New tools for genomic selection in dairy cattle















- Slides: 46

New tools for genomic selection in dairy cattle John B. Cole Animal Improvement Programs Laboratory Agricultural Research Service, USDA Beltsville, MD 20705 -2350 john. cole@ars. usda. gov

Why genomic selection works in dairy Extensive historical data available Well-developed genetic evaluation program Widespread use of AI sires Progeny test programs High-valued animals, worth the cost of genotyping Long generation interval which can be reduced substantially by genomics Department of Animal Sciences, Purdue University, October 23, 2013 (2) Cole

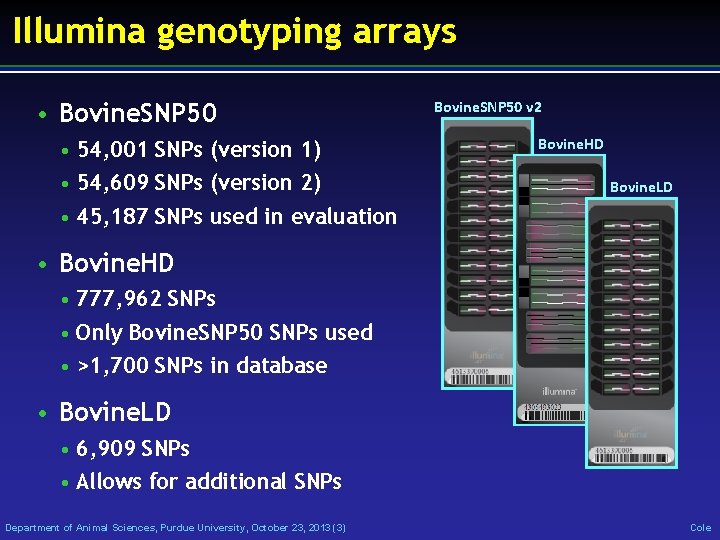
Illumina genotyping arrays • Bovine. SNP 50 • 54, 001 SNPs (version 1) • 54, 609 SNPs (version 2) • 45, 187 SNPs used in evaluation Bovine. SNP 50 v 2 Bovine. HD Bovine. LD • Bovine. HD • 777, 962 SNPs • Only Bovine. SNP 50 SNPs used • >1, 700 SNPs in database • Bovine. LD • 6, 909 SNPs • Allows for additional SNPs Department of Animal Sciences, Purdue University, October 23, 2013 (3) Cole

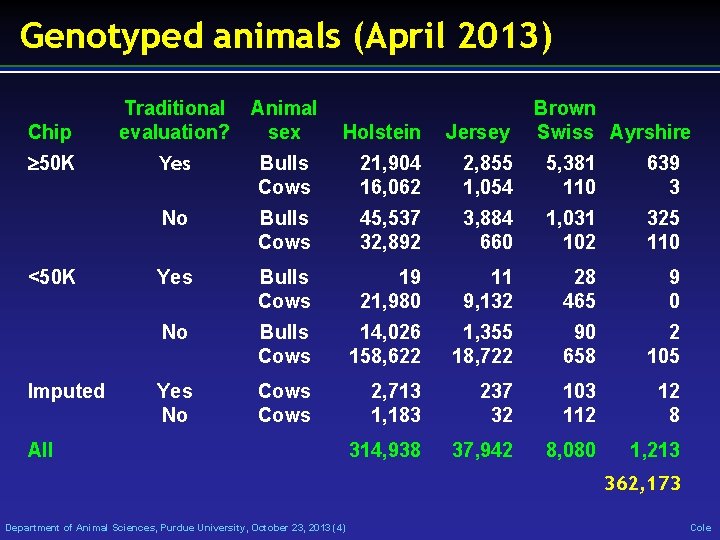
Genotyped animals (April 2013) Chip 50 K <50 K Imputed Traditional Animal evaluation? sex Holstein Jersey Brown Swiss Ayrshire Yes Bulls Cows 21, 904 16, 062 2, 855 1, 054 5, 381 110 639 3 No Bulls Cows 45, 537 32, 892 3, 884 660 1, 031 102 325 110 Yes Bulls Cows 19 21, 980 11 9, 132 28 465 9 0 No Bulls Cows 14, 026 158, 622 1, 355 18, 722 90 658 2 105 Yes No Cows 2, 713 1, 183 237 32 103 112 12 8 314, 938 37, 942 8, 080 1, 213 All 362, 173 Department of Animal Sciences, Purdue University, October 23, 2013 (4) Cole

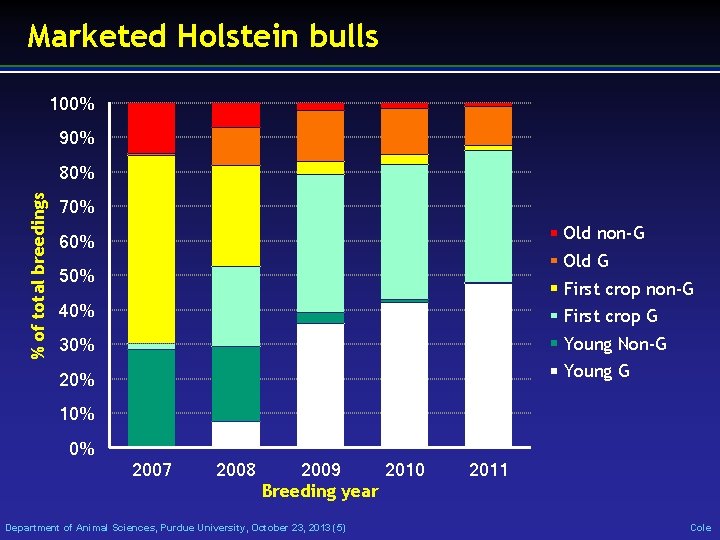
Marketed Holstein bulls 100% 90% % of total breedings 80% 70% Old non-G 60% Old G 50% First crop non-G 40% First crop G 30% Young Non-G 20% Young G 10% 0% 2007 2008 2009 2010 Breeding year Department of Animal Sciences, Purdue University, October 23, 2013 (5) 2011 Cole

What’s a SNP genotype worth? Pedigree is equivalent to information on about 7 daughters For the protein yield (h 2=0. 30), the SNP genotype provides information equivalent to an additional 34 daughters Department of Animal Sciences, Purdue University, October 23, 2013 (6) Cole

What’s a SNP genotype worth? And for daughter pregnancy rate (h 2=0. 04), SNP = 131 daughters Department of Animal Sciences, Purdue University, October 23, 2013 (7) Cole

Genotypes and haplotypes • Genotypes indicate how many copies of each allele were inherited • Haplotypes indicate which alleles are on which chromosome • Observed genotypes partitioned into the two unknown haplotypes • Pedigree haplotyping uses relatives • Population haplotyping finds matching allele patterns Department of Animal Sciences, Purdue University, October 23, 2013 (8) Cole

Haplotyping program – findhap. f 90 • Begin with population haplotyping • Divide chromosomes into segments, ~250 to 75 SNP / segment • List haplotypes by genotype match • Similar to fast. Phase, IMPUTE • End with pedigree haplotyping • Detect crossover, fix noninheritance • Impute nongenotyped ancestors Department of Animal Sciences, Purdue University, October 23, 2013 (9) Cole

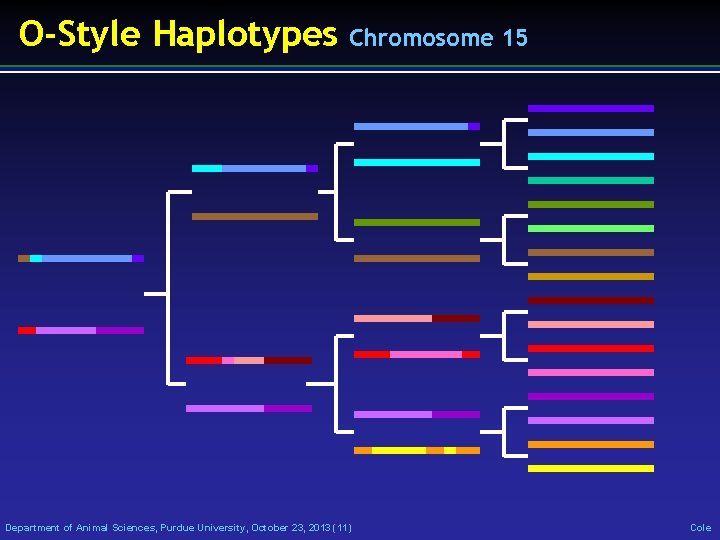
Example Bull: O-Style (USA 137611441) • Read genotypes and pedigrees • Write haplotype segments found • List paternal / maternal inheritance • List crossover locations Department of Animal Sciences, Purdue University, October 23, 2013 (10) Cole

O-Style Haplotypes Chromosome 15 Department of Animal Sciences, Purdue University, October 23, 2013 (11) Cole

Loss-of-function mutations • At least 100 Lo. F per human genome surveyed (Mac. Arthur et al. , 2010) • Of those genes ~20 are completely inactivated • Uncharacterized Lo. F variants likely to have phenotypic effects • How should mating programs deal with this? • Can we find them? Department of Animal Sciences, Purdue University, October 23, 2013 (12) Cole

Recessive defect discovery • Check for homozygous haplotypes • 7 to 90 expected but none observed • 5 of top 11 are potentially lethal • 936 to 52, 449 carrier sire by carrier MGS fertility records • 3. 1% to 3. 7% lower conception rates • Some slightly higher stillbirth rates • Confirmed Brachyspina same way Department of Animal Sciences, Purdue University, October 23, 2013 (13) Cole

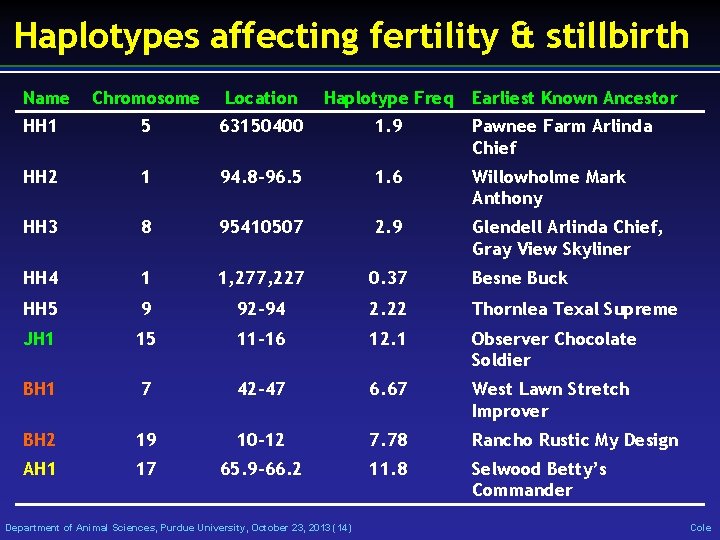
Haplotypes affecting fertility & stillbirth Name Chromosome Location Haplotype Freq HH 1 5 63150400 1. 9 Pawnee Farm Arlinda Chief HH 2 1 94. 8 -96. 5 1. 6 Willowholme Mark Anthony HH 3 8 95410507 2. 9 Glendell Arlinda Chief, Gray View Skyliner HH 4 1 1, 277, 227 0. 37 Besne Buck HH 5 9 92 -94 2. 22 Thornlea Texal Supreme JH 1 15 11 -16 12. 1 Observer Chocolate Soldier BH 1 7 42 -47 6. 67 West Lawn Stretch Improver BH 2 19 10 -12 7. 78 Rancho Rustic My Design AH 1 17 65. 9 -66. 2 11. 8 Selwood Betty’s Commander Department of Animal Sciences, Purdue University, October 23, 2013 (14) Earliest Known Ancestor Cole

Precision mating Eliminate undesirable haplotypes Avoid carrier-to-carrier matings Easy with few recessives, difficult with many recessives Include in selection indices Detection at low allele frequencies Requires many inputs Use a selection strategy for favorable minor alleles (Sun & Van. Raden, 2013) Department of Animal Sciences, Purdue University, October 23, 2013 (15) Cole

Sequencing successes at AIPL/BFGL • Simple loss-of-function mutations • APAF 1 (HH 1) – Spontaneous abortions in Holstein cattle (Adams et al. , 2012) • CWC 15 (JH 1) – Early embryonic death in Jersey cattle (Sonstegard et al. , 2013) • Weaver syndrome – Neurological degeneration and death in Brown Swiss cattle (Mc. Clure et al. , 2013) Department of Animal Sciences, Purdue University, October 23, 2013 (16) Cole

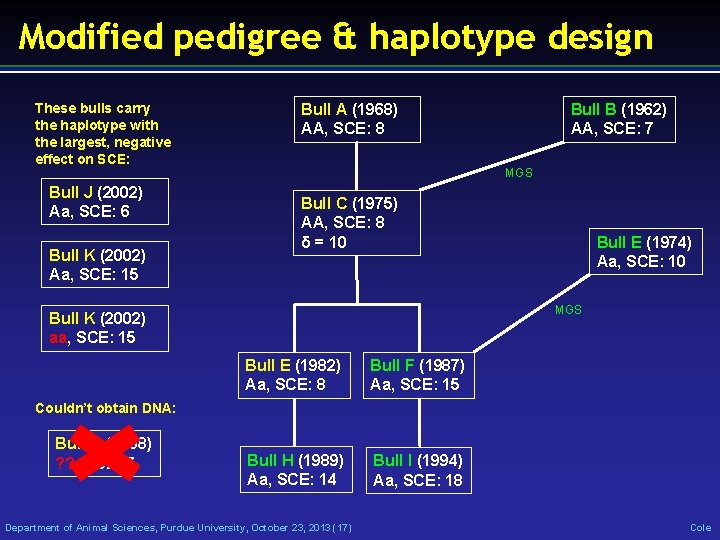
Modified pedigree & haplotype design These bulls carry the haplotype with the largest, negative effect on SCE: Bull J (2002) Aa, SCE: 6 Bull K (2002) Aa, SCE: 15 Bull A (1968) AA, SCE: 8 Bull B (1962) AA, SCE: 7 MGS Bull C (1975) AA, SCE: 8 δ = 10 Bull E (1974) Aa, SCE: 10 MGS Bull K (2002) aa, SCE: 15 Bull E (1982) Aa, SCE: 8 Bull F (1987) Aa, SCE: 15 Bull H (1989) Aa, SCE: 14 Bull I (1994) Aa, SCE: 18 Couldn’t obtain DNA: Bull D (1968) ? ? , SCE: 7 Department of Animal Sciences, Purdue University, October 23, 2013 (17) Cole

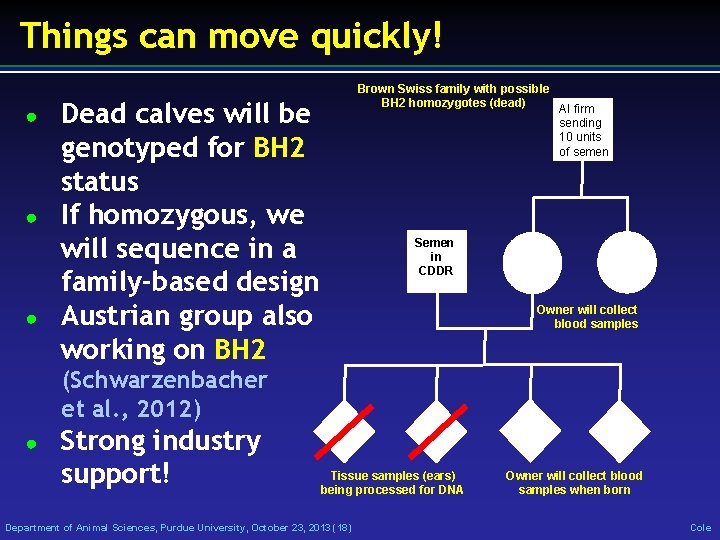
Things can move quickly! ● ● ● Brown Swiss family with possible BH 2 homozygotes (dead) AI firm sending 10 units of semen Dead calves will be genotyped for BH 2 status If homozygous, we will sequence in a family-based design Austrian group also working on BH 2 Semen in CDDR Owner will collect blood samples (Schwarzenbacher et al. , 2012) ● Strong industry support! Tissue samples (ears) being processed for DNA Department of Animal Sciences, Purdue University, October 23, 2013 (18) Owner will collect blood samples when born Cole

Our industry wants new genomic tools Department of Animal Sciences, Purdue University, October 23, 2013 (19) Cole

We already have some tools https: //www. cdcb. us/Report_Data/Marker_Effects/marker_effects. cfm` Department of Animal Sciences, Purdue University, October 23, 2013 (20) Cole

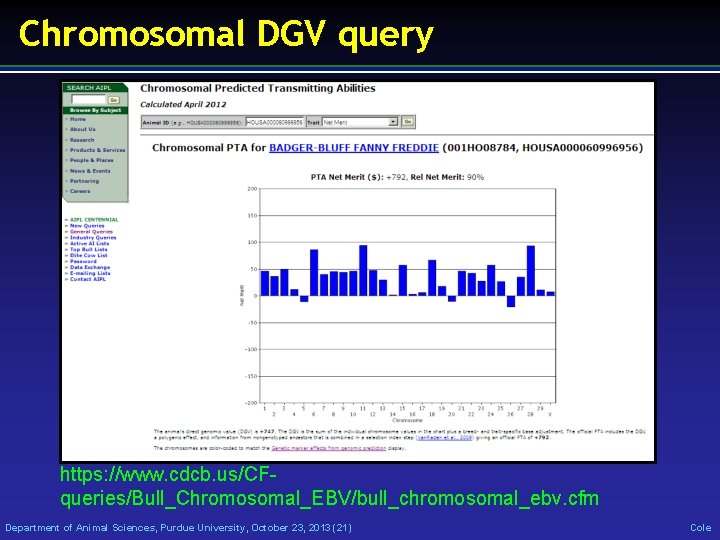
Chromosomal DGV query https: //www. cdcb. us/CFqueries/Bull_Chromosomal_EBV/bull_chromosomal_ebv. cfm Department of Animal Sciences, Purdue University, October 23, 2013 (21) Cole

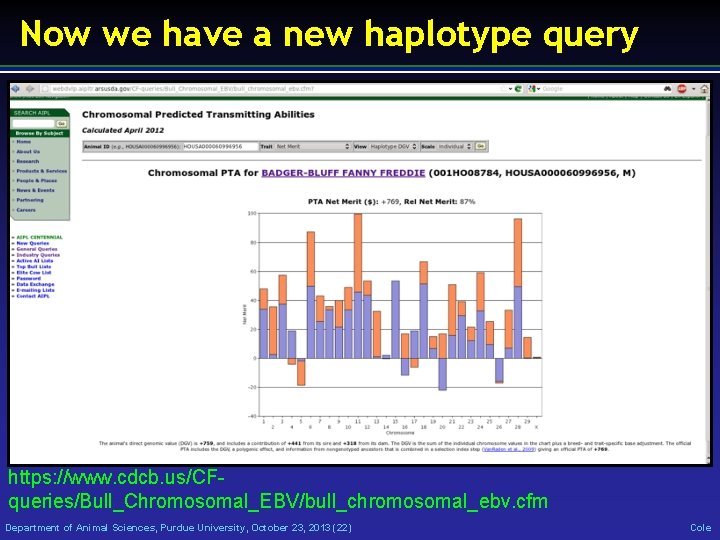
Now we have a new haplotype query https: //www. cdcb. us/CFqueries/Bull_Chromosomal_EBV/bull_chromosomal_ebv. cfm Department of Animal Sciences, Purdue University, October 23, 2013 (22) Cole

Paternal and maternal DGV • Shows the DGV for the paternal and maternal haplotyles • Imputed from 50 K using findhap. f 90 v. 2 • Can we use them to make mating decisions? • People are going to do it – we need to help them! • Who is actually making planned matings? Department of Animal Sciences, Purdue University, October 23, 2013 (23) Cole

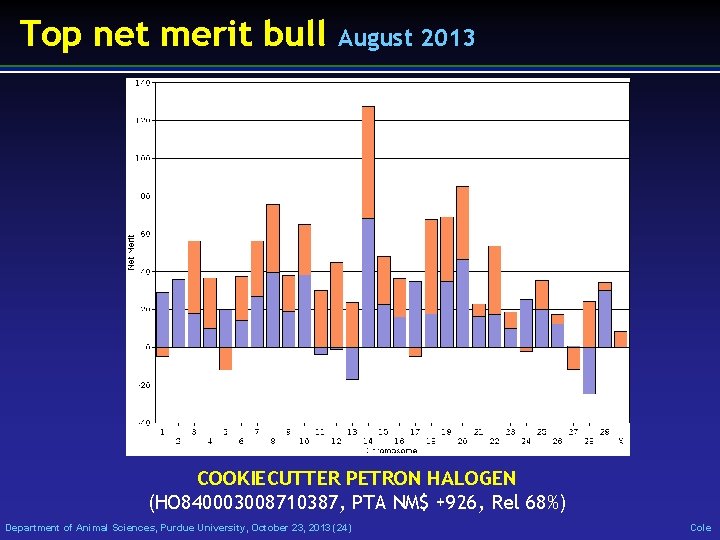
Top net merit bull August 2013 COOKIECUTTER PETRON HALOGEN (HO 840003008710387, PTA NM$ +926, Rel 68%) Department of Animal Sciences, Purdue University, October 23, 2013 (24) Cole

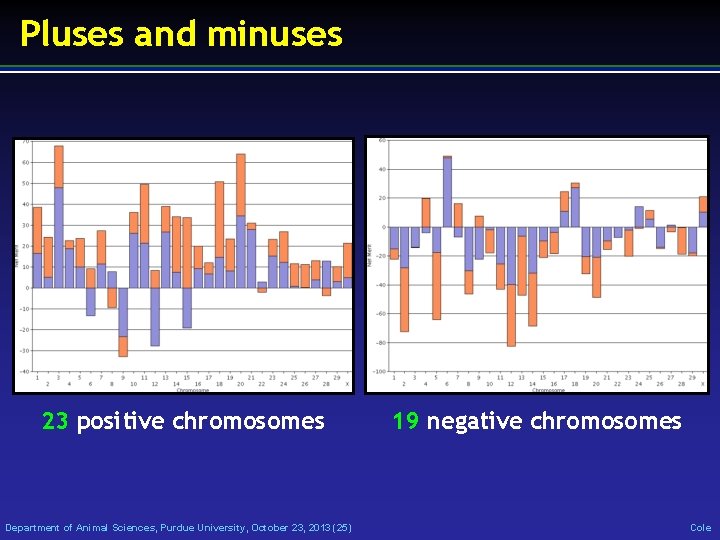
Pluses and minuses 23 positive chromosomes Department of Animal Sciences, Purdue University, October 23, 2013 (25) 19 negative chromosomes Cole

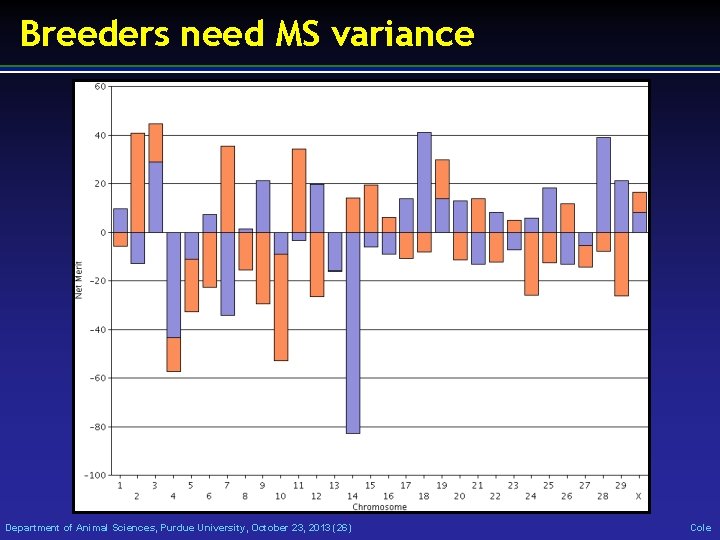
Breeders need MS variance Department of Animal Sciences, Purdue University, October 23, 2013 (26) Cole

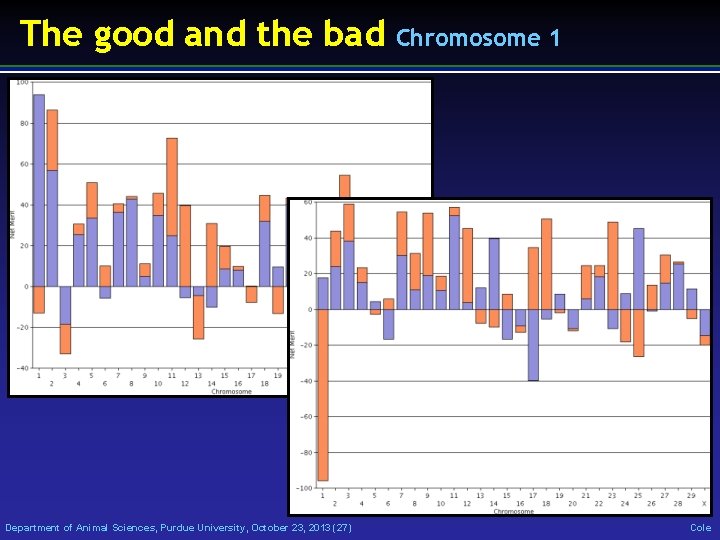
The good and the bad Department of Animal Sciences, Purdue University, October 23, 2013 (27) Chromosome 1 Cole

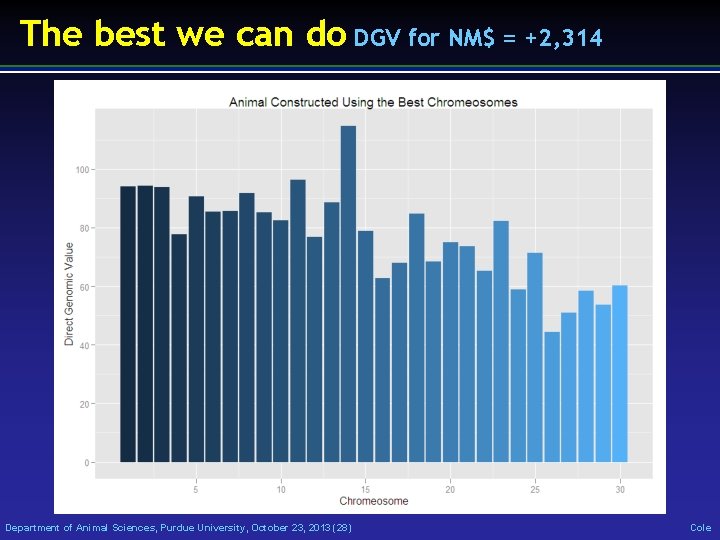
The best we can do DGV for NM$ = +2, 314 Department of Animal Sciences, Purdue University, October 23, 2013 (28) Cole

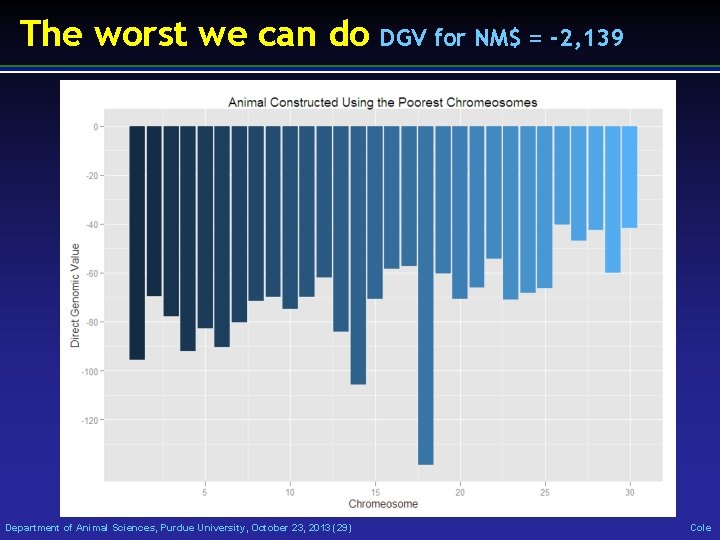
The worst we can do Department of Animal Sciences, Purdue University, October 23, 2013 (29) DGV for NM$ = -2, 139 Cole

Dominance in mating programs Quantitative model Must solve equation for each mate pair Genomic model Compute dominance for each locus Haplotype the population Calculate dominance for mate pairs Most genotyped cows do not yet have phenotypes Department of Animal Sciences, Purdue University, October 23, 2013 (30) Cole

Inbreeding effects Inbreeding alters transcription levels and gene expression profiles (Kristensen et al. , 2005). Moderate levels of inbreeding among active bulls (7. 9 to 18. 2) Are inbreeding effects distributed uniformly across the genome? Can we find genomic regions where heterozygosity is necessary or not using the current population? Department of Animal Sciences, Purdue University, October 23, 2013 (31) Cole

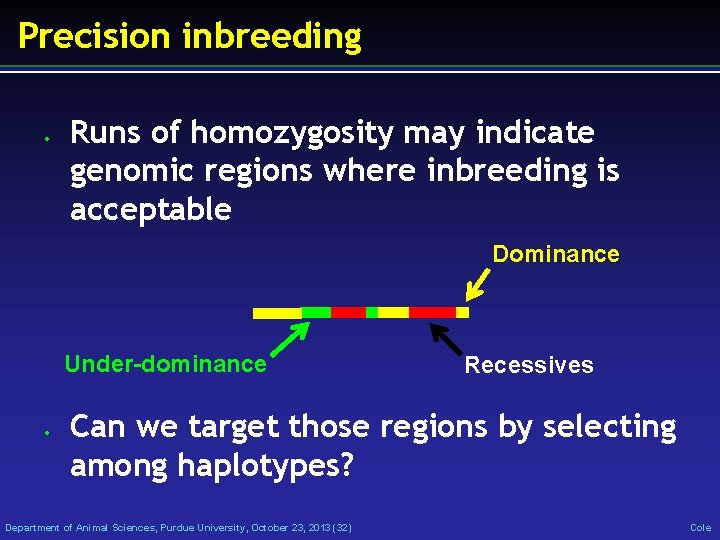
Precision inbreeding • Runs of homozygosity may indicate genomic regions where inbreeding is acceptable Dominance Under-dominance • Recessives Can we target those regions by selecting among haplotypes? Department of Animal Sciences, Purdue University, October 23, 2013 (32) Cole

Challenges with new phenotypes Lack of information Inconsistent trait definitions Often no database of phenotypes Many have low heritabilities Lots of records are needed for accurate evaluation Genetic improvement can be slow Genomics may help with this Department of Animal Sciences, Purdue University, October 23, 2013 (33) Cole

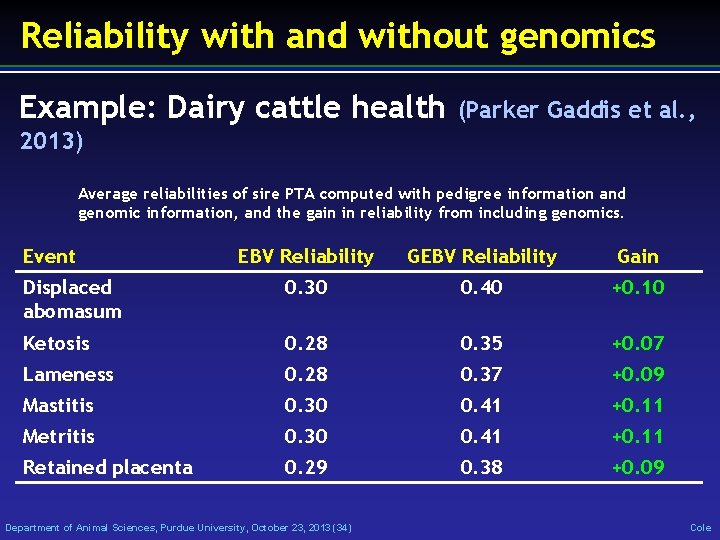
Reliability with and without genomics Example: Dairy cattle health (Parker Gaddis et al. , 2013) Average reliabilities of sire PTA computed with pedigree information and genomic information, and the gain in reliability from including genomics. Event EBV Reliability Gain Displaced abomasum 0. 30 0. 40 +0. 10 Ketosis 0. 28 0. 35 +0. 07 Lameness 0. 28 0. 37 +0. 09 Mastitis 0. 30 0. 41 +0. 11 Metritis 0. 30 0. 41 +0. 11 Retained placenta 0. 29 0. 38 +0. 09 Department of Animal Sciences, Purdue University, October 23, 2013 (34) Cole

Some novel phenotypes being studied Age at first calving (Cole et al. , 2013) Dairy cattle health (Parker Gaddis et al. , 2013) Methane production Milk fatty acid composition Persistency of lactation Rectal temperature Residual feed intake (de Haas et al. , 2011) (Bittante et al. , 2013) (Cole et al. , 2009) (Dikmen et al. , 2013) Department of Animal Sciences, Purdue University, October 23, 2013 (35) (Connor et al. , 2013) Cole

What do we do with novel traits? • Put them into a selection index • • Apply selection for a long time • • Correlated traits are helpful There are no shortcuts Collect phenotypes on many daughters • • Repeated records of limited value Genomics can increase accuracy Department of Animal Sciences, Purdue University, October 23, 2013 (36) Cole

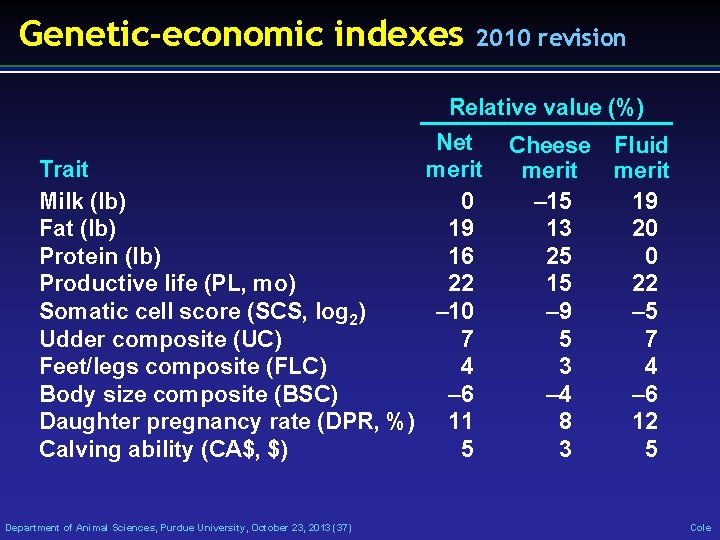
Genetic-economic indexes 2010 revision Relative value (%) Net merit Trait Milk (lb) 0 Fat (lb) 19 Protein (lb) 16 Productive life (PL, mo) 22 Somatic cell score (SCS, log 2) – 10 Udder composite (UC) 7 Feet/legs composite (FLC) 4 Body size composite (BSC) – 6 Daughter pregnancy rate (DPR, %) 11 Calving ability (CA$, $) 5 Department of Animal Sciences, Purdue University, October 23, 2013 (37) Cheese Fluid merit – 15 19 13 20 25 0 15 22 – 9 – 5 5 7 3 4 – 6 8 12 3 5 Cole

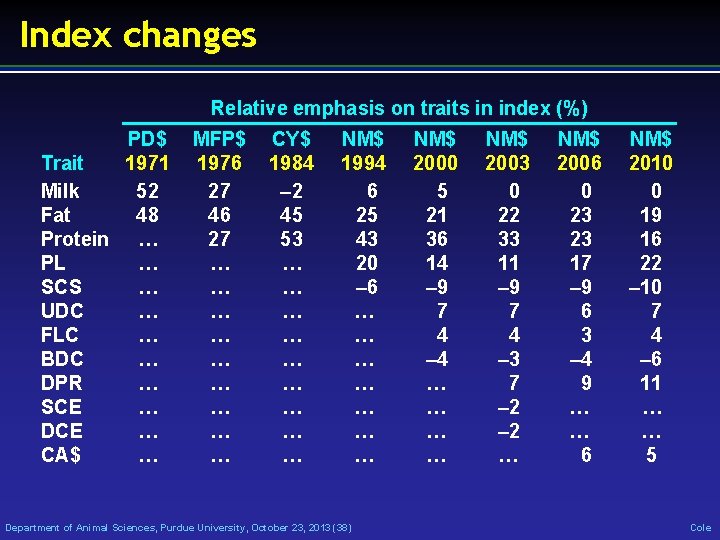
Index changes Relative emphasis on traits in index (%) PD$ 1971 Trait Milk 52 Fat 48 Protein … PL … SCS … UDC … FLC … BDC … DPR … SCE … DCE … CA$ … MFP$ 1976 27 46 27 … … … … … CY$ 1984 – 2 45 53 … … … … … NM$ 1994 6 25 43 20 – 6 … … … … Department of Animal Sciences, Purdue University, October 23, 2013 (38) NM$ 2000 5 21 36 14 – 9 7 4 – 4 … … NM$ 2003 0 22 33 11 – 9 7 4 – 3 7 – 2 … NM$ 2006 0 23 23 17 – 9 6 3 – 4 9 … … 6 NM$ 2010 0 19 16 22 – 10 7 4 – 6 11 … … 5 Cole

What does it mean to be the worst? • Large body size • Eats a lot of expensive feed • Average fertility…or worse! • Begin first lactation with dystocia • Bull calf (sexed semen? ) • Retained placenta, metritis, etc. • Mediocre production • Uses many resources, produces very little Department of Animal Sciences, Purdue University, October 23, 2013 (39) Cole

Dissecting genetic correlations • Compute DGV for 75 -SNP segments • Calculate correlations of DGV for traits of interest for each segment • Is there interesting biology associated with favorable correlations? • …and what about linkage disequilibrium? Department of Animal Sciences, Purdue University, October 23, 2013 (40) Cole

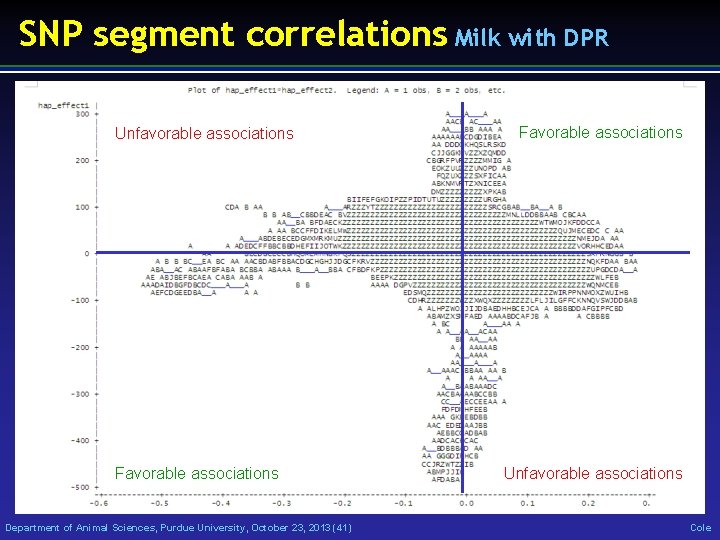
SNP segment correlations Milk with DPR Unfavorable associations Favorable associations Department of Animal Sciences, Purdue University, October 23, 2013 (41) Favorable associations Unfavorable associations Cole

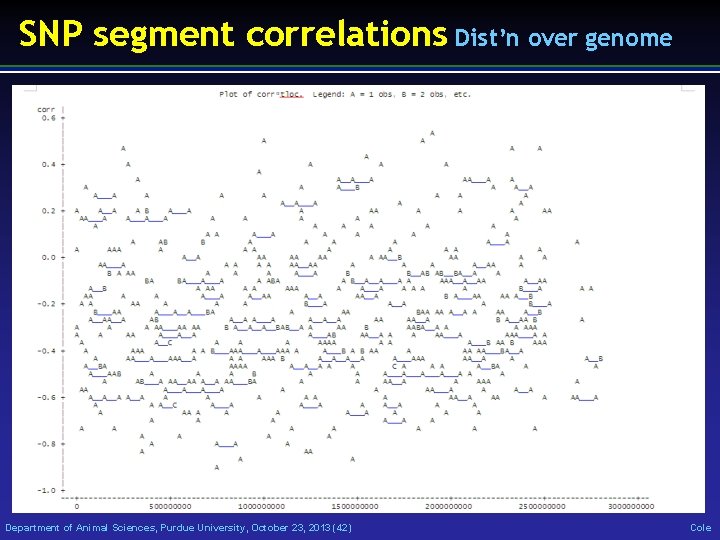
SNP segment correlations Dist’n over genome Department of Animal Sciences, Purdue University, October 23, 2013 (42) Cole

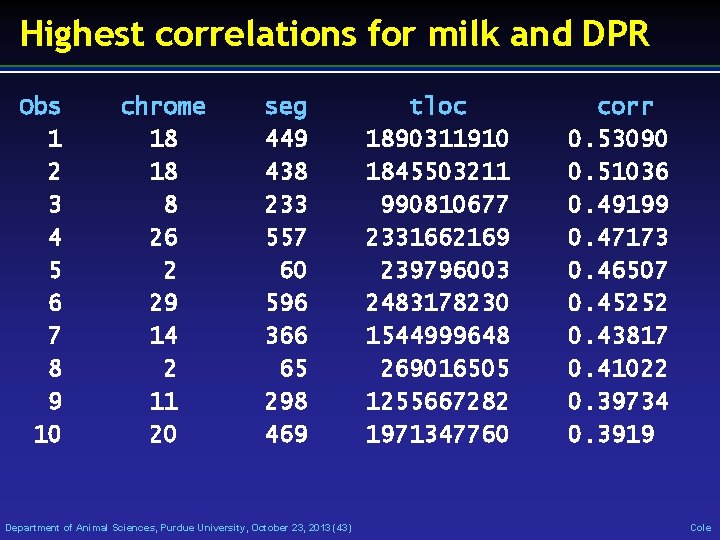
Highest correlations for milk and DPR Obs 1 2 3 4 5 6 7 8 9 10 chrome 18 18 8 26 2 29 14 2 11 20 seg 449 438 233 557 60 596 366 65 298 469 Department of Animal Sciences, Purdue University, October 23, 2013 (43) tloc 1890311910 1845503211 990810677 2331662169 239796003 2483178230 1544999648 269016505 1255667282 1971347760 corr 0. 53090 0. 51036 0. 49199 0. 47173 0. 46507 0. 45252 0. 43817 0. 41022 0. 39734 0. 3919 Cole

Conclusions Non-additive effects may be useful for increasing selection intensity while conserving important heterozygosity Whole-genome sequencing has been very successful at helping economically important loss-of-function mutations Novel phenotypes are necessary to address global food security and a changing climate Department of Animal Sciences, Purdue University, October 23, 2013 (44) Cole

Acknowledgments Paul Van. Raden, George Wiggans, Derek Bickhart, Dan Null, and Tabatha Cooper Animal Improvement Programs Laboratory, ARS, USDA Beltsville, MD Tad Sonstegard, Curt Van Tassell, and Steve Schroeder Bovine Functional Genomics Laboratory, ARS, USDA, Beltsville, MD Chuanyu Sun National Association of Animal Breeders Beltsville, MD Dan Gilbert New Generation Genetics Inc. , Fort Atkinson, WI Department of Animal Sciences, Purdue University, October 23, 2013 (45) Cole

Questions? http: //gigaom. com/2012/05/31/t-mobile-pits-math-against-verizons-the-loser-common-sense/shutterstock_76826245/ Department of Animal Sciences, Purdue University, October 23, 2013 (46) Cole