North American Dairy Cattle Genomic Selection Program G












- Slides: 31

North American Dairy Cattle Genomic Selection Program G. R. Wiggans Animal Improvement Programs Laboratory Agricultural Research Service, USDA Beltsville, MD george. wiggans@ars. usda. gov DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

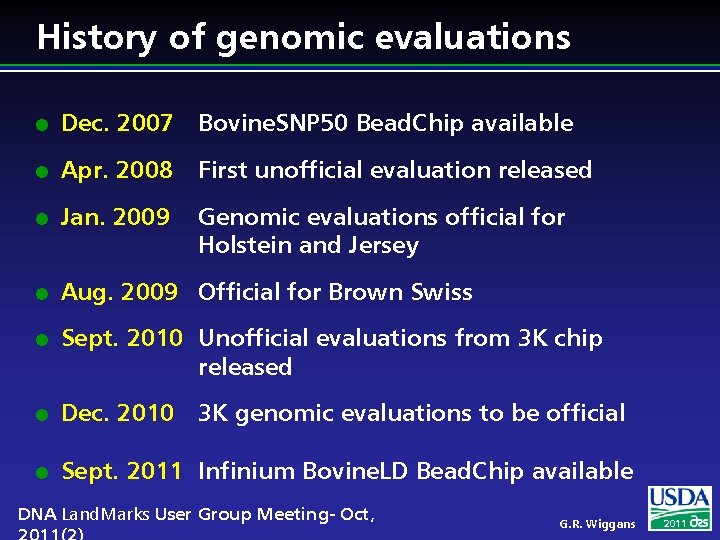
History of genomic evaluations l Dec. 2007 Bovine. SNP 50 Bead. Chip available l Apr. 2008 First unofficial evaluation released l Jan. 2009 Genomic evaluations official for Holstein and Jersey l Aug. 2009 Official for Brown Swiss l Sept. 2010 Unofficial evaluations from 3 K chip released l Dec. 2010 3 K genomic evaluations to be official l Sept. 2011 Infinium Bovine. LD Bead. Chip available DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

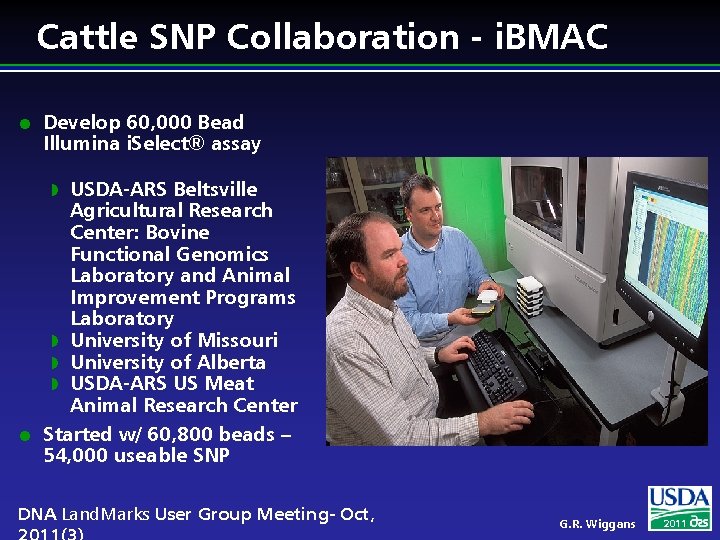
Cattle SNP Collaboration - i. BMAC l Develop 60, 000 Bead Illumina i. Select® assay USDA-ARS Beltsville Agricultural Research Center: Bovine Functional Genomics Laboratory and Animal Improvement Programs Laboratory w University of Missouri w University of Alberta w USDA-ARS US Meat Animal Research Center Started w/ 60, 800 beads – 54, 000 useable SNP w l DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Chips l l l Bovine. SNP 50 w Version 1 54, 001 SNP w Version 2 54, 609 SNP w 45, 187 used in evaluations 50 KV 2 HD LD HD w 777, 962 SNP w Only 50 K SNP used, w >1700 in database LD w 6, 909 SNP w Replaces 3 K DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Use of HD l l Currently only 50 K subset of SNP used Some increase in accuracy from better tracking of QTL possible Potential for across breed evaluations Requires few new HD genotypes once adequate base for imputation developed DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

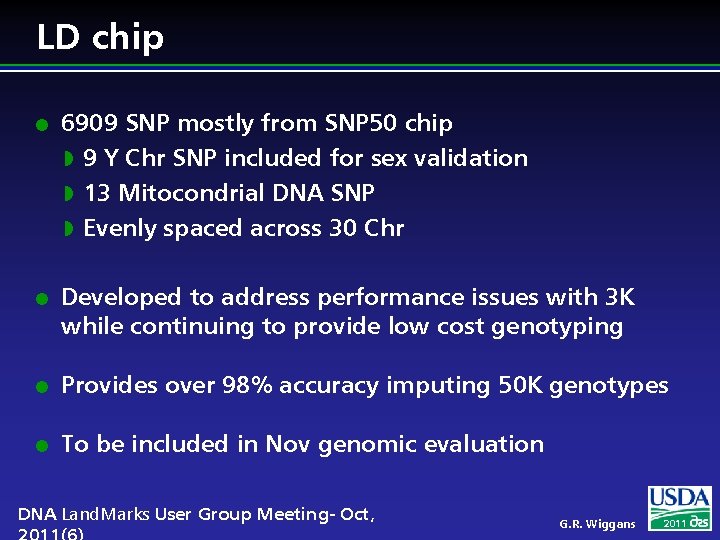
LD chip l l 6909 SNP mostly from SNP 50 chip w 9 Y Chr SNP included for sex validation w 13 Mitocondrial DNA SNP w Evenly spaced across 30 Chr Developed to address performance issues with 3 K while continuing to provide low cost genotyping l Provides over 98% accuracy imputing 50 K genotypes l To be included in Nov genomic evaluation DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Development of LD chip l l Consortium included researchers from USA, AUS and FRA Objective: good imputation performance in dairy breeds w Uniform distribution except heavier at chromosome ends w High MAF, avg MAF over 30% for most breeds w Adequate overlap with 3 K DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Responsibilities of requester l l l Insure animal is properly identified eg HOCANF 000123456789 Enroll animal with breed association or insure pedigree on animal and dam reaches AIPL Collect clean, clearly labeled DNA sample Get sample to lab in time to be included in desired month’s results Resolve parentage conflicts quickly DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Steps to prepare genotypes l Nominate animal for genotyping l Collect blood, hair, semen, nasal swab, or ear punch w Blood may not be suitable for twins l Extract DNA at laboratory l Prepare DNA and apply to Bead. Chip l Do amplification and hybridization, 3 -day process l Read red/green intensities from chip and call genotypes from clusters DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

What can go wrong l l l Sample does not provide adequate DNA quality or quantity Genotype has many SNP that can not be determined (90% call rate required) Parent-progeny conflicts w Pedigree error w Sample ID error (Switched samples) w Laboratory error w Parent-progeny relationship detected that is not in pedigree DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

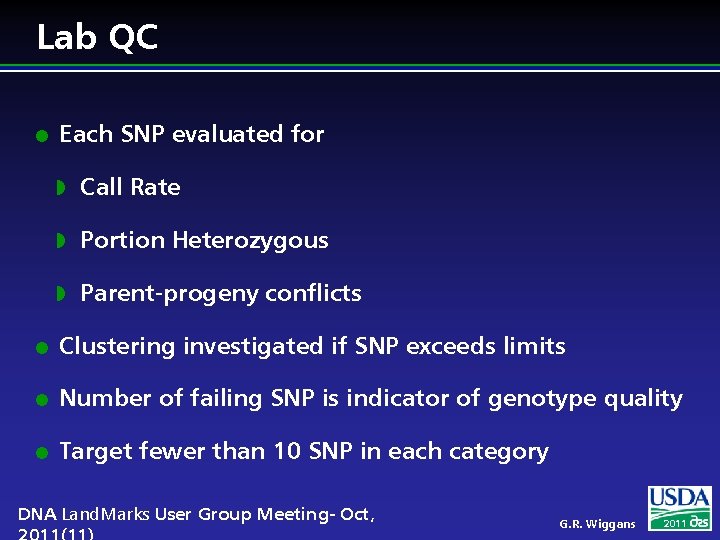
Lab QC l Each SNP evaluated for w Call Rate w Portion Heterozygous w Parent-progeny conflicts l Clustering investigated if SNP exceeds limits l Number of failing SNP is indicator of genotype quality l Target fewer than 10 SNP in each category DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

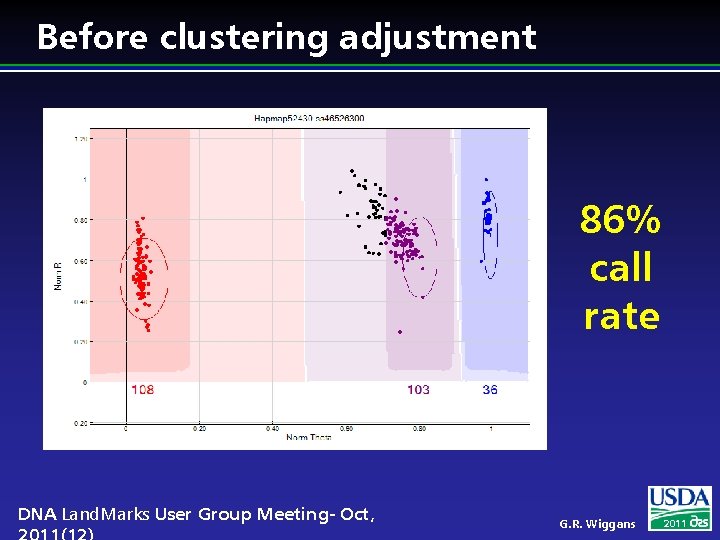
Before clustering adjustment 86% call rate DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

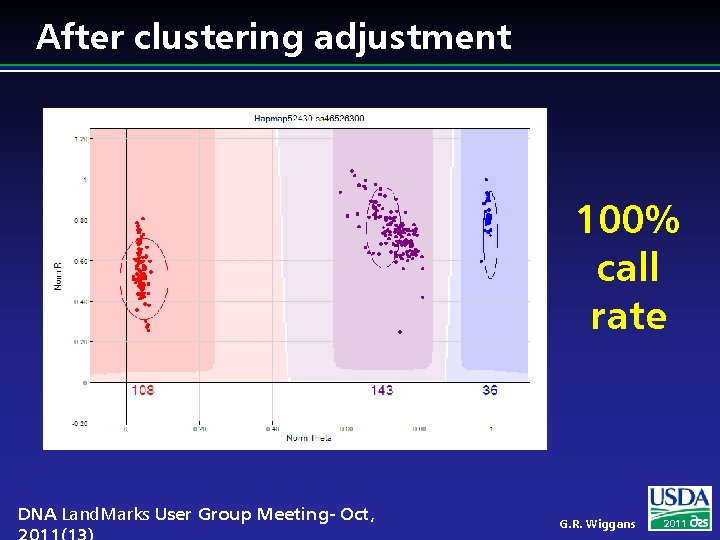
After clustering adjustment 100% call rate DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Parentage validation and discovery l Parent-progeny conflicts detected Animal checked against all other genotypes w Reported to breeds and requesters w Correct sire usually detected w l Maternal Grandsire checking SNP at a time checking w Haplotype checking more accurate w l Breeds moving to accept SNP in place of microsatellites DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Checking facility l l Labs place genotype files on AIPL server Genotypes run through analysis procedures, but not added to database Reports on missing nominations and QC data returned to Lab can Detect sample misidentification w Improve clustering w Applying the same checks used by AIPL w DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Imputation l l Based on splitting the genotype into individual chromosomes (maternal & paternal contributions) Missing SNP assigned by tracking inheritance from ancestors and descendents Imputed dams increase predictor population 3 K, LD, & 50 K genotypes merged by imputing SNP not on LD or 3 K DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

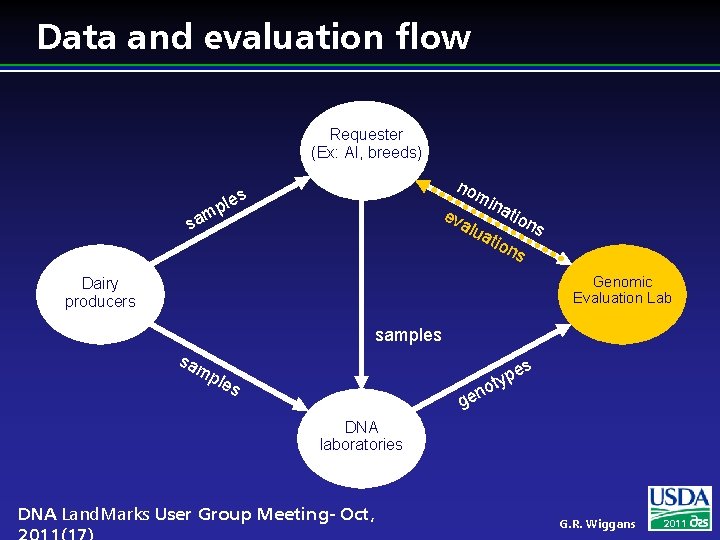
Data and evaluation flow Requester (Ex: AI, breeds) mp sa no les ev mi na alu tio ns ati on s Genomic Evaluation Lab Dairy producers samples sa es mp les p oty n ge DNA laboratories DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Collaboration l Full sharing of genotypes with Canada w l Trading of Brown Swiss genotypes with Switzerland, Germany, and Austria w l CDN calculates genomic evaluations on Canadian base Interbull may facilitate sharing Agreements with Italy and Great Britain provide genotypes for Holstein w Negotiations underway with other countries DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

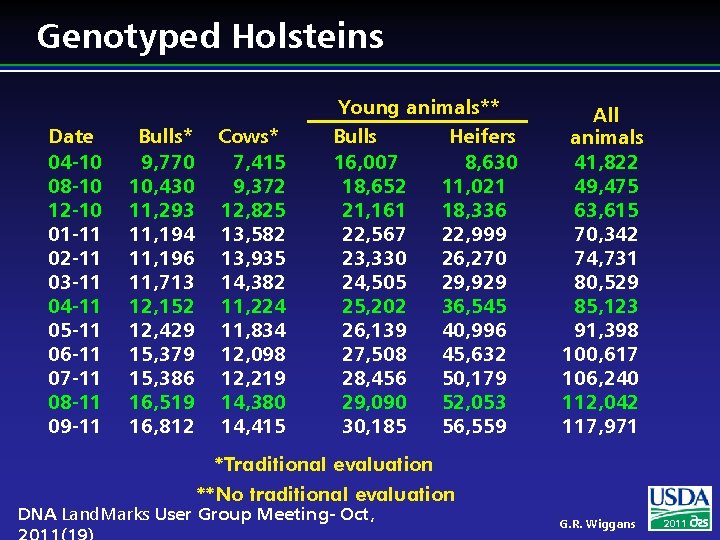
Genotyped Holsteins Date 04 -10 08 -10 12 -10 01 -11 02 -11 03 -11 04 -11 05 -11 06 -11 07 -11 08 -11 09 -11 Bulls* 9, 770 10, 430 11, 293 11, 194 11, 196 11, 713 12, 152 12, 429 15, 379 15, 386 16, 519 16, 812 Cows* 7, 415 9, 372 12, 825 13, 582 13, 935 14, 382 11, 224 11, 834 12, 098 12, 219 14, 380 14, 415 Young animals** Bulls Heifers 16, 007 8, 630 18, 652 11, 021 21, 161 18, 336 22, 567 22, 999 23, 330 26, 270 24, 505 29, 929 25, 202 36, 545 26, 139 40, 996 27, 508 45, 632 28, 456 50, 179 29, 090 52, 053 30, 185 56, 559 All animals 41, 822 49, 475 63, 615 70, 342 74, 731 80, 529 85, 123 91, 398 100, 617 106, 240 112, 042 117, 971 *Traditional evaluation **No traditional evaluation DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

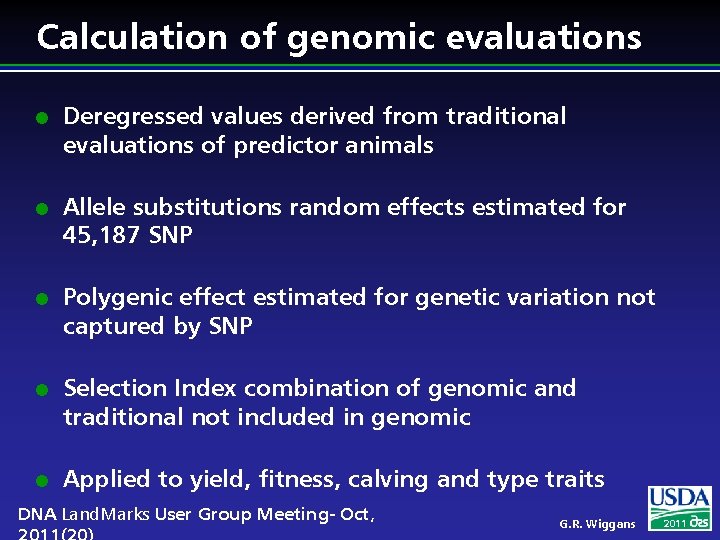
Calculation of genomic evaluations l l l Deregressed values derived from traditional evaluations of predictor animals Allele substitutions random effects estimated for 45, 187 SNP Polygenic effect estimated for genetic variation not captured by SNP Selection Index combination of genomic and traditional not included in genomic Applied to yield, fitness, calving and type traits DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

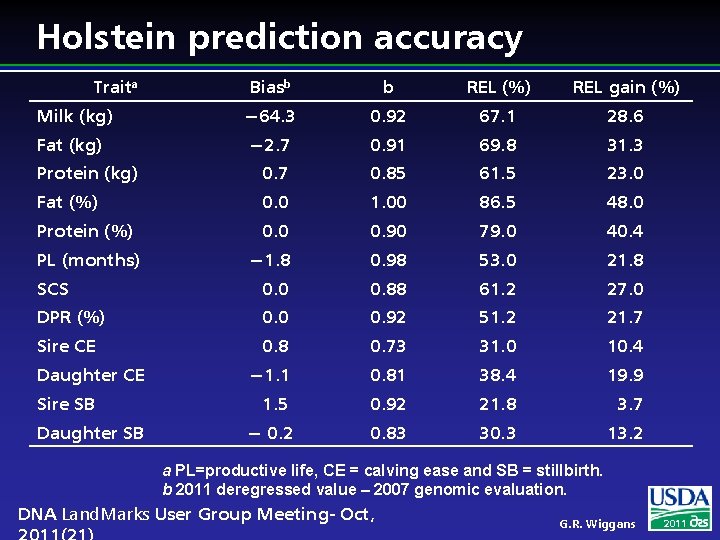
Holstein prediction accuracy Traita Biasb b REL (%) REL gain (%) Milk (kg) − 64. 3 0. 92 67. 1 28. 6 Fat (kg) − 2. 7 0. 91 69. 8 31. 3 Protein (kg) 0. 7 0. 85 61. 5 23. 0 Fat (%) 0. 0 1. 00 86. 5 48. 0 Protein (%) 0. 0 0. 90 79. 0 40. 4 − 1. 8 0. 98 53. 0 21. 8 SCS 0. 0 0. 88 61. 2 27. 0 DPR (%) 0. 0 0. 92 51. 2 21. 7 Sire CE 0. 8 0. 73 31. 0 10. 4 − 1. 1 0. 81 38. 4 19. 9 1. 5 0. 92 21. 8 3. 7 − 0. 2 0. 83 30. 3 13. 2 PL (months) Daughter CE Sire SB Daughter SB a PL=productive life, CE = calving ease and SB = stillbirth. b 2011 deregressed value – 2007 genomic evaluation. DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

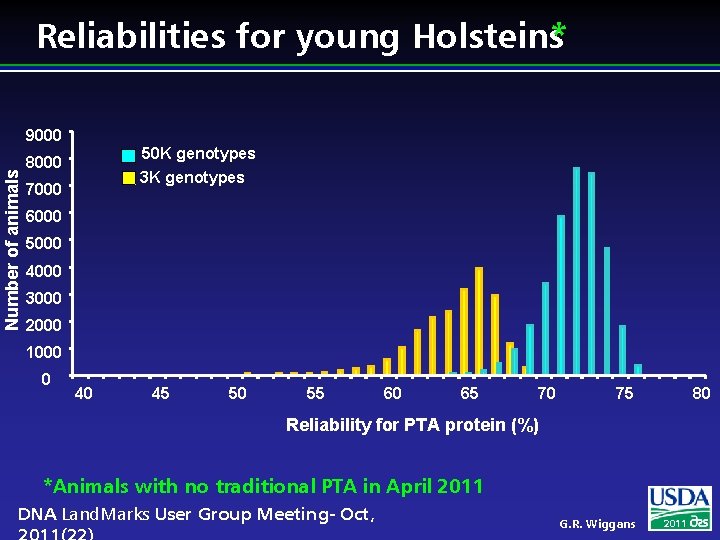
Reliabilities for young Holsteins* Number of animals 9000 50 K genotypes 8000 3 K genotypes 7000 6000 5000 4000 3000 2000 1000 0 40 45 50 55 60 65 70 75 80 Reliability for PTA protein (%) *Animals with no traditional PTA in April 2011 DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

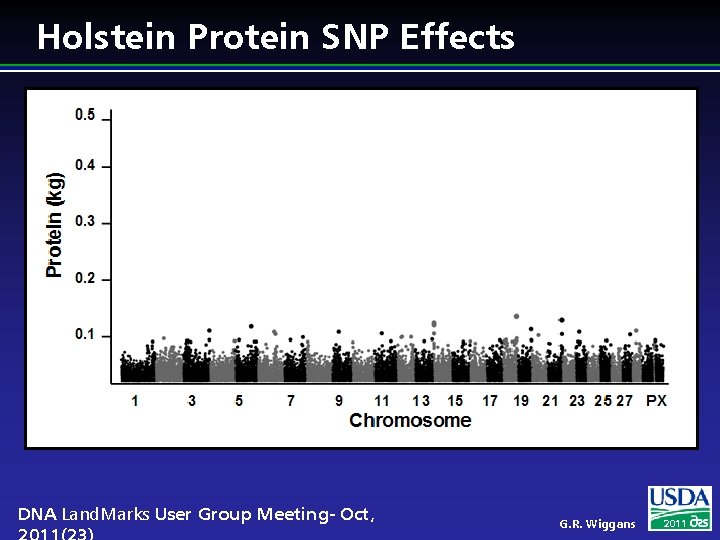
Holstein Protein SNP Effects DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Use of genomic evaluations l Determine which young bulls to bring into AI service l Use to select mating sires l Pick bull dams l Market semen from 2 -year-old bulls DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Use of LD genomic evaluations l Sort heifers for breeding w Flush w Sexed semen w Beef bull l Confirm parentage to avoid inbreeding l Predict inbreeding depression better l Precision mating considering genomics(future) DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Ways to increase accuracy l l Automatic addition of traditional evaluations of genotyped bulls when reach 5 years of age Possible genotyping of 10, 000 bulls with semen in CDDR l Collaboration with more countries l Use of more SNP from HD chips l Full sequencing DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Application to more traits l l l Animal’s genotype is good for all traits Traditional evaluations required for accurate estimates of SNP effects Traditional evaluations not currently available for heat tolerance or feed efficiency Research populations could provide data for traits that are expensive to measure Will resulting evaluations work in target population? DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Impact on producers l l l Young-bull evaluations with accuracy of early 1 st crop evaluations AI organizations marketing genomically evaluated 2 year-olds Genotype usually required for cow to be bull dam Rate of genetic improvement likely to increase by up to 50% Studs reducing progeny-test programs DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Why Genomics works in Dairy l Extensive historical data available l Well developed genetic evaluation program l Widespread use of AI sires l Progeny test programs l High valued animals, worth the cost of genotyping l Long generation interval which can be reduced substantially by genomics DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

Summary l l l Extraordinarily rapid implementation of genomic evaluations Chips provide genotypes of high accuracy Comprehensive checking insures quality of genotypes stored Young-bull acquisition and marketing now based on genomic evaluations Genotyping of many females because of Low density chips DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011

DNA Land. Marks User Group Meeting- Oct, G. R. Wiggans 2011