Macromolecular SmallAngle Xray Scattering What is SAXS Small
















- Slides: 64

Macromolecular Small-Angle X-ray Scattering

What is SAXS? • • Small Angle X-ray Scattering proportional to /Molecular size Typical x-ray wavelengths ~ 0. 1 nm Typical molecular dimensions 1 -100 nm Scattering angles are small 0 -2 o historically. Now 0 -15 o range is of increasing experimental interest

Why SAXS ? • Atomic level structures from crystallography or NMR “gold standard” for structural inferences • Crystallography, by definition, studies static structures • Most things crystallize only under rather specific, artificial conditions • Kinetics of molecular interactions frequently of interest • SAXS can provide useful, although limited, information on relatively fast time scales

Why not visible light? • Diffraction effects limit to ~ /50 (5 ) • i. e. 14 nm visible light vs 1 - 2 nm RG for small proteins • Water absorbs many wavelengths of light strongly • 0. 2 < < 160 nm experimentally inaccessible

What is SAXS Used for? • • Estimating sizes of particulates Interactions in fluids Sizes of micelles etc in emulsions Size distributions of subcomponents in materials • Structure and dynamics of biological macromolecules

SAXS and Biological Macromolecules • How well does the crystal structure represents the native structure in solution? • Can we get even some structural from large proportion of macro-molecules that do not crystallize? • How can we test hypotheses concerning large scale structural changes on ligand binding etc. in solution • SAXS can frequently provide enough information for such studies • May even be possible to deduce protein fold solely from SAXS data

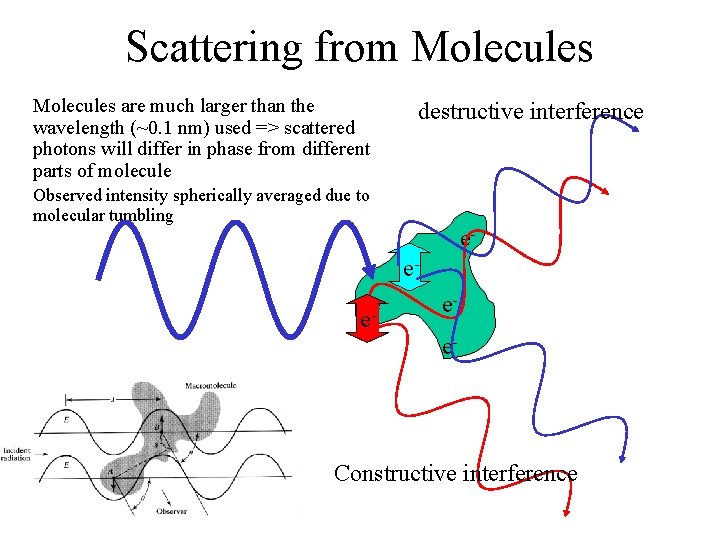
Scattering from Molecules are much larger than the wavelength (~0. 1 nm) used => scattered photons will differ in phase from different parts of molecule destructive interference Observed intensity spherically averaged due to molecular tumbling eee- e ee- Constructive interference

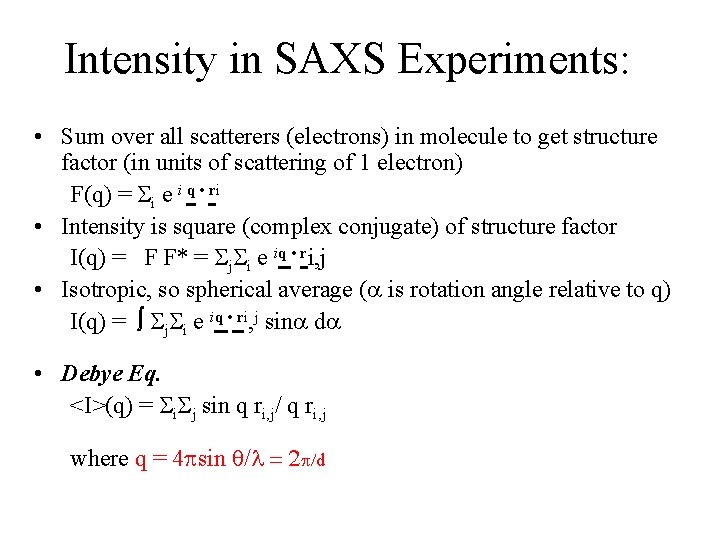
Intensity in SAXS Experiments: • Sum over all scatterers (electrons) in molecule to get structure factor (in units of scattering of 1 electron) F(q) = i e i q • ri • Intensity is square (complex conjugate) of structure factor I(q) = F F* = j i e iq • ri, j • Isotropic, so spherical average ( is rotation angle relative to q) I(q) = j i e iq • ri, j sin d • Debye Eq. <I>(q) = i j sin q ri, j/ q ri, j where q = 4 sin q/ = 2 /d

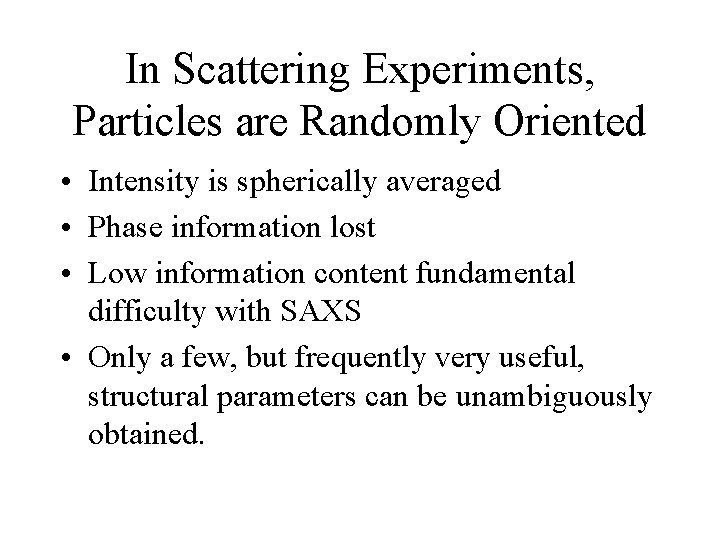
In Scattering Experiments, Particles are Randomly Oriented • Intensity is spherically averaged • Phase information lost • Low information content fundamental difficulty with SAXS • Only a few, but frequently very useful, structural parameters can be unambiguously obtained.

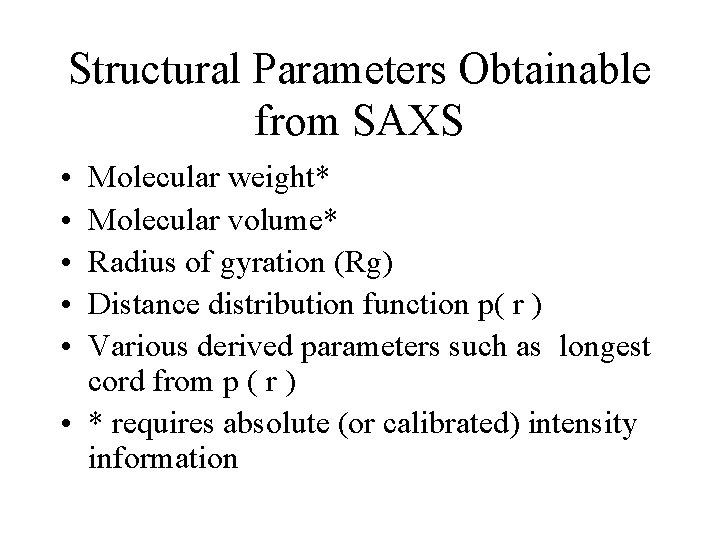
Structural Parameters Obtainable from SAXS • • • Molecular weight* Molecular volume* Radius of gyration (Rg) Distance distribution function p( r ) Various derived parameters such as longest cord from p ( r ) • * requires absolute (or calibrated) intensity information

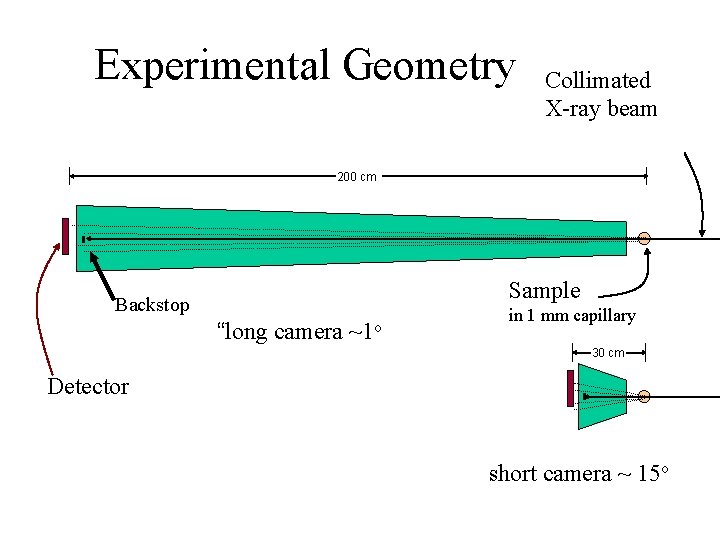
Experimental Geometry Collimated X-ray beam 200 cm Sample Backstop “long camera ~1 o in 1 mm capillary 30 cm Detector short camera ~ 15 o

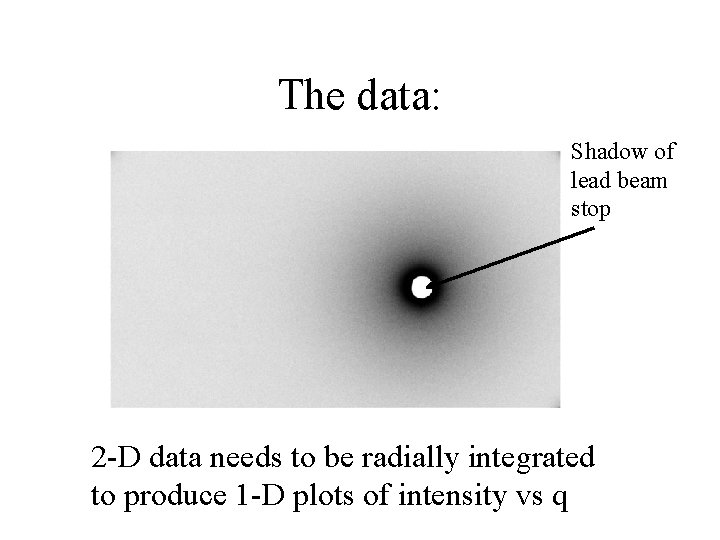
The data: Shadow of lead beam stop 2 -D data needs to be radially integrated to produce 1 -D plots of intensity vs q

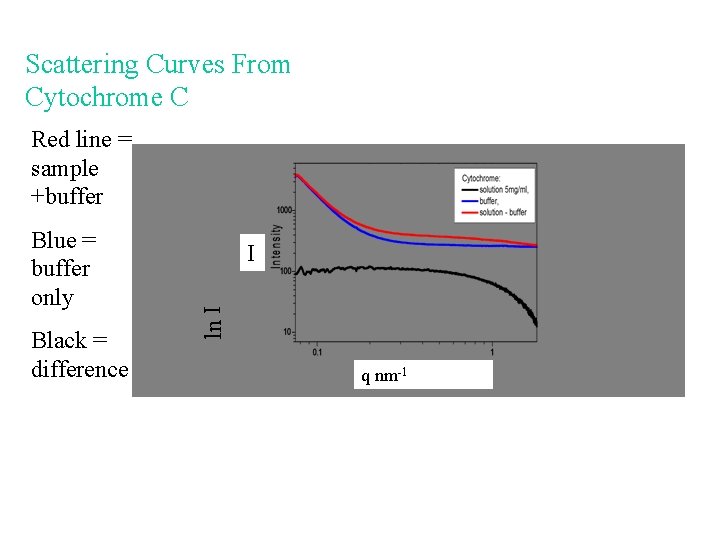
Scattering Curves From Cytochrome C Red line = sample +buffer Black = difference I ln I Blue = buffer only q nm-1

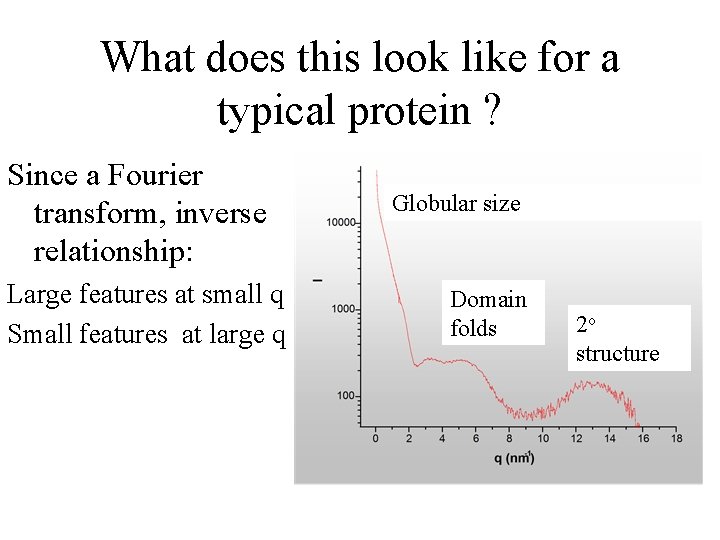
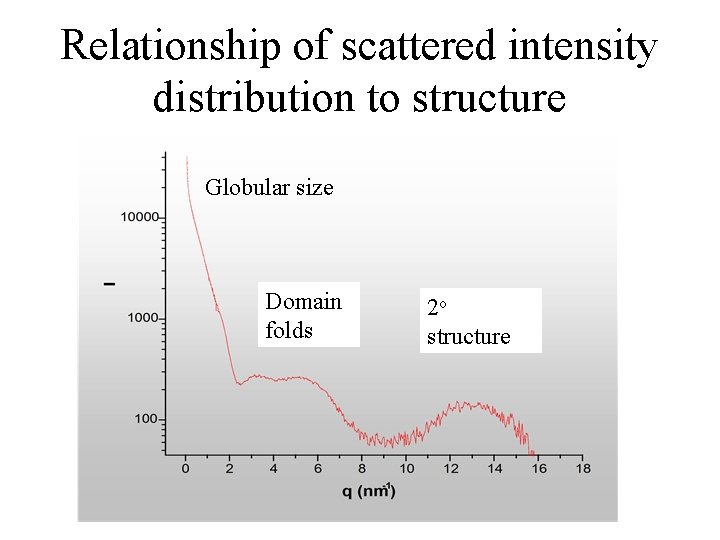
What does this look like for a typical protein ? Since a Fourier transform, inverse relationship: Large features at small q Small features at large q Globular size Domain folds 2 o structure

What’s Rg? • Analogous to moment of inertia in mechanics • Rg 2 = p(r) r 2 d. V p(r) d. V

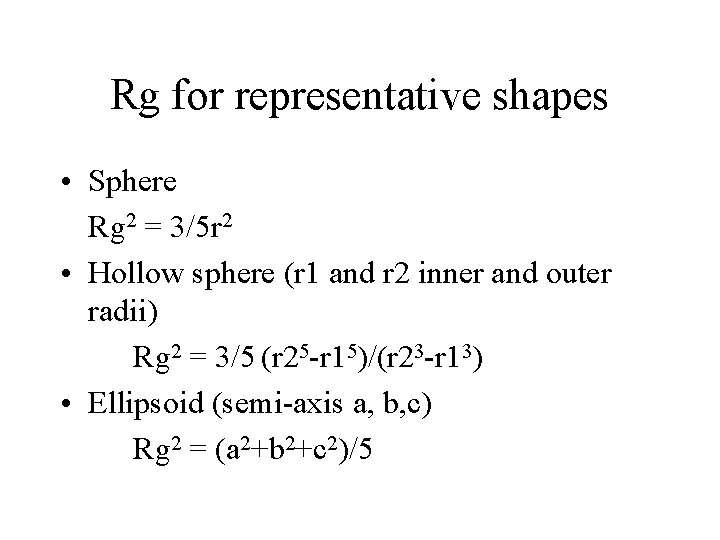
Rg for representative shapes • Sphere Rg 2 = 3/5 r 2 • Hollow sphere (r 1 and r 2 inner and outer radii) Rg 2 = 3/5 (r 25 -r 15)/(r 23 -r 13) • Ellipsoid (semi-axis a, b, c) Rg 2 = (a 2+b 2+c 2)/5

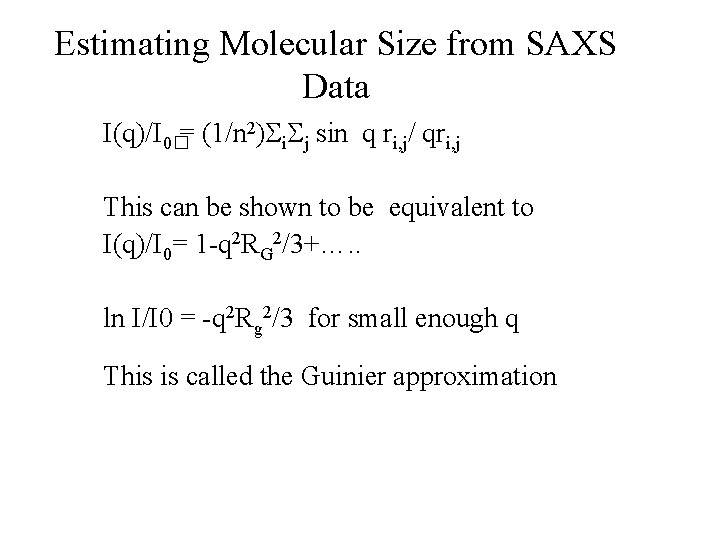
Estimating Molecular Size from SAXS Data I(q)/I 0�= (1/n 2) i j sin q ri, j/ qri, j This can be shown to be equivalent to I(q)/I 0= 1 -q 2 RG 2/3+…. . ln I/I 0 = -q 2 Rg 2/3 for small enough q This is called the Guinier approximation

Guinier Plot ln I vs. q 2 Inner part will be a straight line Slope proportional to Rg 2 – Only valid near q = 0 (i. e. where third term is insignificant) – For spherical objects, Guinier approximation holds even in the third term… so the Guinier region is larger for more globular proteins – Usual limit: Rg qmax <1. 3

Need a good example Guiner plot

Configuration Changes in Plasminogen EACA Bz

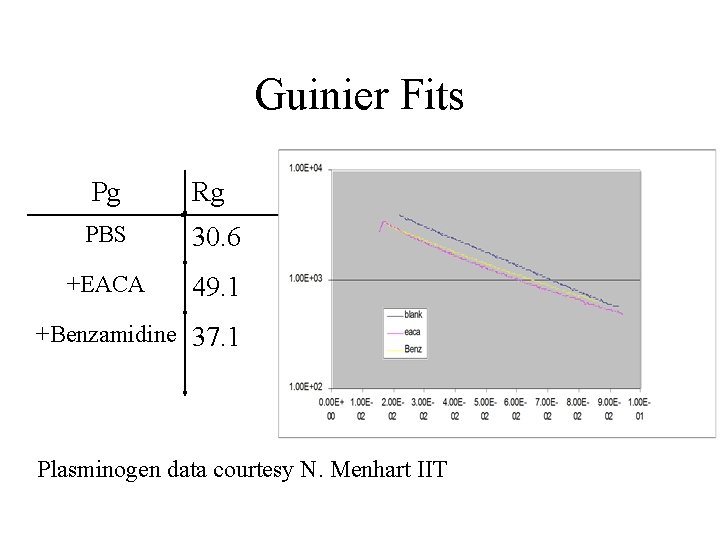
Guinier Fits Pg Rg PBS 30. 6 +EACA 49. 1 +Benzamidine 37. 1 Plasminogen data courtesy N. Menhart IIT

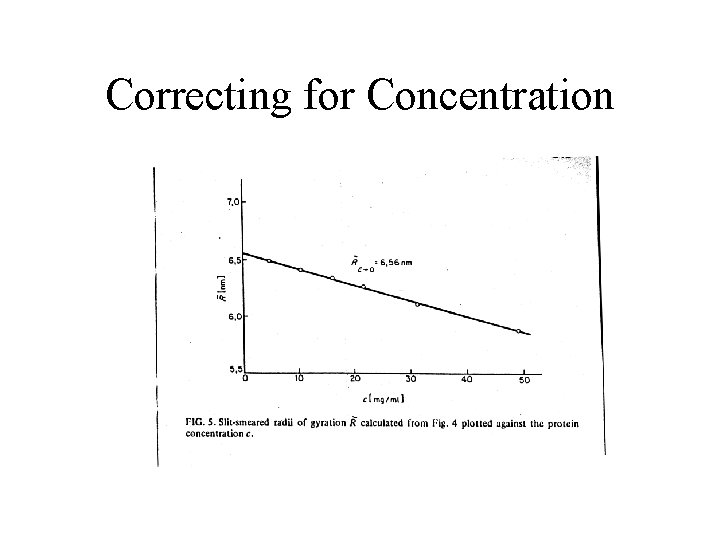
Need for Series of Concentrations • SAXS intensity equations valid only at infinite dilution • Excess density of protein over H 2 O very low • Need a non-negligible concentration ( > 1 mg/ml) to get enough signal. • In practice use a concentration series from ~ 3 - 30 mg/ml and extrapolate to zero by various means • Only affects low angle regime • Can use much higher concentrations for high angle region (where scattering weak anyway)

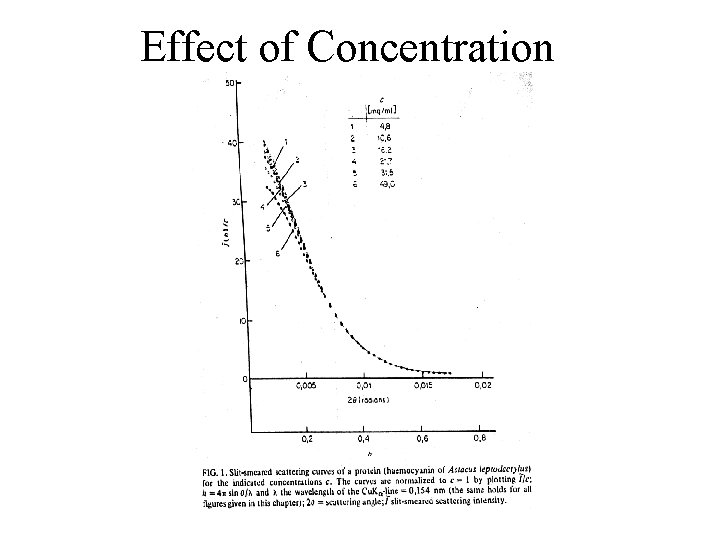
Effect of Concentration

Correcting for Concentration

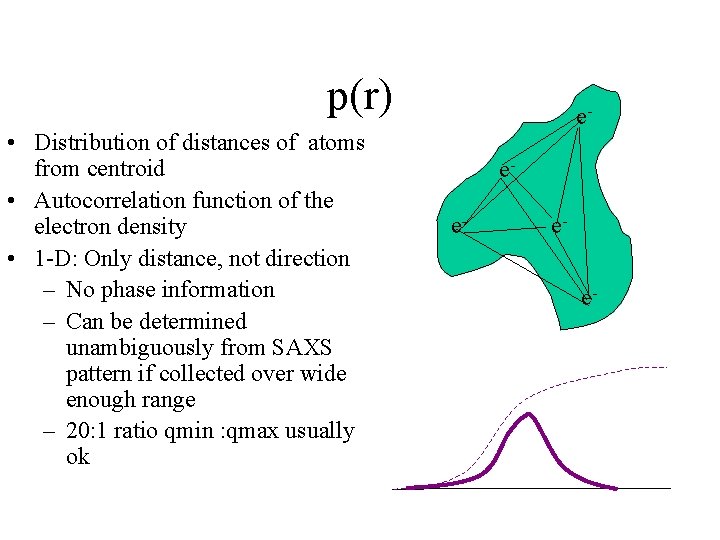
Shape information • SAXS patterns have relatively low information content • Sources of information loss: – Spherical averaging – X-ray phase loss, so can’t invert Fourier transform • In general cannot recover full shape, but can unambiguously compute distribution of distance s within molecule: i. e. p(r) function

p(r) • Distribution of distances of atoms from centroid • Autocorrelation function of the electron density • 1 -D: Only distance, not direction – No phase information – Can be determined unambiguously from SAXS pattern if collected over wide enough range – 20: 1 ratio qmin : qmax usually ok eee-

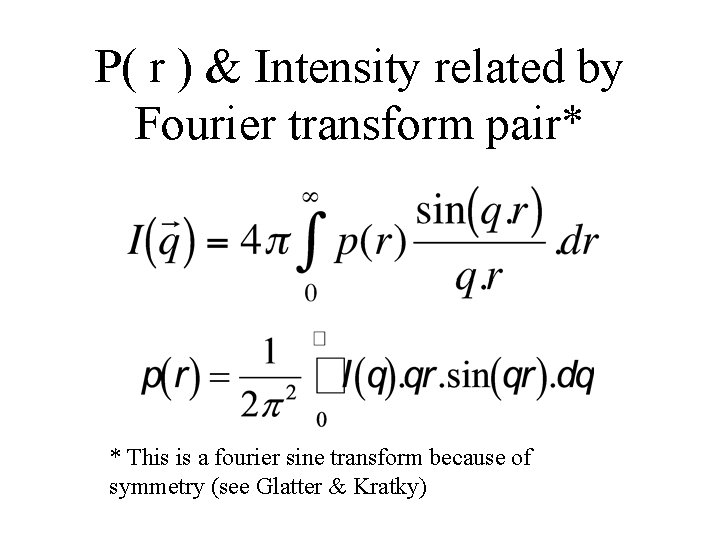
P( r ) & Intensity related by Fourier transform pair* * This is a fourier sine transform because of symmetry (see Glatter & Kratky)

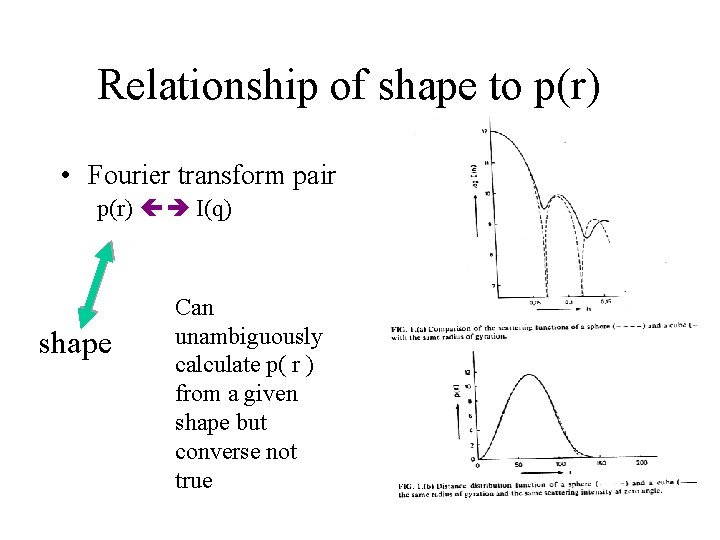
Relationship of shape to p(r) • Fourier transform pair p(r) I(q) shape Can unambiguously calculate p( r ) from a given shape but converse not true

Inversion intensity equation not trivial • Need to worry about termination effects, experimental noise and various smearing effects • Inversion of intensity equation requires use of various “regularization approaches” • One popular approach implemented in program GNOM (Svergun et al. J. Appl. Cryst. 25: 495)

Example of p(r ) Analysis

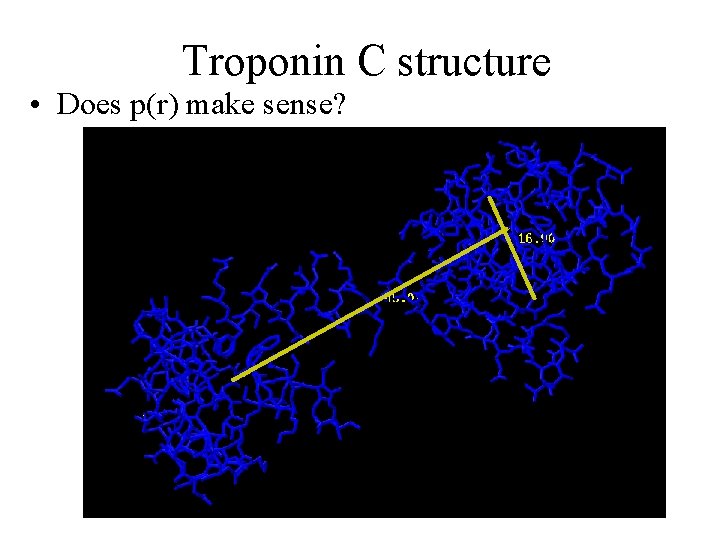
Troponin C structure • Does p(r) make sense?

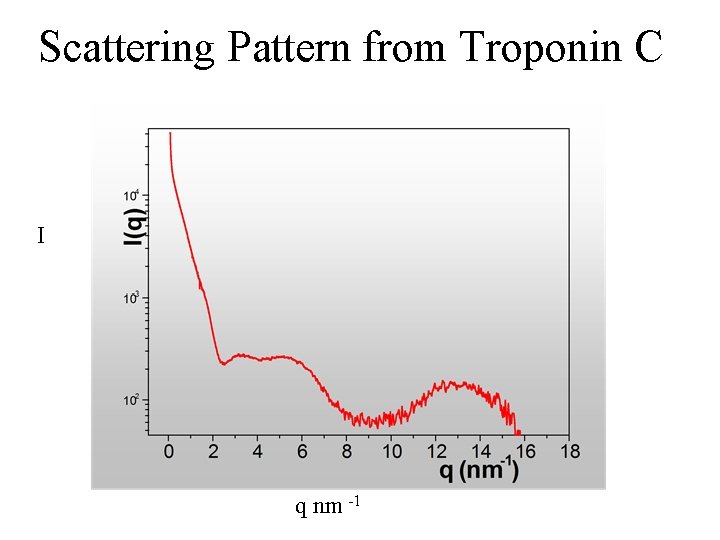
Scattering Pattern from Troponin C I q nm -1

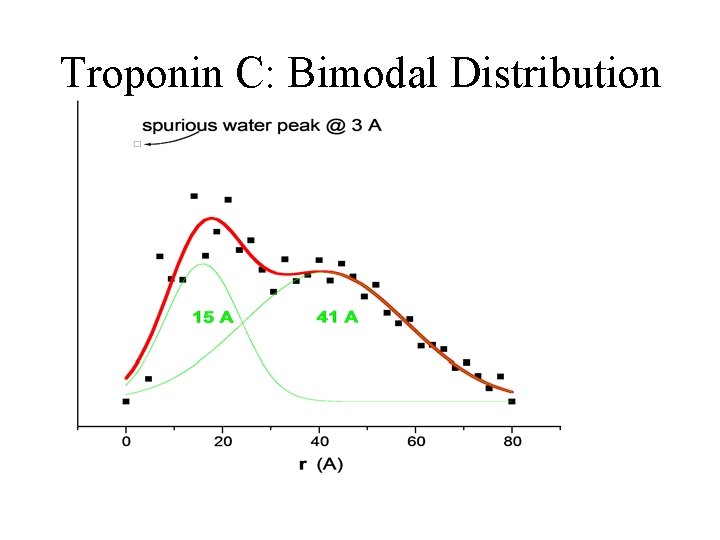
Troponin C: Bimodal Distribution

Hypothesis Testing with SAXS • p (r ) gives an alternative measure of Rg and also “longest cord” • Predict Rg and p( r ) from native crystal structure (tools exist for pdb data) and from computer generated hypothetical structures under conditions of interest • Are the hypothesized structures consistent with SAXS data?

SAXS Data Alone Cannot Yield an Unambiguous Structure • One can combine Rg and P( r ) information with: – Simulations based on other knowledge (i. e. partial structures by NMR or X-ray) – Rigid body refinement Or Whole pattern simulations using various physical criteria: – Positive e density, – finite extent, – Connectivity – chemically meaningful density distributions

Reconstruction of Molecular Envelopes • Very active area of research • 3 main approaches: • Spherical harmonic-based algorithms (Svergun, & Stuhrmann, 1991, Acta Crystallogr. A 47, 736), genetic algorithms (Chacon et al, 1998, Biophys. J. 74, 2760), simulated annealing (Svergun, 1999 Biophys. J. 76, 2879), and “give ‘n take” algorithms (Walter et al, 2000, J. Appl. Cryst 33, 350). • Latter three make use of “Dummy atom approach” using the Debye formula.

Configuration Changes in Plasminogen EACA Bz

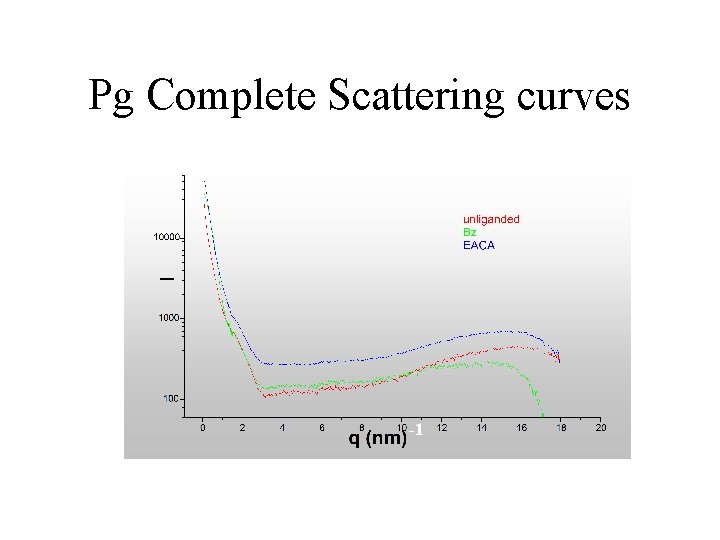
Pg Complete Scattering curves -1

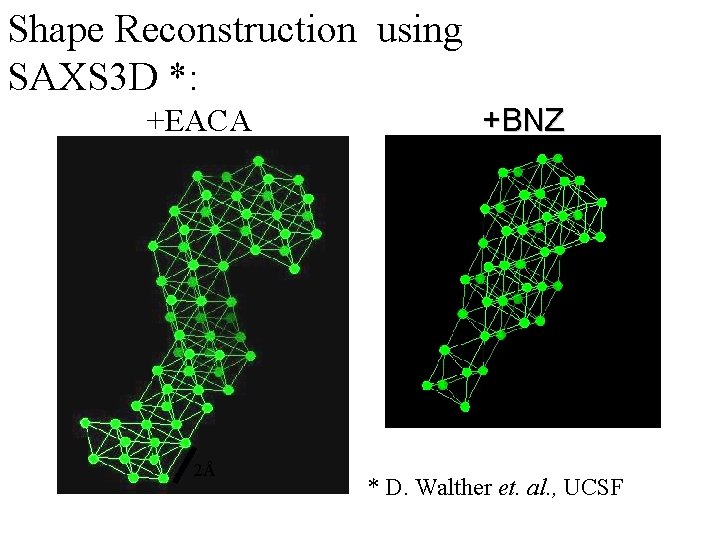
Shape Reconstruction using SAXS 3 D *: +EACA +BNZ 2Å 2Å * D. Walther et. al. , UCSF

Relationship of scattered intensity distribution to structure Globular size Domain folds 2 o structure

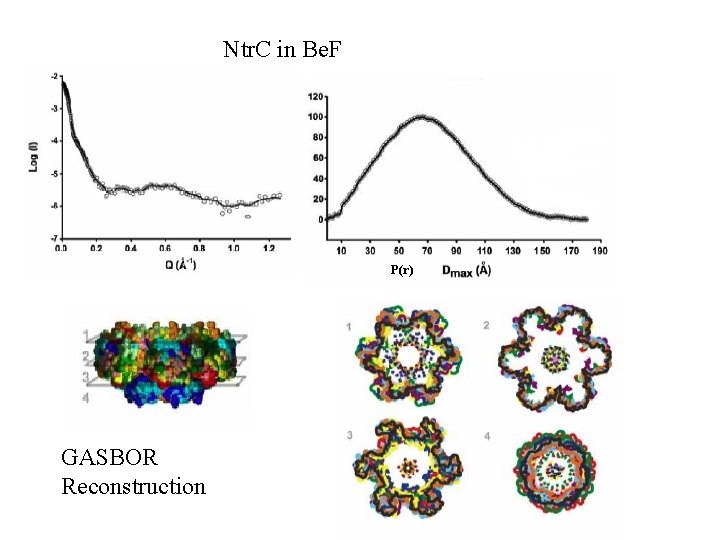
Ntr. C in Be. F P(r) GASBOR Reconstruction

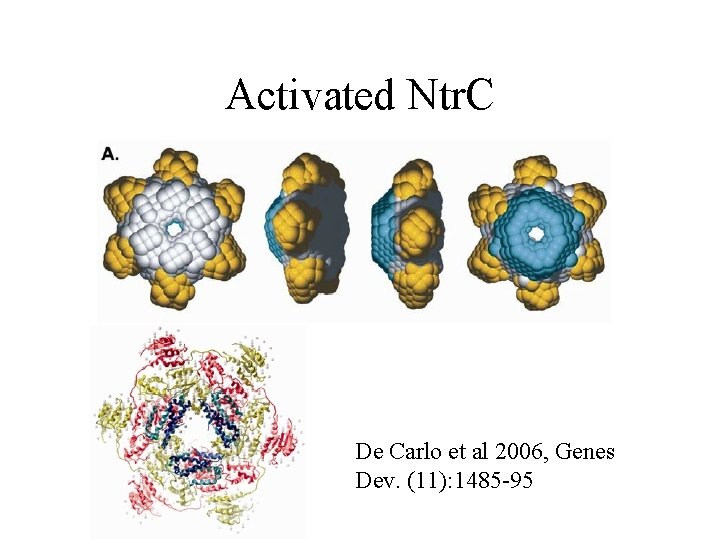
Activated Ntr. C De Carlo et al 2006, Genes Dev. (11): 1485 -95

Ntr. C in various nucleotide states Courtesy B. T. Nixon Penn State

Technical Requirements for SAXS • • Monodispersed sample (usually) Very stable, very well collimated beam Very mechanically stable apparatus Methods to assess and control radiation damage and radiation induced aggregation (flow techniques) • Ability to accurately measure and correct for variations in incident and transmitted beam intensity • High dynamic range, high sensitivity and low noise detector

Detectors For SAXS • 1 -D or 2 D position sensitive gas proportional counters – Pros: High dynamic range, zero read noise – Cons: limited count rate capability typically 105 - 106 cps, 1 -D detectors very inefficient high q range • 2 D CCD detectors – Pros: integrating detectors - no intrinsic count rate limit, 2 -D so can efficiently collect high q data – Cons: Significant read noise, finite dynamic range – Most commercial detectors designed for crystallography too high read noise – Bio. CAT has special purpose, high sensitivity, low noise CCD detectors – Now commercially available from Aviex

Why Do You Need a Third Generation Source for SAXS? • Time resolved protein folding studies using SAXS => The “Protein Folding Problem” • High throughout molecular envelope determinations using SAXS => “Structural genomics” Combined techniques => Liquid chromatography and SAXS

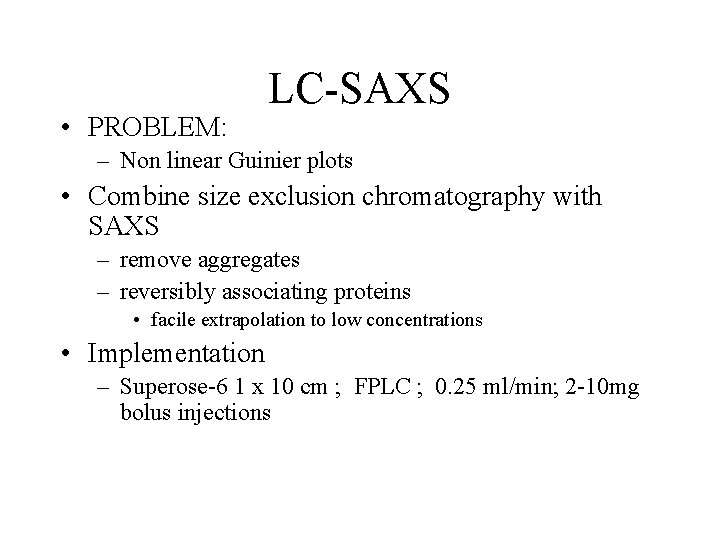
• PROBLEM: LC-SAXS – Non linear Guinier plots • Combine size exclusion chromatography with SAXS – remove aggregates – reversibly associating proteins • facile extrapolation to low concentrations • Implementation – Superose-6 1 x 10 cm ; FPLC ; 0. 25 ml/min; 2 -10 mg bolus injections

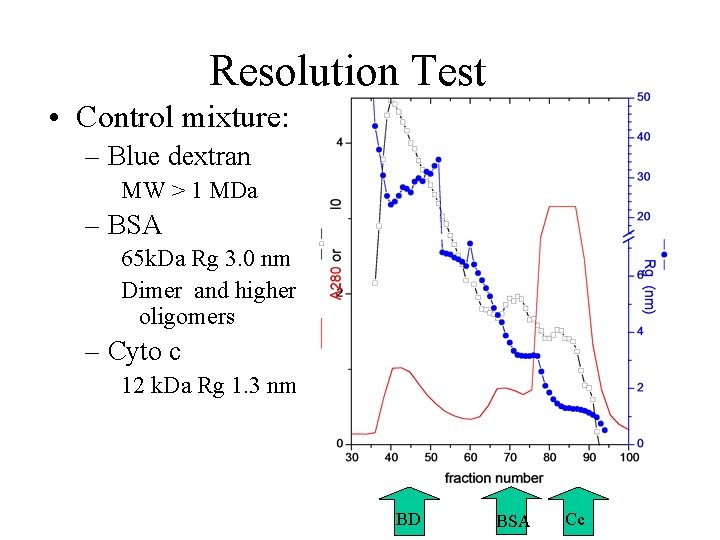
Resolution Test • Control mixture: – Blue dextran MW > 1 MDa – BSA 65 k. Da Rg 3. 0 nm Dimer and higher oligomers – Cyto c 12 k. Da Rg 1. 3 nm BD BSA Cc

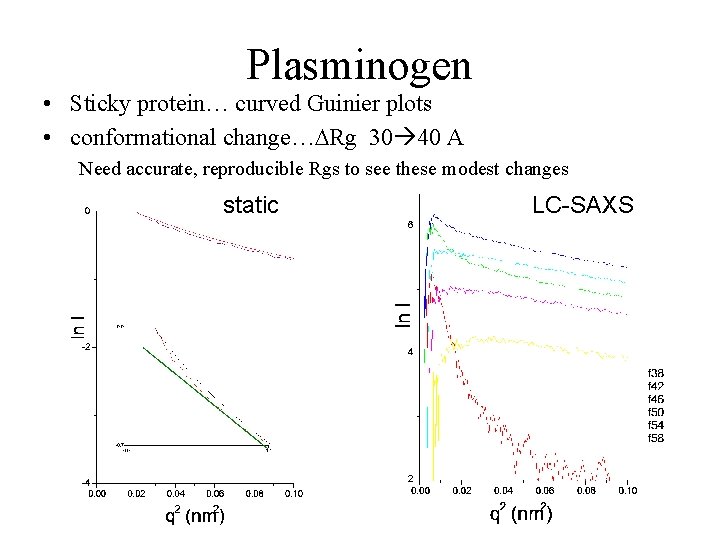
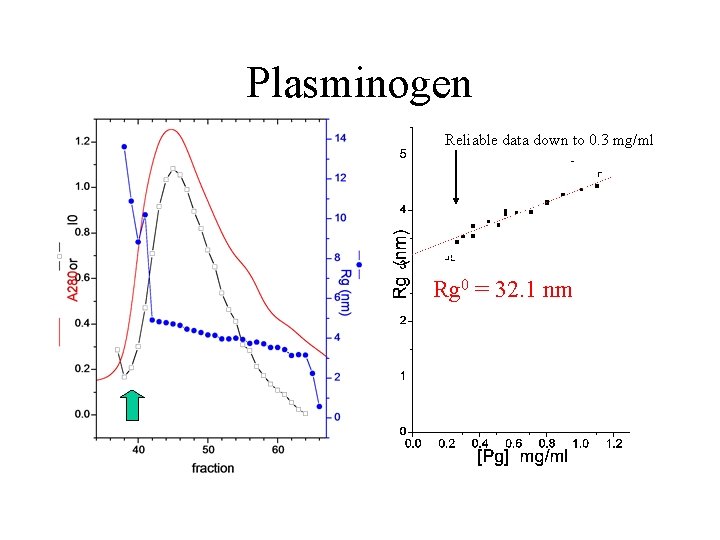
Plasminogen • Sticky protein… curved Guinier plots • conformational change…DRg 30 40 A Need accurate, reproducible Rgs to see these modest changes static LC-SAXS

Plasminogen Reliable data down to 0. 3 mg/ml Rg 0 = 32. 1 nm

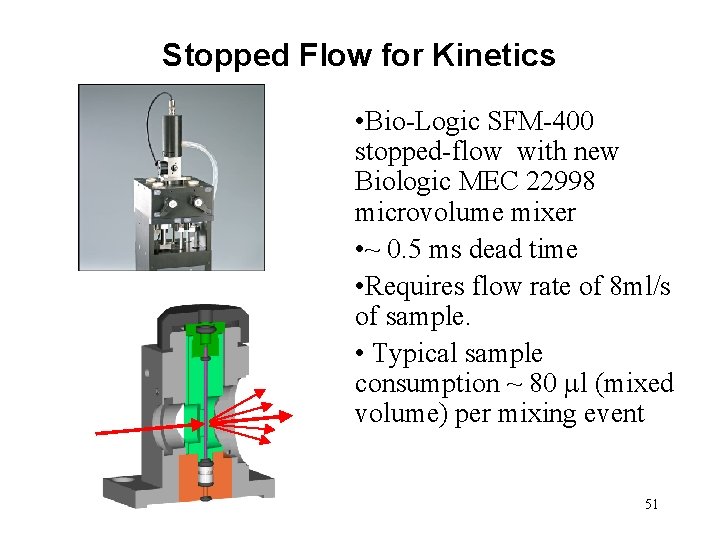
Stopped Flow for Kinetics • Bio-Logic SFM-400 stopped-flow with new Biologic MEC 22998 microvolume mixer • ~ 0. 5 ms dead time • Requires flow rate of 8 ml/s of sample. • Typical sample consumption ~ 80 l (mixed volume) per mixing event 51

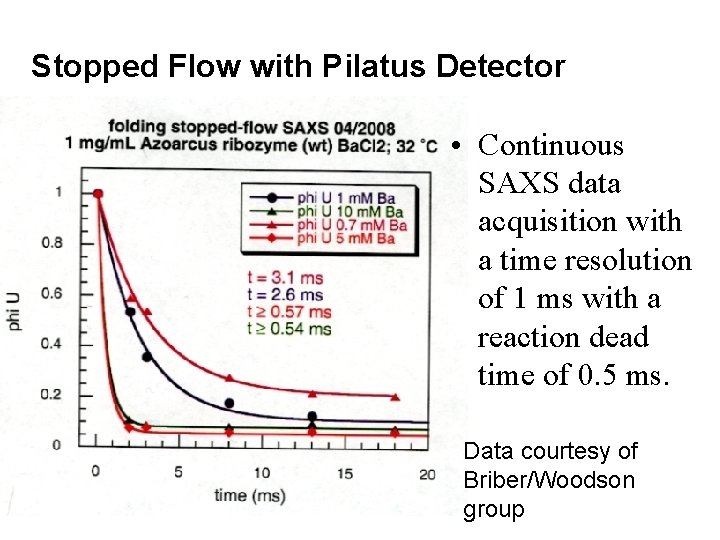
Stopped Flow with Pilatus Detector • Continuous SAXS data acquisition with a time resolution of 1 ms with a reaction dead time of 0. 5 ms. Data courtesy of Briber/Woodson group

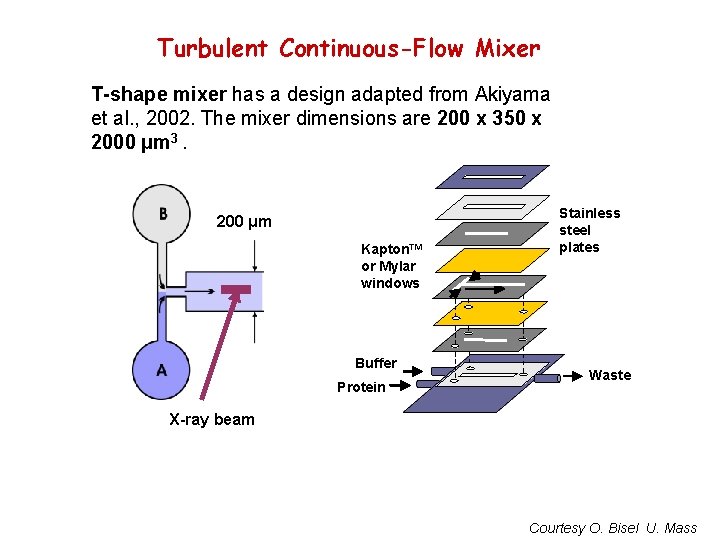
Turbulent Continuous-Flow Mixer T-shape mixer has a design adapted from Akiyama et al. , 2002. The mixer dimensions are 200 x 350 x 2000 μm 3. 200 μm Kapton™ or Mylar windows Buffer Protein Stainless steel plates Waste X-ray beam Courtesy O. Bisel U. Mass

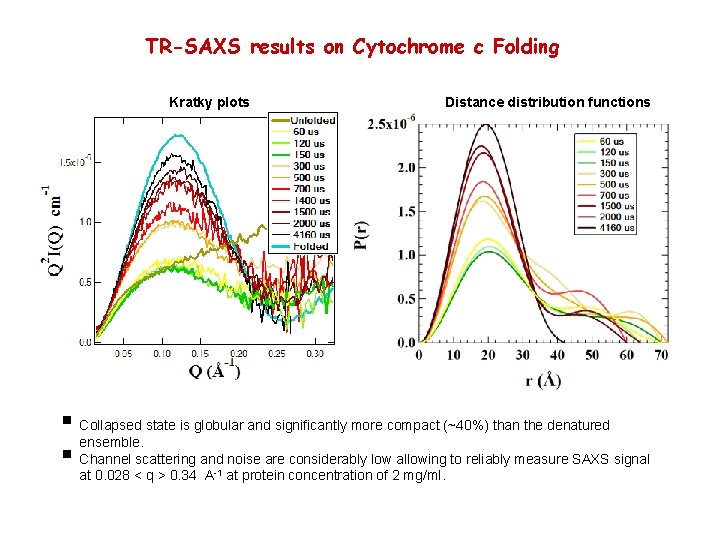
TR-SAXS results on Cytochrome c Folding Kratky plots Distance distribution functions § Collapsed state is globular and significantly more compact (~40%) than the denatured ensemble. § Channel scattering and noise are considerably low allowing to reliably measure SAXS signal at 0. 028 < q > 0. 34 A-1 at protein concentration of 2 mg/ml.

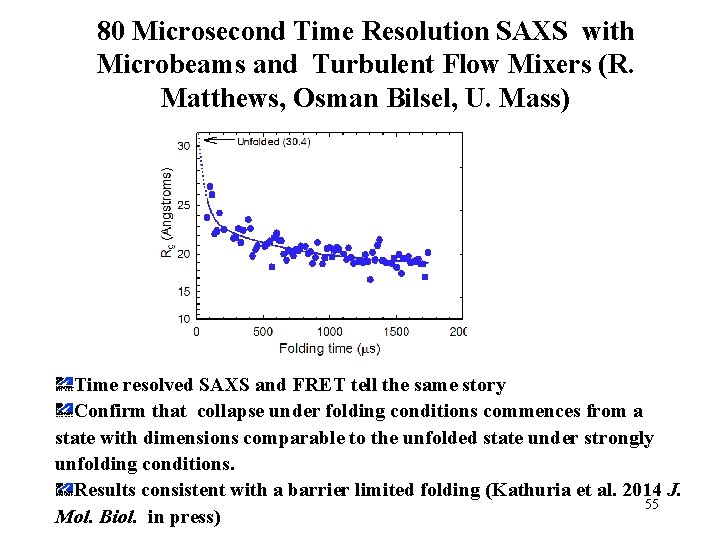
80 Microsecond Time Resolution SAXS with Microbeams and Turbulent Flow Mixers (R. Matthews, Osman Bilsel, U. Mass) Time resolved SAXS and FRET tell the same story Confirm that collapse under folding conditions commences from a state with dimensions comparable to the unfolded state under strongly unfolding conditions. Results consistent with a barrier limited folding (Kathuria et al. 2014 J. 55 Mol. Biol. in press)









For further reading…. . • A Guinier “X-ray Diffraction in Crsytals, Imperfect Crystals and Amorphous Bodies” Freeman, 1963 • C. Cantor and P. Schimmel “Biophysical Chemistry part II: Techniques for the study of Biological Structure and Function” Freeman, 1980 • O. Glatter and O. Kratky “Small-angle X-ray Scattering” Academic Press 1982 • See Dmitri Svergun’s web site at http: //www. emblhamburg. de/Externalinfo/Research/Sax