Genomic Selection in Dairy Cattle Paul Van Raden



- Slides: 15

Genomic Selection in Dairy Cattle Paul Van. Raden, Curt Van Tassell, George Wiggans, Tad Sonstegard, and Jeff O’Connell Animal Improvement Programs Laboratory and Bovine Functional Genomics Laboratory, USDA Agricultural Research Service, Beltsville, MD, USA Paul. Van. Raden@ars. usda. gov 2008 2007

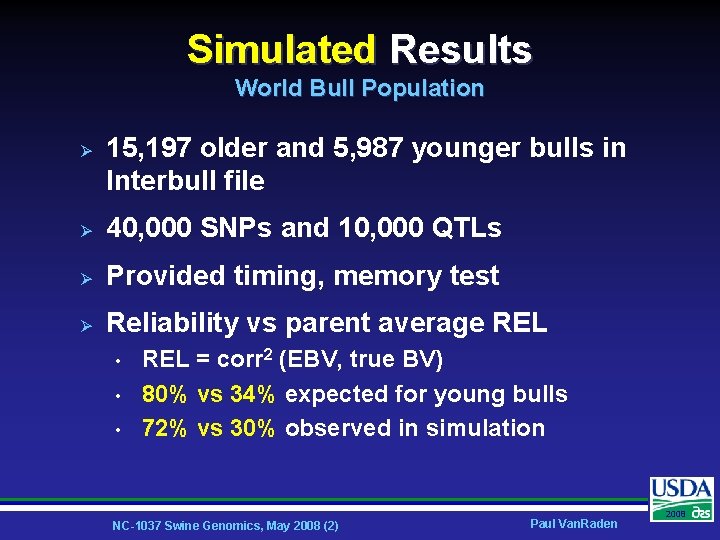
Simulated Results World Bull Population Ø 15, 197 older and 5, 987 younger bulls in Interbull file Ø 40, 000 SNPs and 10, 000 QTLs Ø Provided timing, memory test Ø Reliability vs parent average REL • • • REL = corr 2 (EBV, true BV) 80% vs 34% expected for young bulls 72% vs 30% observed in simulation NC-1037 Swine Genomics, May 2008 (2) Paul Van. Raden 2008

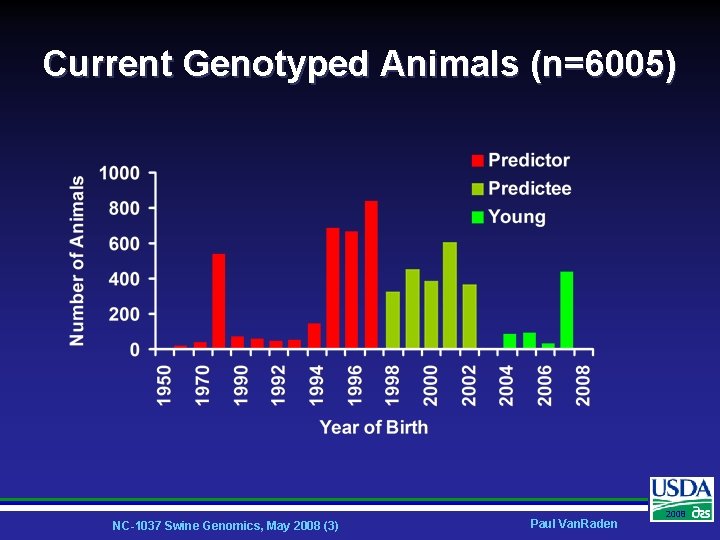
Current Genotyped Animals (n=6005) NC-1037 Swine Genomics, May 2008 (3) Paul Van. Raden 2008

Genomic Methods Ø Direct genomic evaluation • • Ø Inversion for linear prediction, REL Iteration for nonlinear prediction Combined genomic evaluation • • Traditional PA or PTA, subset PA or PTA, and direct genomic combined by REL in 3 x 3 selection index Nonlinear genomic predictions used NC-1037 Swine Genomics, May 2008 (4) Paul Van. Raden 2008

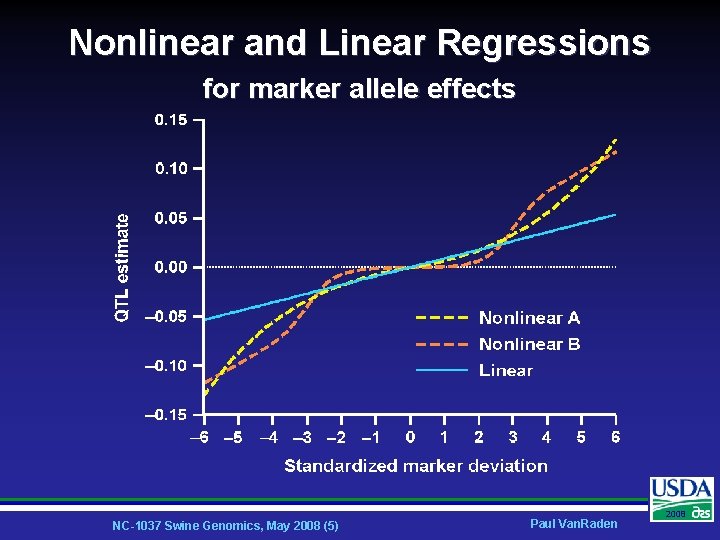
Nonlinear and Linear Regressions for marker allele effects NC-1037 Swine Genomics, May 2008 (5) Paul Van. Raden 2008

Experimental Design Actual North American Data Ø Ø Ø August 2003 PTAs for 3, 576 older bulls to predict January 2008 daughter deviations for 1, 759 younger bulls (total = 5, 335 bulls) Results computed for 27 traits: 5 yield, 5 health, 16 conformation, and Net Merit Nonlinear A used, B didn’t work NC-1037 Swine Genomics, May 2008 (6) Paul Van. Raden 2008

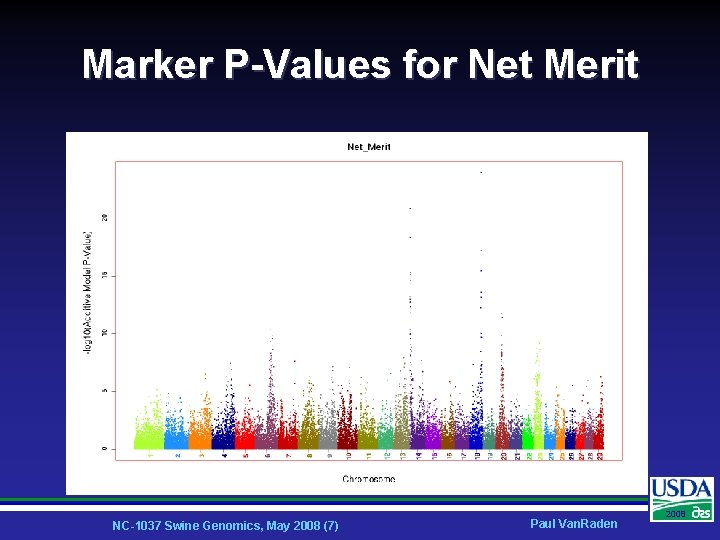
Marker P-Values for Net Merit NC-1037 Swine Genomics, May 2008 (7) Paul Van. Raden 2008

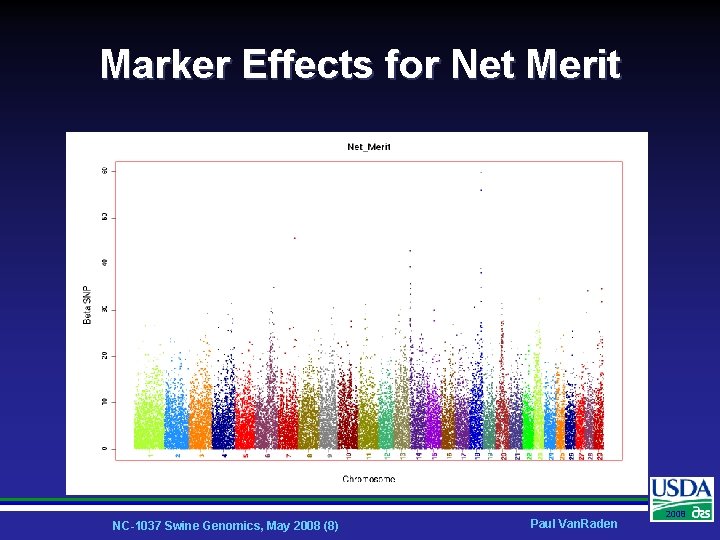
Marker Effects for Net Merit NC-1037 Swine Genomics, May 2008 (8) Paul Van. Raden 2008

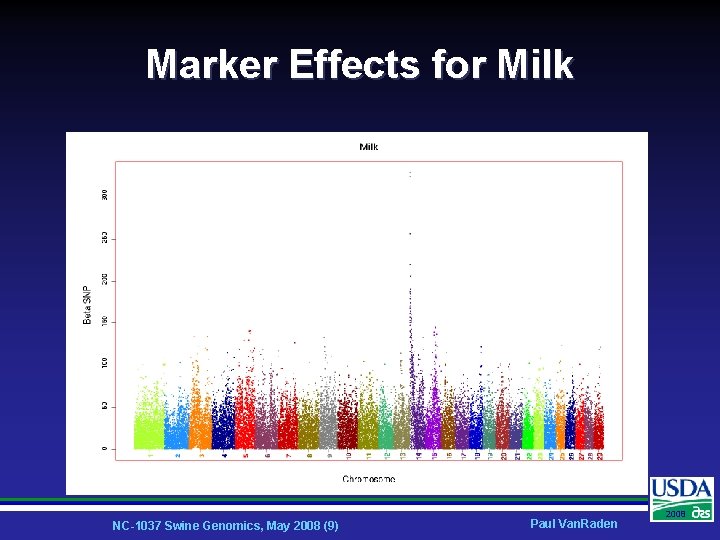
Marker Effects for Milk NC-1037 Swine Genomics, May 2008 (9) Paul Van. Raden 2008

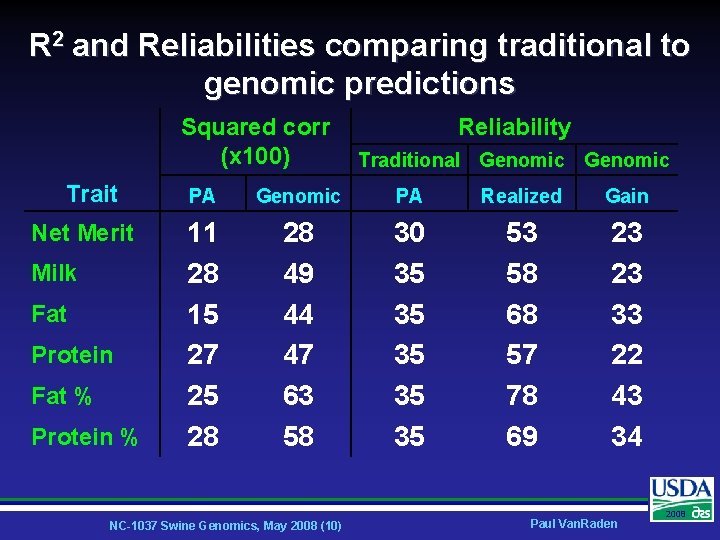
R 2 and Reliabilities comparing traditional to genomic predictions Squared corr (x 100) Trait Net Merit Milk Fat Protein Fat % Protein % Reliability Traditional Genomic PA Realized Gain 11 28 15 27 25 28 28 49 44 47 63 58 30 35 35 35 53 58 68 57 78 69 23 23 33 22 43 34 NC-1037 Swine Genomics, May 2008 (10) Paul Van. Raden 2008

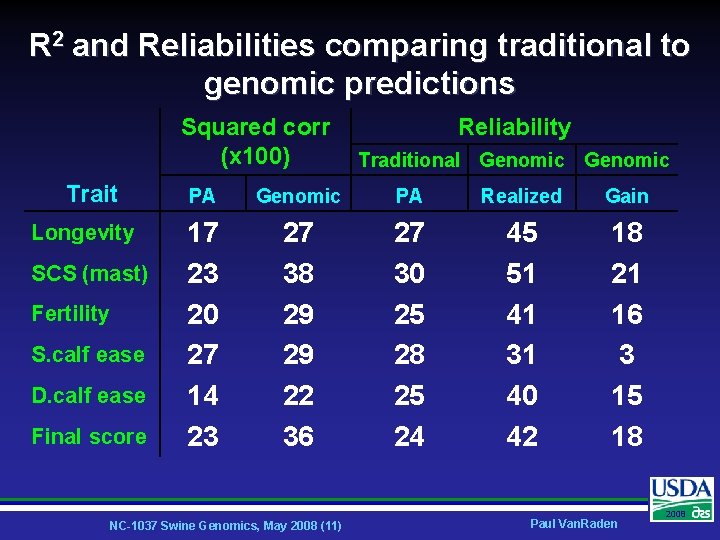
R 2 and Reliabilities comparing traditional to genomic predictions Squared corr (x 100) Trait Longevity SCS (mast) Fertility S. calf ease D. calf ease Final score Reliability Traditional Genomic PA Realized Gain 17 23 20 27 14 23 27 38 29 29 22 36 27 30 25 28 25 24 45 51 41 31 40 42 18 21 16 3 15 18 NC-1037 Swine Genomics, May 2008 (11) Paul Van. Raden 2008

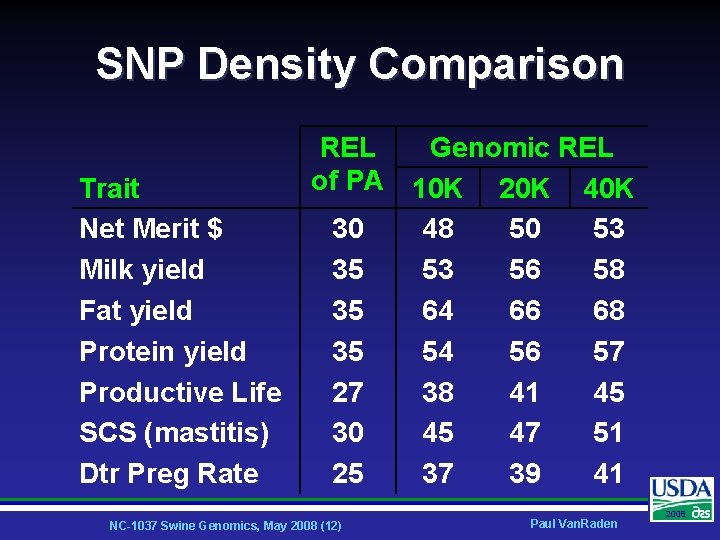
SNP Density Comparison Trait Net Merit $ Milk yield Fat yield Protein yield Productive Life SCS (mastitis) Dtr Preg Rate REL Genomic REL of PA 10 K 20 K 40 K 30 48 50 53 35 53 56 58 35 64 66 68 35 54 56 57 27 38 41 45 30 45 47 51 25 37 39 41 NC-1037 Swine Genomics, May 2008 (12) Paul Van. Raden 2008

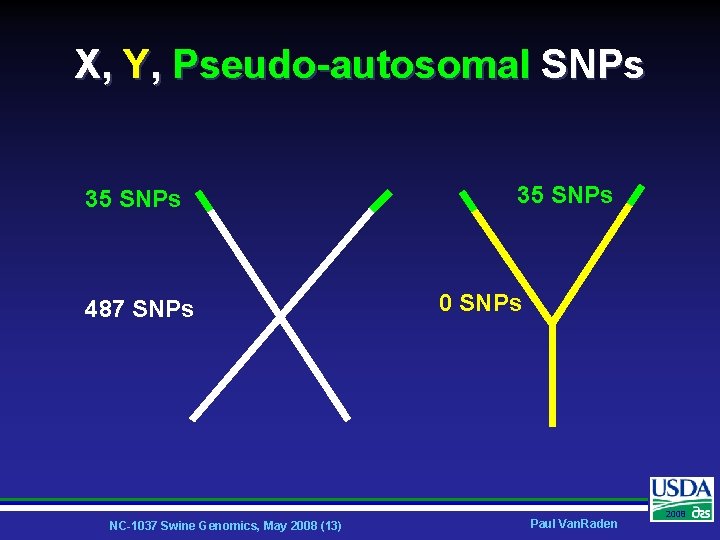
X, Y, Pseudo-autosomal SNPs 35 SNPs 487 SNPs NC-1037 Swine Genomics, May 2008 (13) 35 SNPs 0 SNPs Paul Van. Raden 2008

Conclusions Ø Ø Genomic predictions significantly better than parent average (P <. 0001) for all 26 traits tested Gains in reliability equivalent on average to 11 daughters with records • • Ø Analysis used 3, 576 historical bulls Current data includes 5, 285 proven bulls Larger populations require more SNPs NC-1037 Swine Genomics, May 2008 (14) Paul Van. Raden 2008

Acknowledgments Ø Funding: • • Ø Genotyping and DNA extraction: • Ø NRI grants 2006 -35205 -16888, 16701 CDDR Contributors (NAAB, Semex) BFGL, U. Missouri, U. Alberta, Gene. Seek, GIFV, and Illumina Computing from AIPL staff NC-1037 Swine Genomics, May 2008 (15) Paul Van. Raden 2008