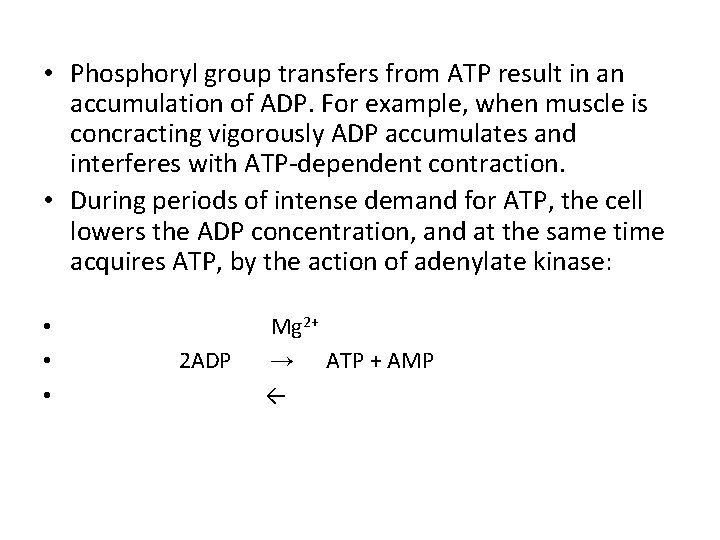
• Phosphoryl group transfers from ATP result in an accumulation of ADP. For example, when muscle is concracting vigorously ADP accumulates and interferes with ATP-dependent contraction. • During periods of intense demand for ATP, the cell lowers the ADP concentration, and at the same time acquires ATP, by the action of adenylate kinase: • • • 2 ADP Mg 2+ → ATP + AMP ←

• This reaction is fully reversible, so after the intense demand for ATP ends, the enzyme can recycle AMP by converting it to ADP which can then be phosphorylated to ATP in mitochondria • A similar enzyme guanylate kinase, converts GMP to GDP at the expense of ATP. By-pathways such as these, energy conserved in the catabolic production of ATP is used to supply the cell with all required NTPs and d. NTPs

6. Some nucleotides are involved in signal transduction • c. AMP and c. GMP

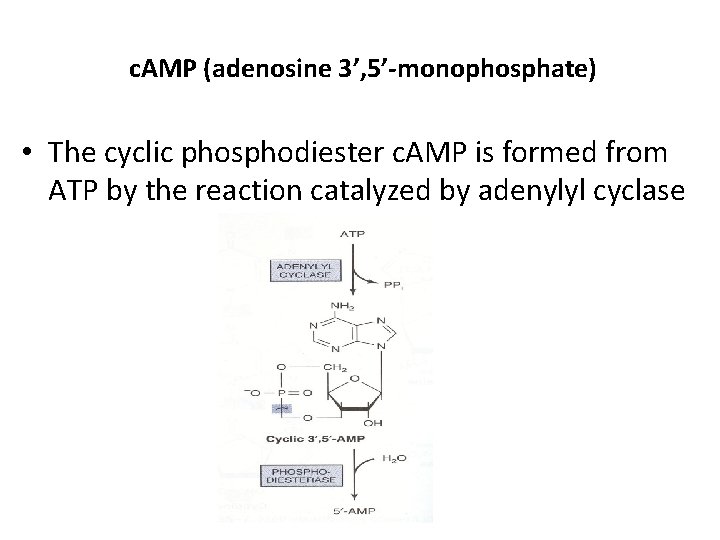
c. AMP (adenosine 3’, 5’-monophosphate) • The cyclic phosphodiester c. AMP is formed from ATP by the reaction catalyzed by adenylyl cyclase

• c. AMP is a second messenger in signal transduction • Adenylyl cyclase activity is regulated by complex interactions, many of which involve hormon receptors • As a second messenger, c. AMP participates numerous regulatory functions by activating c. AMP dependent protein kinases • c. AMP is broken down by c. AMP phosphodiesterase

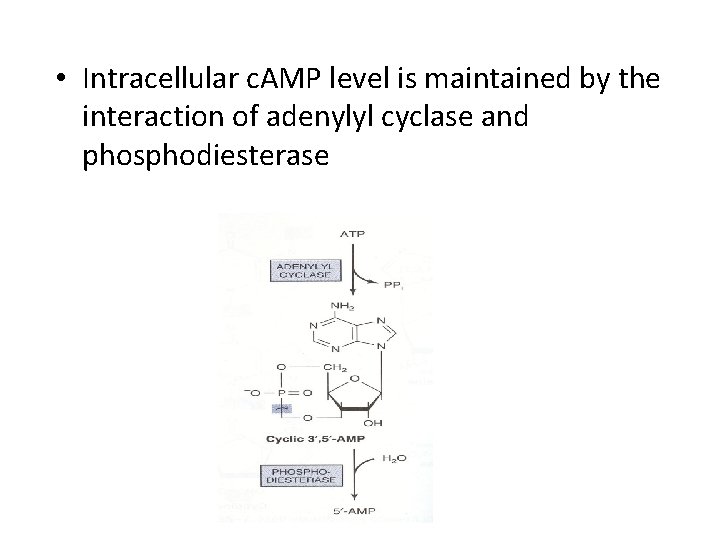
• Intracellular c. AMP level is maintained by the interaction of adenylyl cyclase and phosphodiesterase

c. GMP (guanosine 3’, 5’-monophosphate) • c. GMP is a second messenger in signal transduction that can act antagonistically to c. AMP • c. GMP is formed from GMP by guanylyl cyclase • Both adenylyl and guanylyl cyclases are regulated by effectors that include hormones • A phosphodiesterase hydrolyzes c. GMP to GMP

• An increase in the level of the c. GMP as response to the nitric oxide serves as the main second messenger during events that characterize the relaxation of smooth muscle

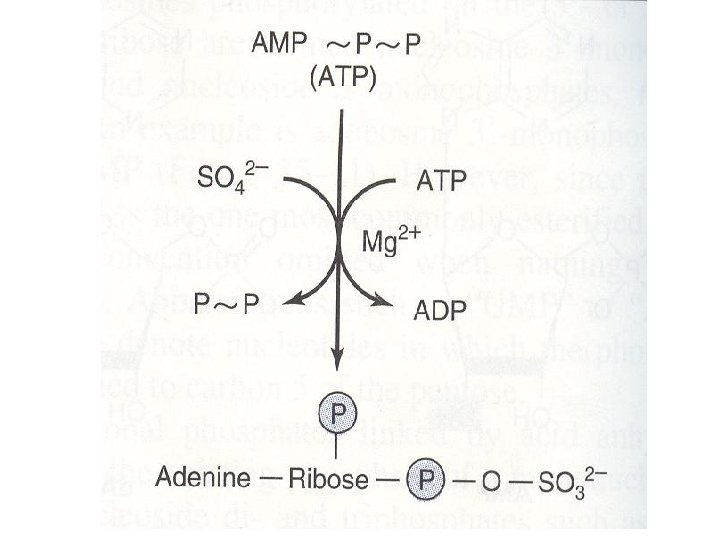
Other functions of free nucleotides • 1. ADENOSINE DERIVATIVE NUCLEOTIDES • Adenosine 3’-phosphate 5’-phosulfate (active sulfate) • This is the sulfate donor for the formation of the sulfated proteoglycans or urinary metabolites of drugs excreted as sulfate conjugates


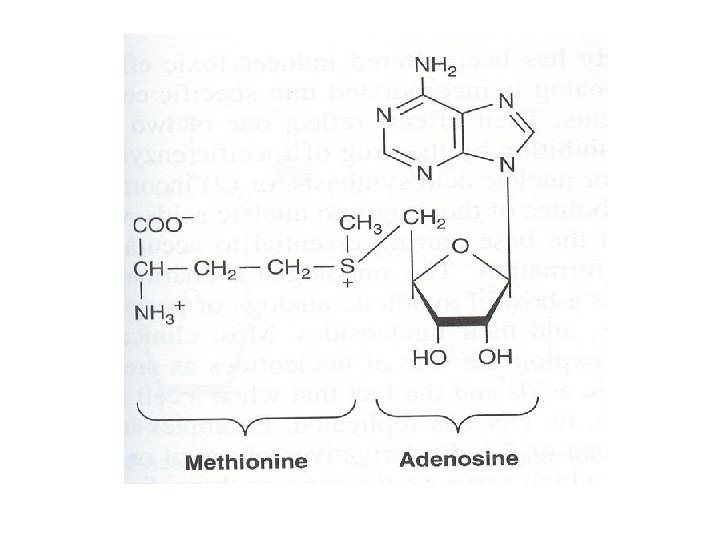
S-adenosylmethionine (SAM) • This is a form of active methionine • SAM serves as a methyl group donor for methylation reactions • SAM forms propylamine which is required in polyamine synthesis


2. GUANOSINE DERIVATIVE NUCLEOTIDES • Guanosine nucleotides participate in the conversion of succinyl-Co. A to succinate, a reaction that is coupled to the substrate level phosphorylation of GDP to GTP • GTP is required for • * activation of adenylyl cyclase (as a allosteric regulator) • **protein synthesis (as a energy source)

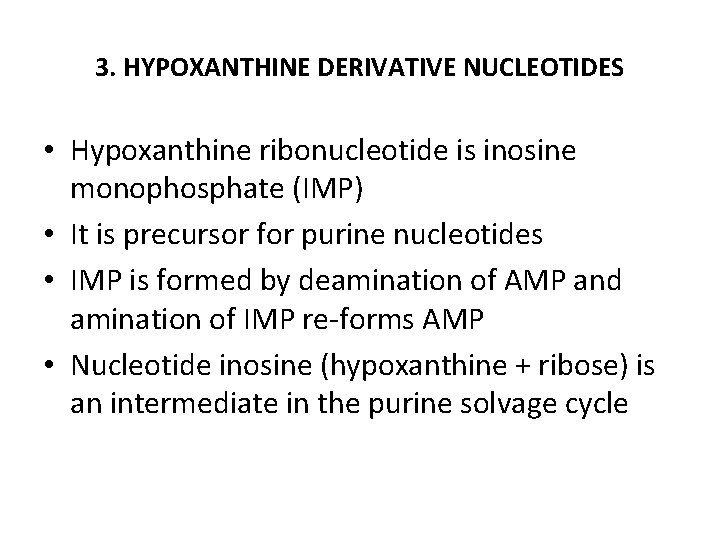
3. HYPOXANTHINE DERIVATIVE NUCLEOTIDES • Hypoxanthine ribonucleotide is inosine monophosphate (IMP) • It is precursor for purine nucleotides • IMP is formed by deamination of AMP and amination of IMP re-forms AMP • Nucleotide inosine (hypoxanthine + ribose) is an intermediate in the purine solvage cycle

4. URACYL DERIVATIVE NUCLEOTIDES • UDP-sugar derivatives participate in sugar epimerization • UDP-glucose is the glucosyl donor for biosynthesis of glycogen and glucosyldisaccharides • Other UDP-sugars act as sugar donors for biosynthesis of oligosaccharides of glycoproteins and proteoglycans • UDP-glucuronic acid is the glucuronyl group donor for conjugation reactions

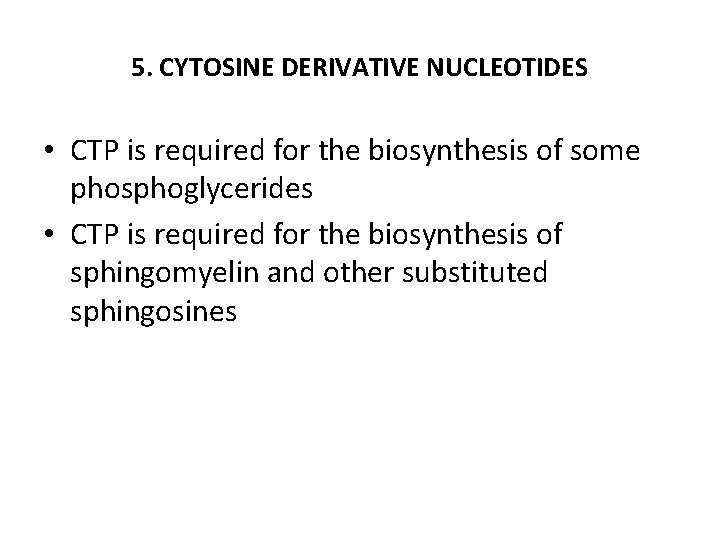
5. CYTOSINE DERIVATIVE NUCLEOTIDES • CTP is required for the biosynthesis of some phosphoglycerides • CTP is required for the biosynthesis of sphingomyelin and other substituted sphingosines

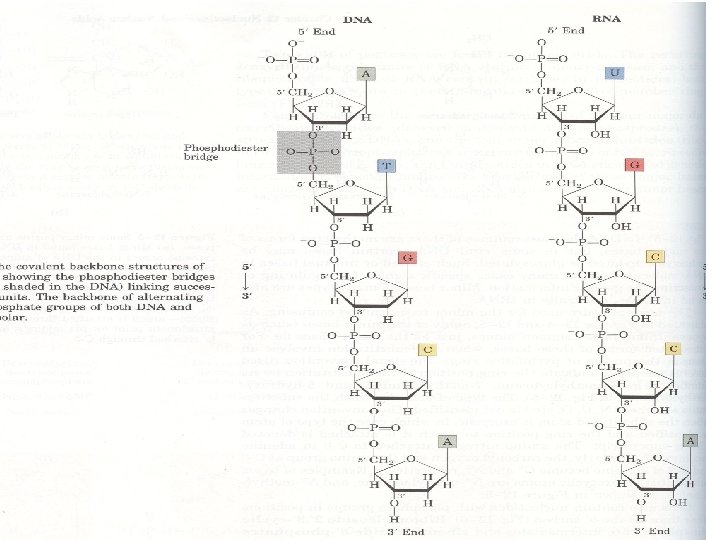
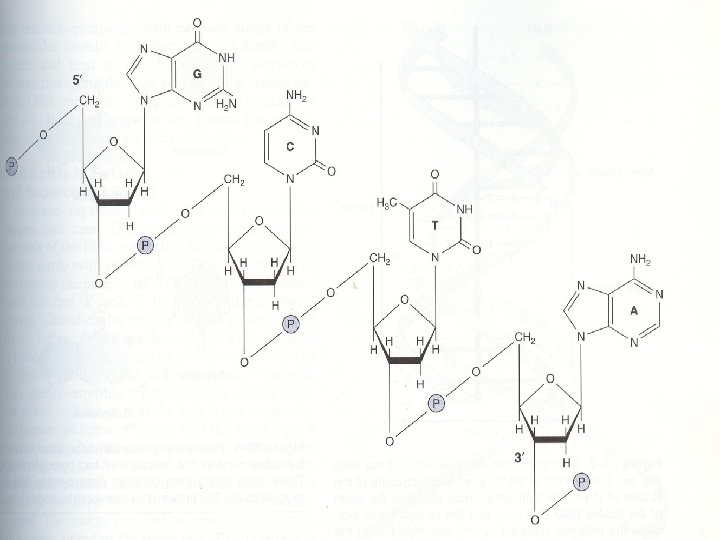
Polynucleotides • Mononucleotides are covalently linked through phosphate-group ‘bridges’ • Specifically, the 5’-OH group of one nucleotide unit is joined to the 3’-OH group of the next nucleotide by a phosphodiester linkage resulting to release of one molecule water • This results in a dinucleotide • The term oligonucleotide is used for polymers containing 50 or fewer nucleotides


• Polynucleotides are directional macromolecules • Since the phosphodiester bond links 3’- and 5’carbons of adjacent monomers, each end of a polimers is distinct. They are reffered as ‘ 3’-end’ or ‘ 5’-end’ of polynucleotides • In the most representations displaying only the base sequences 5’ end is shown on the left and 3’ end is shown on the right: • 5’ GTATTGC 3’ •

• Nucleic acids are the polimers containing many nucleotides linked by phosphodiester linkage • Hydrolysis of polynucleotides has a large energy barrier and this reaction is too slow in the absence of phosphodiesterases • Phosphodiester linkes are hydrolyzed by phosphodiesterases

Nucleic acid structure and function • Genetic information is coded by DNA • Genes control the synthesis of various types of RNA which are involved in protein synthesis • Nucleic acids (DNA and RNA) are the polimers containing nucleotides as monomers. • Covalent backbones of nucleic acids consist of alternating phosphate and pentose residues, and the characteristic bases may be regarded as side groups joint to the backbone at regular intervals

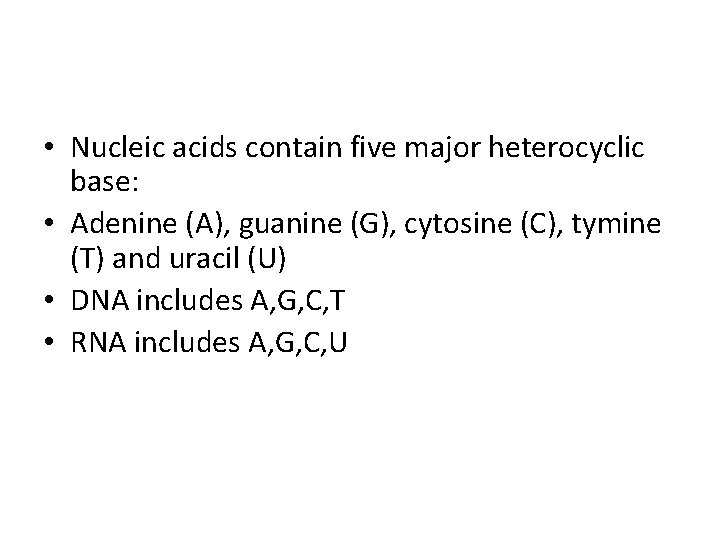
• Nucleic acids contain five major heterocyclic base: • Adenine (A), guanine (G), cytosine (C), tymine (T) and uracil (U) • DNA includes A, G, C, T • RNA includes A, G, C, U


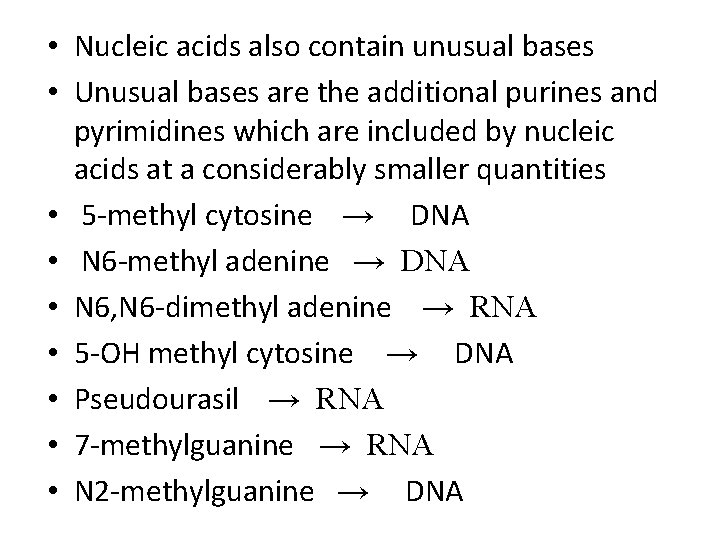
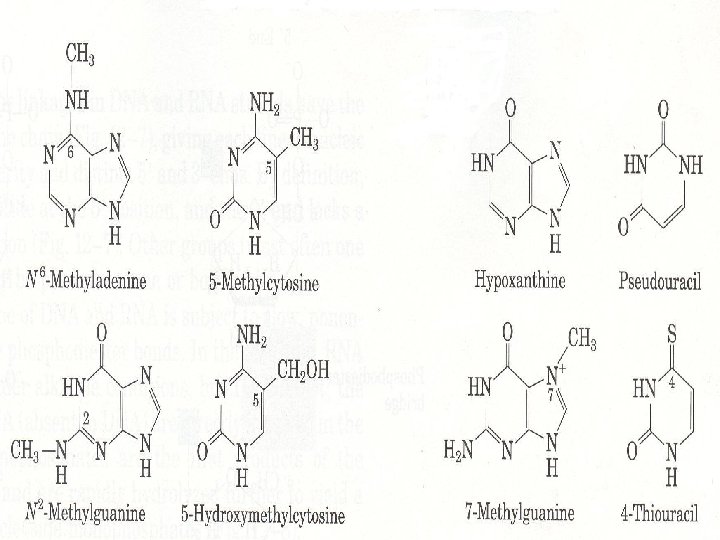
• Nucleic acids also contain unusual bases • Unusual bases are the additional purines and pyrimidines which are included by nucleic acids at a considerably smaller quantities • 5 -methyl cytosine → DNA • N 6 -methyl adenine → DNA • N 6, N 6 -dimethyl adenine → RNA • 5 -OH methyl cytosine → DNA • Pseudourasil → RNA • 7 -methylguanine → RNA • N 2 -methylguanine → DNA


• They serve important functions in oligonucleotide recognition in DNA and RNA. • The presence of specific methylated nucleotide bases allows the DNAses arising from infection to distinguish between native and foreign oligonucleotides • Methylated cytosine in the promoter region is responsible for gene expression. • Unusual bases play role in regulation of halflife of RNAs

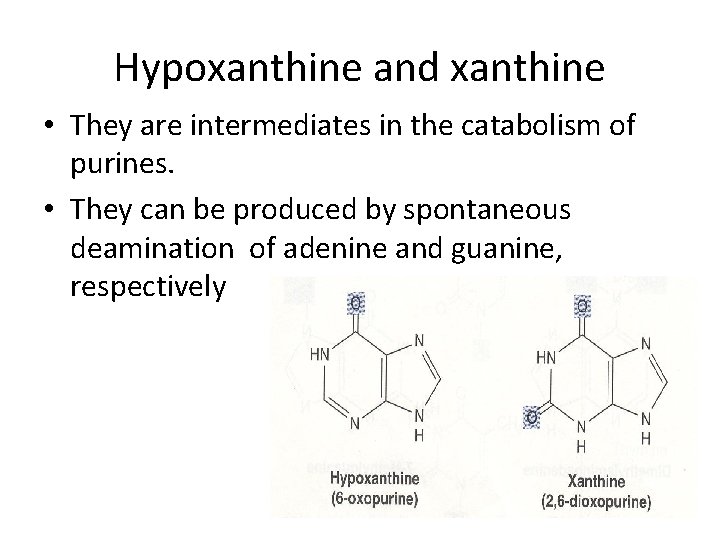
Hypoxanthine and xanthine • They are intermediates in the catabolism of purines. • They can be produced by spontaneous deamination of adenine and guanine, respectively

• Hypoxanthine is a mutagenic lesion, it pairs with C unless repaired • Methylated purines of the plants: • Theophylline → Dimethylxanthine → Tea • Caffeine → Trimethylxanthine → Coffee

• Backbones of DNA and RNA are hydrophilic because of the; • 1. hydrogen bonds between OH groups of the sugars and surrounding water • 2. Phosphate groups which are completely ionized and negatively charged at p. H 7 • These negative charges are generally neutralized by ionic interactions with positive charges on proteins, metal ions and polyamines

• Monomeric units of DNA helded in the polimeric form constitutes a single strand • The informational content of DNA resides in the sequence in which these monomers are ordered • The demonstration that DNA contained the genetic information was first made in 1944 in a series of experiments • In early 1950 s Watson, Crick and Wilkins determined a model of double-stranded DNA


• This model was determined based on x-ray diffraction data from the DNA molecule and the observations of Chargaff • Chargaff rule: The concentration of deoxyadenine nucleotides equals that of thymidine nucleotides (A=T), while the concentration of deoxyguanosine nucleotides equals that of deoxycytidine nucleotides (G=C)

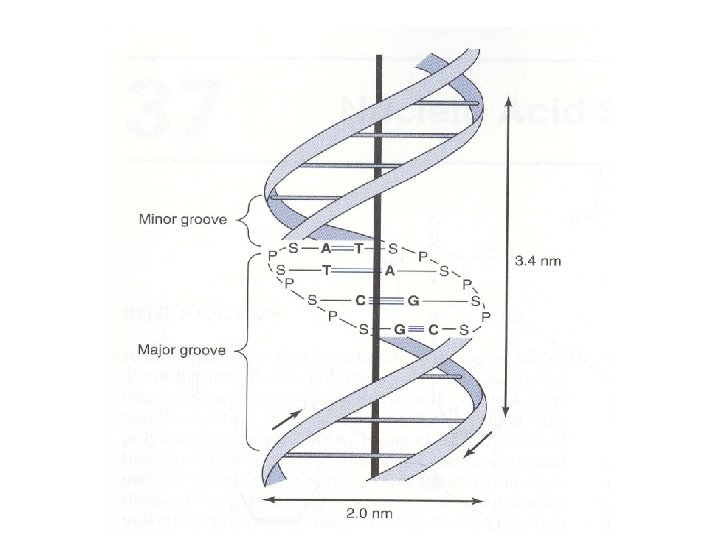
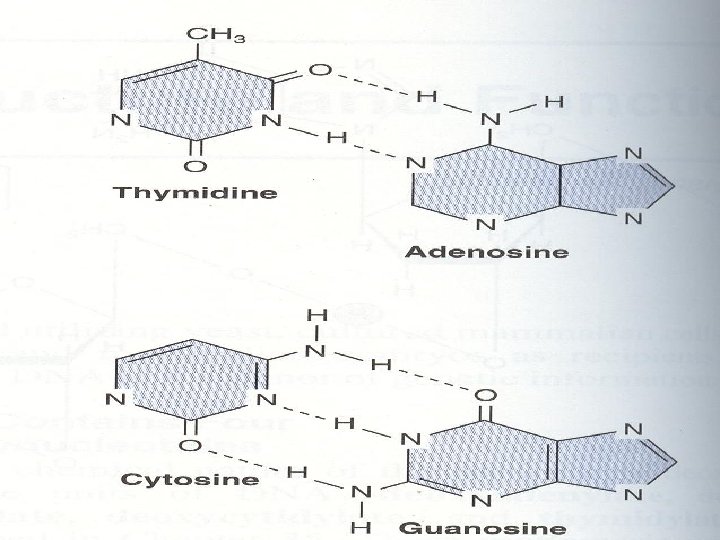
• The two strands of this double-stranded helix are held in register by hydrogen bonds between the purine and pyrimidine bases of the respective linear molecules • The pairings between the purine and pyrimidine nucleotides on the opposite strands are very specific and are present between A and T; C and G


• This common form of DNA is said to be righthanded because as one looks down the double helix the base residues form a spiral in a clockwise direction

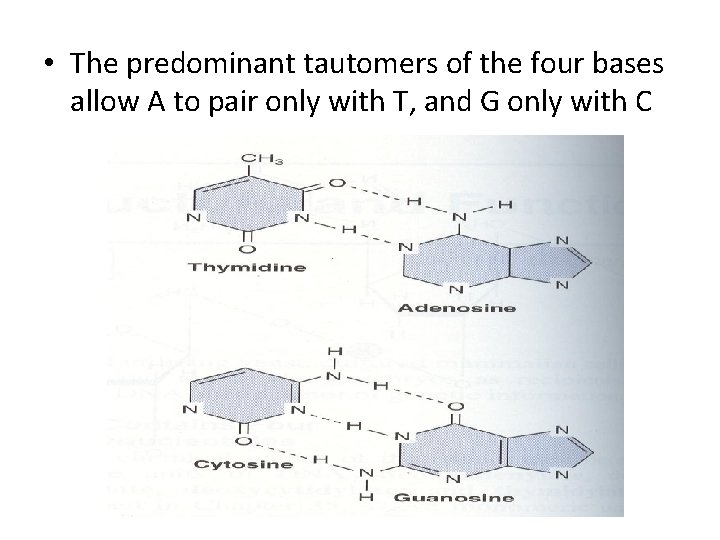
• The predominant tautomers of the four bases allow A to pair only with T, and G only with C

• The two strands of the double-helical molecule are antiparallel; one strand runs in the 3’-5’ direction while the other in the 5’-3’ direction • In the double-stranded DNA molecules, genetic information resides in the sequence of nucleotides on one strand, template strand • This is the strand of DNA that is copied during nucleic acid synthesis (noncoding strand) • The opposite strand is considered the coding strand because it matches the RNA transcript that encodes the protein

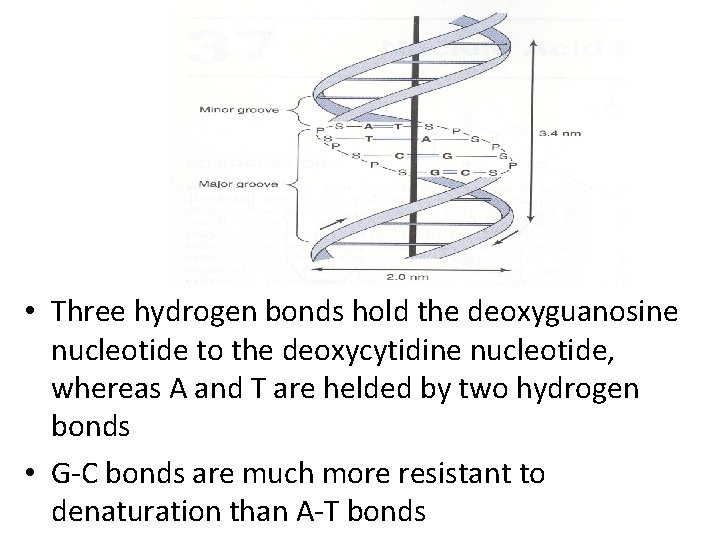
• The two strands wind around a central axis in the form of a double helix • Double-stranded DNA exists in at least six forms (A-E and Z) • The B form is usually found under physiologic conditions • A single turn of B-form DNA about the axis of the molecule contains ten base pairs • The distance spanned by one turn of B-form DNA is 3. 4 nm, the helical diameter of the double helix is 2 nm

• Three hydrogen bonds hold the deoxyguanosine nucleotide to the deoxycytidine nucleotide, whereas A and T are helded by two hydrogen bonds • G-C bonds are much more resistant to denaturation than A-T bonds

The denaturation of DNA (melting) • Double stranded DNA can be separated into two component strands in solution by increasing the temperature or decreasing the salt concentration • As a result of denaturation, optical absorbance of bases increases • Double stranded DNA molecule exhibits properties of a rigid rod and in solution is a viscous material because of the stacking of the bases and hydrogen bonding but it loses its viscosity upon denaturation

• DNA rich in G-C pairs melts at a higher temperature than that rich in A-T pairs • Separated strands of DNA will renature when physiologic temperature and salt conditions are achieved • The rate of reassociation depends upon the concentration of the complementary strands • There are grooves in the DNA molecule • A major groove and a minor groove wind along the molecule parallel to the phosphodiester backbones.

• In these grooves, regulatory proteins can interact specifically with exposed atoms of the nucleotides • Therefore these proteins recognize and bind to specific nucleotide sequences without disturbing the base pairing • Regulatory proteins can control the expression of specific genes via such interactions

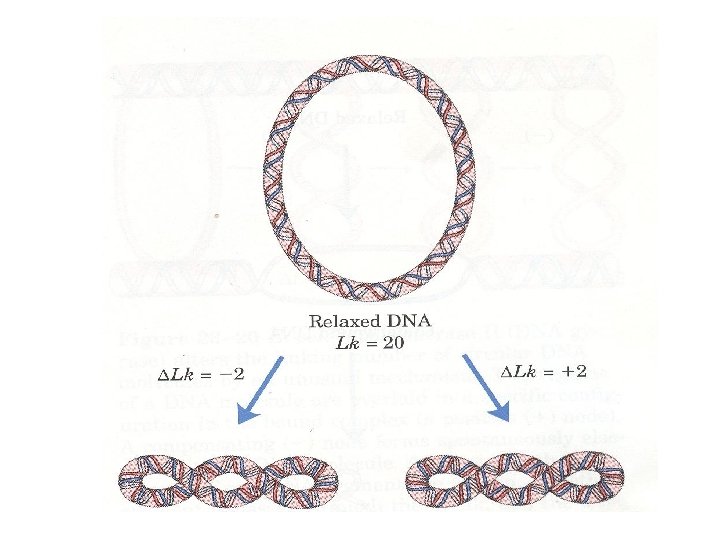
• DNA exists in relaxed and supercoiled forms • In bacteria and many DNA containing animal viruses the ends of the DNA molecules are joined to create a closed circle with no terminal • This structure does not destroy polarity but eliminates all free 3’ and 5’ hydroxyl and phosphoryl groups • Closed circles may exist in relaxed or supercoiled form


• Supercoils are introduced when a closed circle is twisted around its own axis or when a linear piece of dublex DNA is twisted • This is an energy-requiring process and puts the DNA under stress • Negative supercoils are formed when the molecule is twisted in the opposide direction of clockwise turns of the right-handed double helix found in B-DNA. Such DNA is said to be underwound

• The transition to another form is required energy • One such transition occurs during the strand separation just before the replication • Enzymes that catalyze topologic changes of DNA are called as topoisomerases • Topoisomerases can relax or insert supercoils using ATP • The best characterized is bacterial gyrase which induced negative supercoilig in DNA

• DNA provides a template for replication and transcription • The genetic information stored in DNA serves for two purposes: • a)It provides the information for the synthesis of all protein molecules of the organism • b)It provides the information inherited by daughter cells • Both these functions require that the DNA molecule serves as a template for the transcription of the information into RNA and for the replication of information into daughter DNA molecules

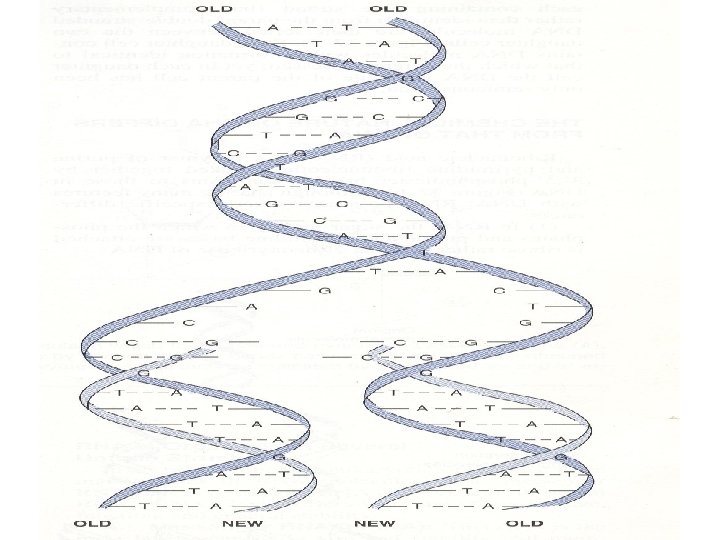
• Replication of the DNA molecule occurs in a semiconservative manner. Thus, when each strand of the double-stranded parent DNA molecule separated during replication, each serves as a template on which a new complementary strand is synthesized • Each daughter cell contains DNA molecules with information identical to that which the parent possessed; • Each daughter cell the DNA molecule of the parent cell has been only semiconserved


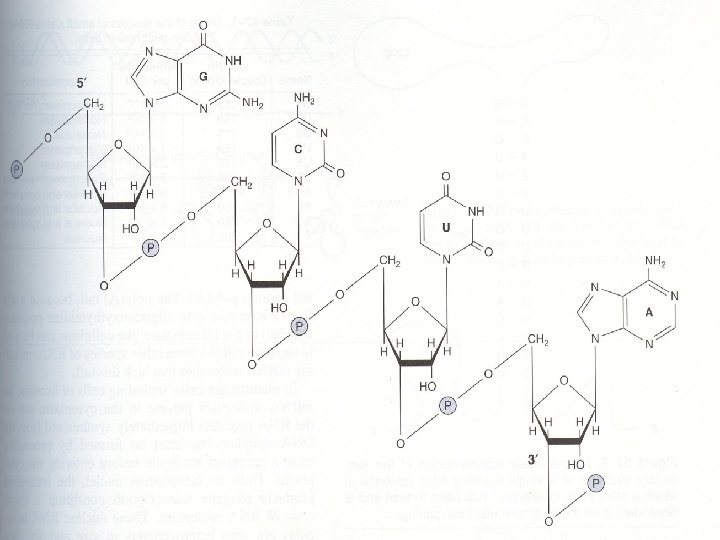
Chemical structure of RNA • The chemical nature of RNA differs from that of DNA • RNA is also formed by purines and pyrimidines linked by 3’-5’ phosphodiester bonds • Although sharing many features with DNA , RNA possesses several specific differences: • 1. In RNA, the sugar moiety to which the phosphates and purine and pyrimidines are attached is ribose instead of deoxyribose of DNA • 2. The pyrimidine components of RNA is differ from those of DNA. RNA contains the A, G, C but does not contain T (with a rare exception), instead of T, U is present in RNA


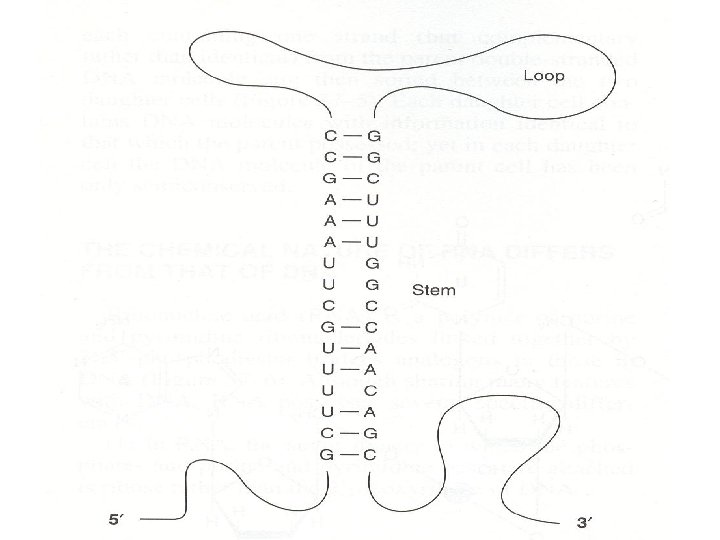
• 3. RNA exist as a single strand. However, given the proper complementary base sequence with opposite polarity, single strand RNA is capable of folding back on itself like a hairpin thus acquiring double-stranded structure


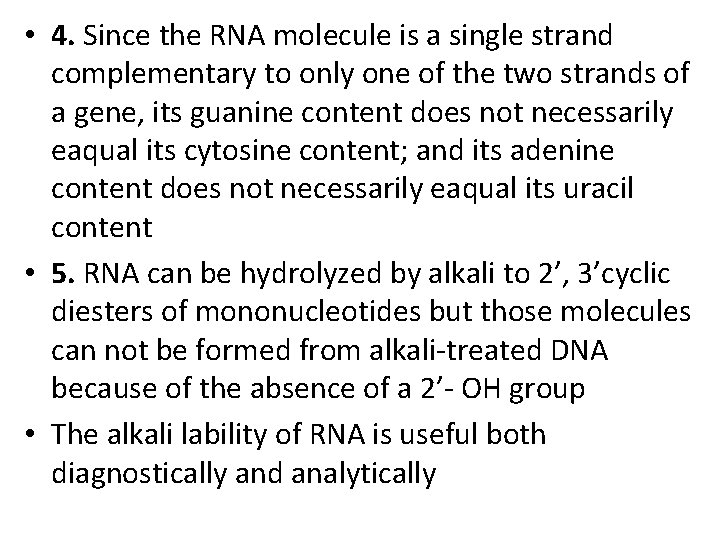
• 4. Since the RNA molecule is a single strand complementary to only one of the two strands of a gene, its guanine content does not necessarily eaqual its cytosine content; and its adenine content does not necessarily eaqual its uracil content • 5. RNA can be hydrolyzed by alkali to 2’, 3’cyclic diesters of mononucleotides but those molecules can not be formed from alkali-treated DNA because of the absence of a 2’- OH group • The alkali lability of RNA is useful both diagnostically and analytically

• Information within the RNA is contained in its sequence of purine and pyrimidine nucleotides. • The sequence is complementary to the template strand of the gene that was transcribed • Because of this complementarity, a RNA molecule can bind specifically via the basepairing rules to its template DNA strand; but it does not bind to coding strand • The nucleotide sequence of the RNA molecule is the same as that of the coding strand of the gene, except for U replacing T

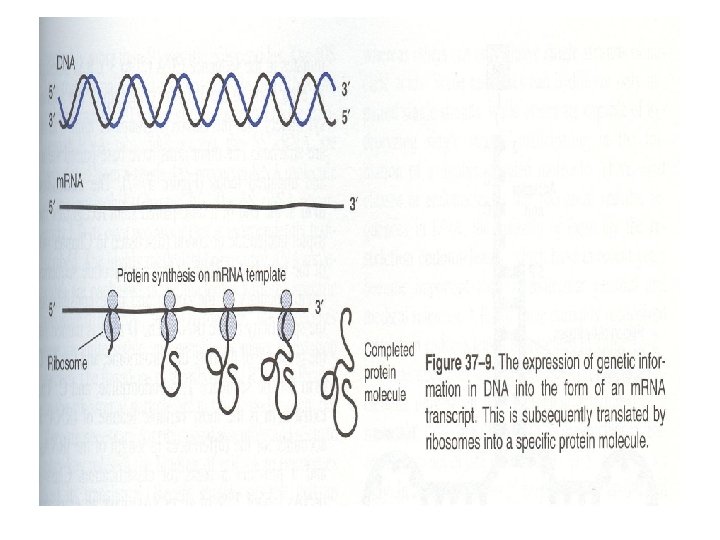
RNA types and their funtions • Cytoplasmic RNA molecules that serve as a templates for protein synthesis are designated as messnger RNAs (m. RNA). m. RNA molecules transfer genetic information from DNA to protein-synthesizing machinary • Many other cytoplasmic RNA molecules have structural roles. They contribute to the formation of ribosomes (ribosomal RNA, r. RNA) or serve as adapter molecules (transfer RNA, t. RNA) for the translation of RNA information into specific sequences of polymerized amino acids

• Some RNA molecules have intrinsic catalytic activity. The activity of these ribozymes often involved in the cleavage of a nucleic acid • In human cells there are small nuclear RNA (sn. RNA) species. These are not directly involved in protein synthesis but that may have roles in RNA processing • These relatively small molecules vary in size from 90 to 300 nucleotides

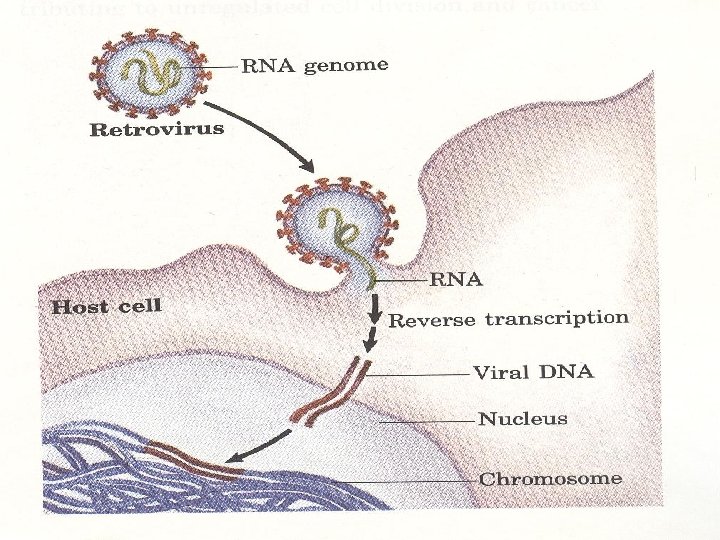
• The genetic material for some animal and plant viruses is RNA rather than DNA • Many animal RNA viruses are, retroviruses, transcribed by an RNA-dependent DNA polymerase , the so-called reverse transcriptase, to produce a double- stranded DNA copy of their RNA genome • In many cases, the resulting double- stranded DNA transcript is integrated into the host genome and subsequently serves as a template for gene expression and from which new viral RNA genomes can be transcribed


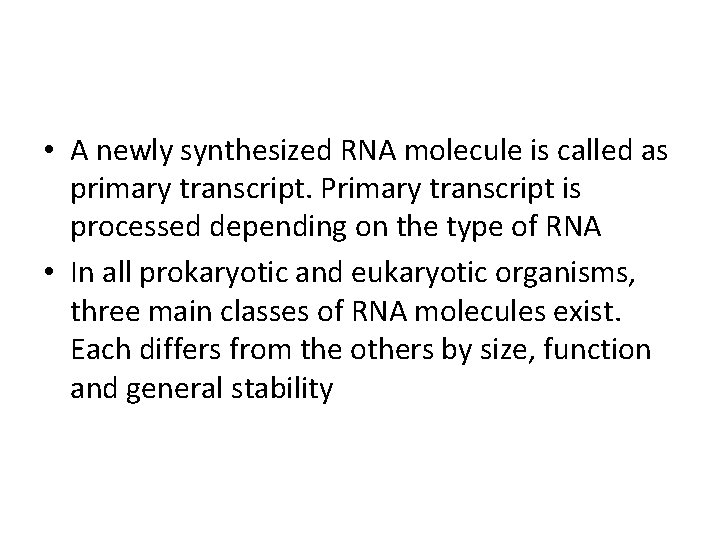
• A newly synthesized RNA molecule is called as primary transcript. Primary transcript is processed depending on the type of RNA • In all prokaryotic and eukaryotic organisms, three main classes of RNA molecules exist. Each differs from the others by size, function and general stability

1. Messenger RNA (m. RNA) • This is the most heterogenous class in size and stability • All members of the class function as messengers conveying the information in a gene to the protein-synthesizing machinery • Each serves as a template for a specific sequence of amino acids that is polimerized to form a specific protein


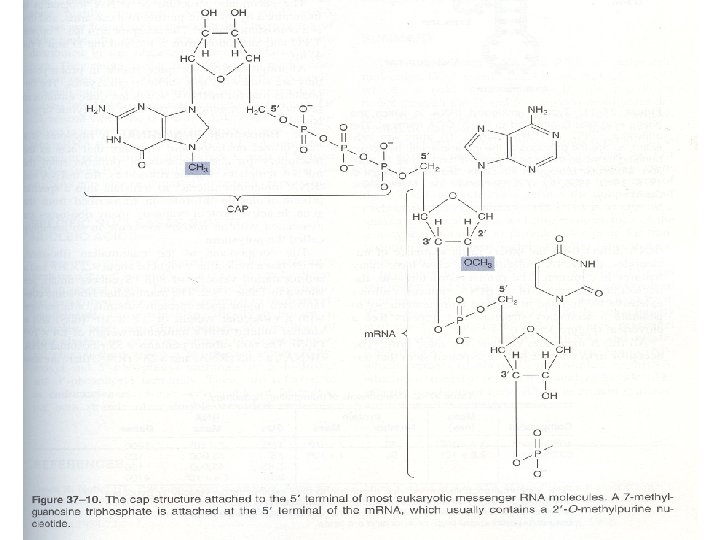
• m. RNAs have some unique chemical characteristics: • The 5’ terminal of m. RNA is capped by a 7 methylguanosine triphosphate • The cap is involved in the recognition of m. RNA by translating machinery, and it probably helps stabilize the m. RNA by preventing the attack of 5’-exonucleases • The protein synthesis begins translating the m. RNA into proteins at the 5’or capped terminal


• The other end of most m. RNAs has attached a polymer of adenylate residues 20 -250 nucleotides in lenght so called the poly A tail • The funtion of the poly A tail seems that it maintains the stability of the m. RNA by preventing the attack of 3’-exonucleases

2. Transfer RNA (t. RNA) • t. RNA molecules vary in lenght from 74 to 95 nucleotides • t. RNA molecules serve as a adapters for the translation of the information in the sequence of nucleotides of m. RNA into specific amino acids • There at least 20 species of t. RNA molecules in every cell. At least one, often several corresponding to each of the 20 amino acid are required for protein synthesis

• Although each specific t. RNA differs from the others in its sequence of nucleotides, the t. RNA molecules as a class have many features in common • The primary structure of all t. RNA molecules allows extensive folding and intrastrand complementarity to generate a secondary structure • This structure appears like a cloverleaf


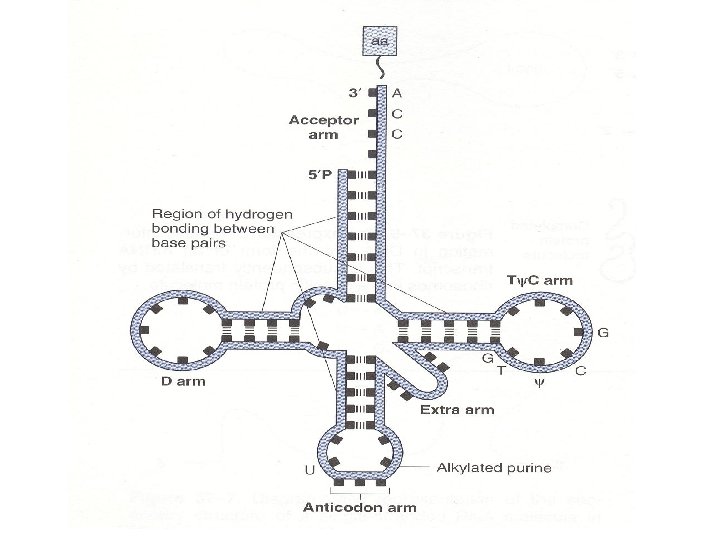
• All t. RNA molecules contain four main arms • The acceptor arm consists of a base-paired stem that terminates in the sequence CCA (5’ to 3’). It is through an ester bond to the 3’ hydroxyl group of the adenosyl moiety that the carboxyl groups of amino acids are attached

• The other arms have base-paired stems and unpaired loops • The anticodon arm at the end of a base-paired stem recognizes the triplet nucleotide or codon of the template m. RNA. It has a nucleotide sequence complementary to the codon and is responsible for the specifity of the t. RNA

• The D arm is named for the presence of the base dihydrouridine, and the TΨC arm for the sequence T, pseudouridine, and C

• The extra arm is the most variable feature of t. RNA. It accounts for the differences in lenght of the t. RNAs; and it provides a basis for classification. • Class 1 t. RNAs have an extra arm that is 3 -5 bp long • Class 2 t. RNAs have an extra arm that is 13 -21 bp long

• The secondary structure of t. RNA molecules is maintained by the base pairing in these arms and this is a consistent feature : • The TΨC and anticodon arms have 5 bp • The D arm has 3 -4 bp • The acceptor arm has 7 bp

3. Ribosomal RNA (r. RNA) • A ribosome is a cytoplasmic nucleoprotein structure that acts as the machinery for the synthesis of proteins from the m. RNA templates • On the ribosomes, the m. RNA and t. RNA molecules interact to translate into a specific protein molecule information transcribed from the gene • During the active protein synthesis, many ribosomes are associated with an m. RNA molecule in an assembly called the polysome

• The mammalian ribosome contains two major nucleoprotein subunits, a larger one with a molecular weight of 2. 8 x 106 (sedimentation velocity is 60 S*) and a smaller subunit with a molecular weight of 1. 4 x 106 (40 S) • The 60 S subunit contains a 5 S ribosomal RNA, a 5. 8 S r. RNA and a 28 S r. RNA; there also probably more than 50 specific polypeptides • *Svedberg unit (sedimentation coefficient)

• The 40 S subunit is smaller and contains 18 S r. RNA and approximately 30 polypeptide chains • All of the r. RNA molecules, except the 5 S r. RNA are processed from a single 45 S precursor RNA molecule in the nucleus • 5 S r. RNA has its own precursor that is independently transcribed • The highly methylated r. RNA molecules are packaged in the nucleus with the specific ribosomal proteins; but in the cytoplasm, ribosomes remain quite stable and capable of many translaton cycles

• Ribosomal RNA molecules are necessary for ribosomal assembly and seem to play key roles in the binding of m. RNA to ribosomes and its translation

Small stable RNA (sn. RNA) • A large number of discrete, highly conserved and small stable RNA species are found in the mammalian cells • The majority of these molecules exist as ribonucleoproteins and are distributed in the nucleus, cytoplasm or in both • They are involved in m. RNA processing and gene regulation

• Of the several sn. RNAs, U 1, U 2, U 4, U 5 and U 6 are involved in intron removaland the processing of primary transcript into m. RNA. The U 4 and U 6 sn. RNAs are required for poly (A) processing

Nucleases • Nucleic acids are digested by nucleases • Nucleases exhibit specifity to deoxyribonucleic acids are referred to as deoxyribonucleases • Those which specifically hydrolyze ribonucleic acids are ribonucleases • Enzymes capable of cleaving internal phosphodiester bonds are referred to as endonucleases

• Some of the endonucleases are capable of hydrolyzing both strands of a double-stranded molecule, whereas others can only clevage single strands of nucleic acids • Endonuclease classes that recognize specific sequences in DNA are present. They are referred as restriction endonucleases and are widely used in molecular genetic investigations

• Some nucleases are capable of hydrolyzing a nucleotide only when it is present at a terminal of a nucleic acid. These enzymes are referred to as exonucleases • Exonucleases can act in one direction only; 3’→ 5’ or 5’→ 3’