Assessment of macromolecular interactions and identification of macromolecular








- Slides: 30

Assessment of macromolecular interactions and identification of macromolecular assemblies in crystalline state (PISA software) Eugene Krissinel Macromolecular Structure Database keb@ebi. ac. uk EBI is an Outstation of the European Molecular Biology Laboratory.

http: //www. ebi. ac. uk/msd www. pdb. org www. ebi. ac. uk/msd www. pdbj. org

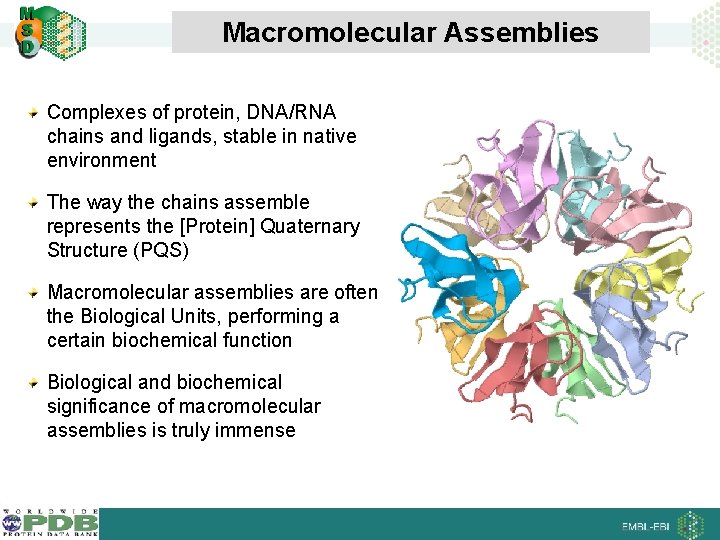
Macromolecular Assemblies Complexes of protein, DNA/RNA chains and ligands, stable in native environment The way the chains assemble represents the [Protein] Quaternary Structure (PQS) Macromolecular assemblies are often the Biological Units, performing a certain biochemical function Biological and biochemical significance of macromolecular assemblies is truly immense

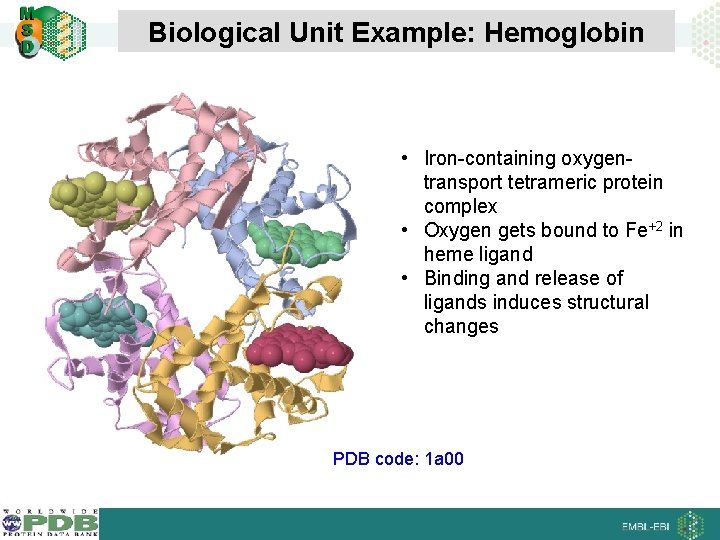
Biological Unit Example: Hemoglobin • Iron-containing oxygentransport tetrameric protein complex • Oxygen gets bound to Fe+2 in heme ligand • Binding and release of ligands induces structural changes PDB code: 1 a 00

Biological Units are Hard to Study Light / Neutron / X-ray / Small angle scatterings: mainly composition and multimeric state may be found. 3 D shape may be guessed from mobility measurements. Electron microscopy: not a fantastic resolution and not applicable to all objects NMR is not good for big chains, even less so for protein assemblies. Very few quaternary structures have been identified experimentally.

Asymmetric Unit vs. Biological Unit Crystal = translated Unit Cells More than 80% of macromolecular structures are solved by means of X-ray diffraction on crystals. It is reasonable to expect that Biological Units make construction blocks for the crystal. An X-ray diffraction experiment produces atomic coordinates of the Asymmetric Unit, which is stored as a PDB file. Unit Cell = all space symmetry group mates of ASU In general, neither Asymmetric Unit nor Unit Cell has any direct relation to Biological Unit may be made of • a single ASU • part of ASU • several ASU parts PDB file (ASU) Biological Unit

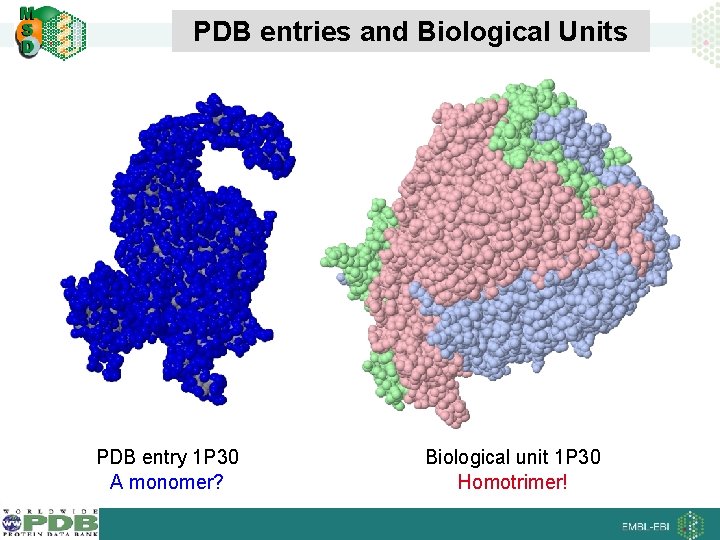
PDB entries and Biological Units PDB entry 1 P 30 A monomer? Biological unit 1 P 30 Homotrimer!

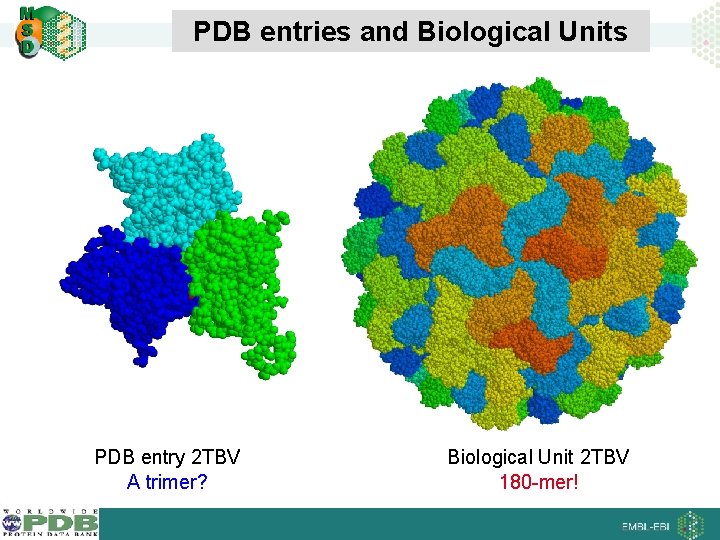
PDB entries and Biological Units PDB entry 2 TBV A trimer? Biological Unit 2 TBV 180 -mer!

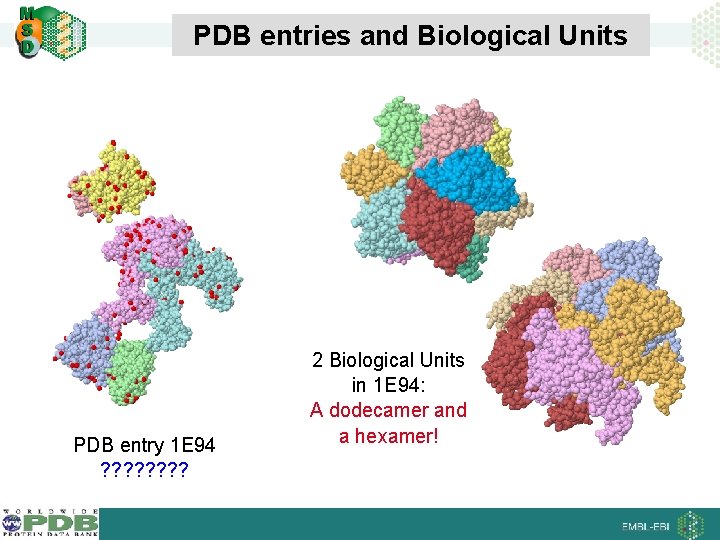
PDB entries and Biological Units PDB entry 1 E 94 ? ? ? ? 2 Biological Units in 1 E 94: A dodecamer and a hexamer!

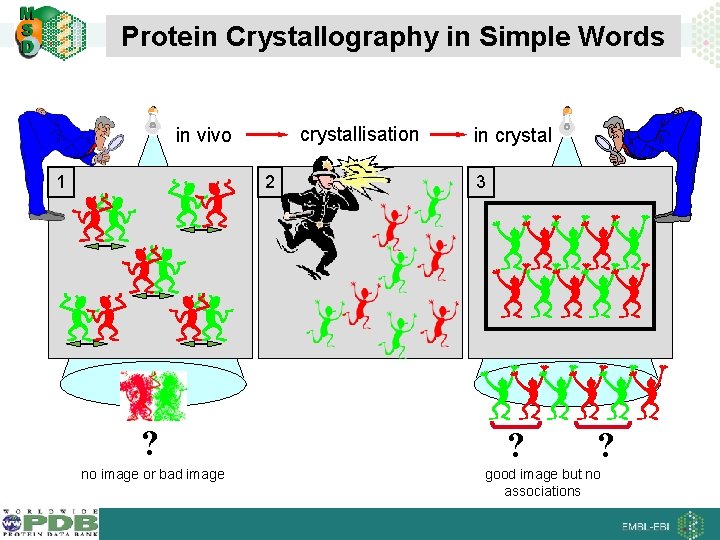
Protein Crystallography in Simple Words crystallisation in vivo 1 2 ? no image or bad image in crystal 3 ? ? good image but no associations

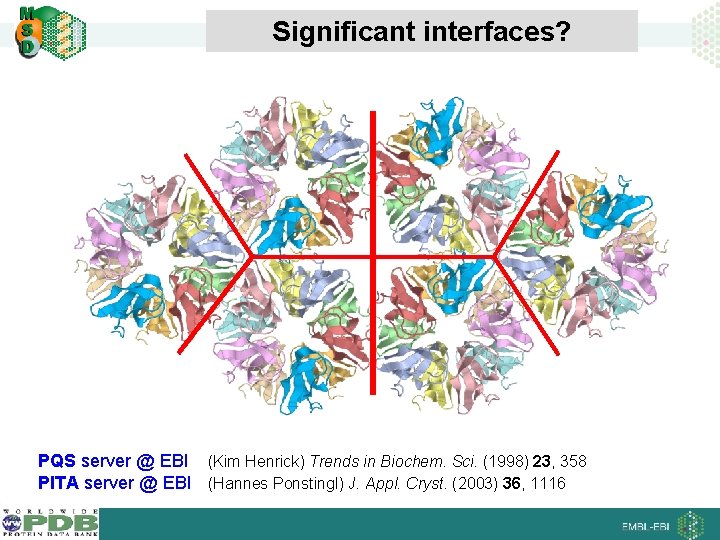
Significant interfaces? PQS server @ EBI (Kim Henrick) Trends in Biochem. Sci. (1998) 23, 358 PITA server @ EBI (Hannes Ponstingl) J. Appl. Cryst. (2003) 36, 1116

Significant interfaces or artefacts? “No single parameter absolutely differentiates the interfaces from all other surface patches” Jones, S. & Thornton, J. M. (1996) Principles of protein-protein interactions, Proc. Natl. Acad. Sci. USA, 93, 13 -20. Formation of N>2 -meric complexes is most probably a corporate process involving a set of interfaces. Therefore significance of an interface should not be detached from the context of protein complex “…the type of complexes need to be taken into account when characterizing interfaces between them. ” Jones, S. & Thornton, J. M. , ibid.

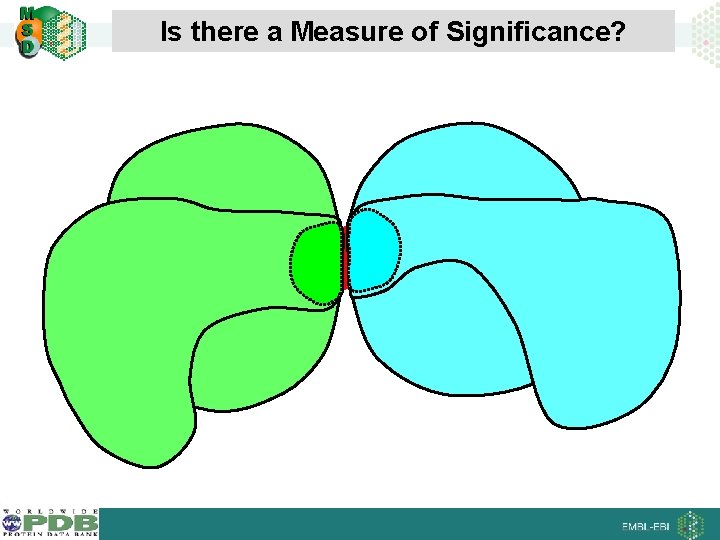
Is there a Measure of Significance?

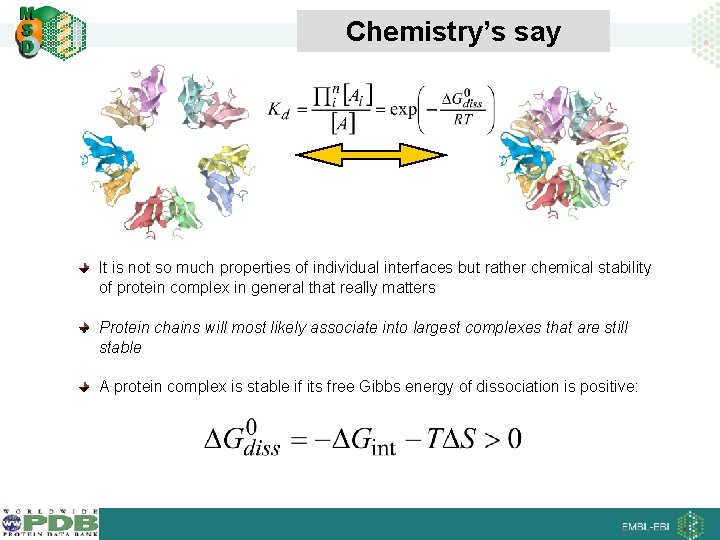
Chemistry’s say It is not so much properties of individual interfaces but rather chemical stability of protein complex in general that really matters Protein chains will most likely associate into largest complexes that are still stable A protein complex is stable if its free Gibbs energy of dissociation is positive:

Detection of Biological Units in Crystals Method Summary 1. Enumerate all possible assemblies in crystal, subject to crystal structure: space symmetry group, geometry and composition of the Asymmetric Unit (Graph Theory used) 2. Evaluate assemblies for chemical stability 3. Leave only stable assemblies in the list and range them by chances to be a Biological Unit : • Larger assemblies take preference • Single-assembly solutions take preference • Otherwise, assemblies with higher Gdiss take preference

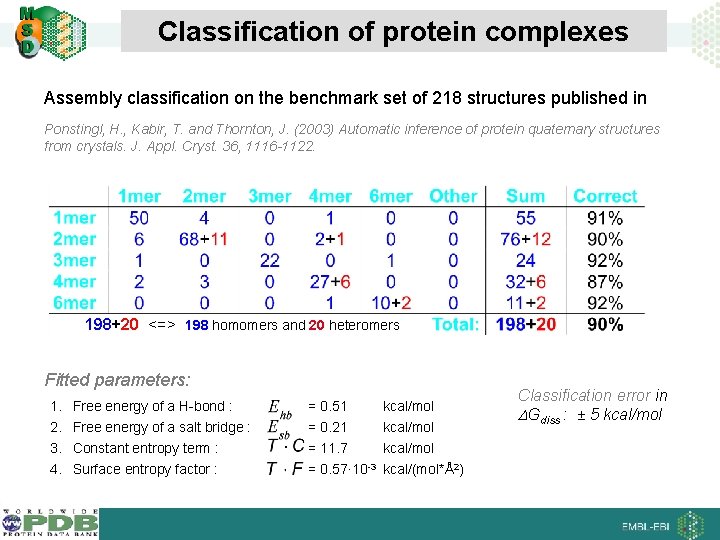
Classification of protein complexes Assembly classification on the benchmark set of 218 structures published in Ponstingl, H. , Kabir, T. and Thornton, J. (2003) Automatic inference of protein quaternary structures from crystals. J. Appl. Cryst. 36, 1116 -1122. 198+20 <=> 198 homomers and 20 heteromers Fitted parameters: 1. 2. 3. 4. Free energy of a H-bond : = 0. 51 kcal/mol Free energy of a salt bridge : = 0. 21 kcal/mol Constant entropy term : Surface entropy factor : = 11. 7 kcal/mol = 0. 57· 10 -3 kcal/(mol*Å2) Classification error in Gdiss : ± 5 kcal/mol

Classification of protein-DNA complexes Assembly classification on the benchmark set of 212 protein – DNA complexes published in Luscombe, N. M. , Austin, S. E. , Berman H. M. and Thornton, J. M. (2000) An overview of the structures of protein-DNA complexes. Genome Biol. 1, 1 -37. 2 mer 1 3 mer 6 4 mer 0 5 mer 0 6 mer 1 10 mer 0 3 mer 0 96 2 0 0 0 4 mer 0 0 83 2 0 0 5 mer 0 0 0 3 0 0 Classification error in 6 mer 10 mer Other 0 0 0 1 0 2 0 0 0 13 0 1 0 Total: : ± 5 kcal/mol Sum 1 105 85 5 15 1 212 Correct 100% 91% 98% 60% 87% 100% 93%

Free energy of misclassifications

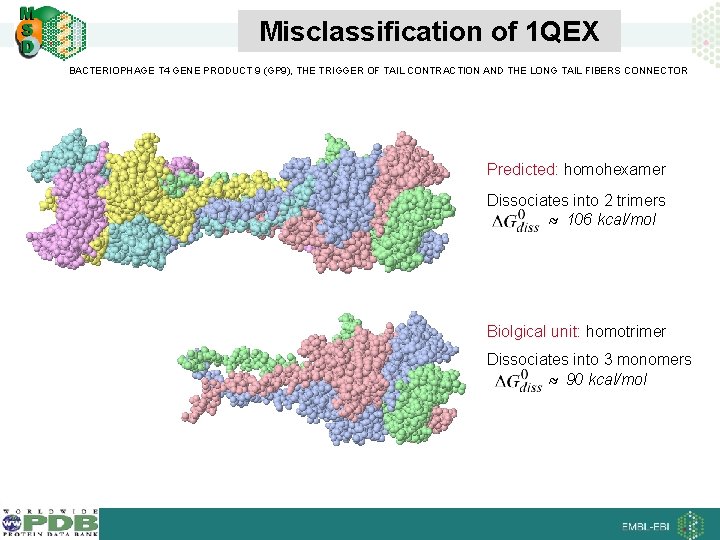
Misclassification of 1 QEX BACTERIOPHAGE T 4 GENE PRODUCT 9 (GP 9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR Predicted: homohexamer Dissociates into 2 trimers 106 kcal/mol Biolgical unit: homotrimer Dissociates into 3 monomers 90 kcal/mol

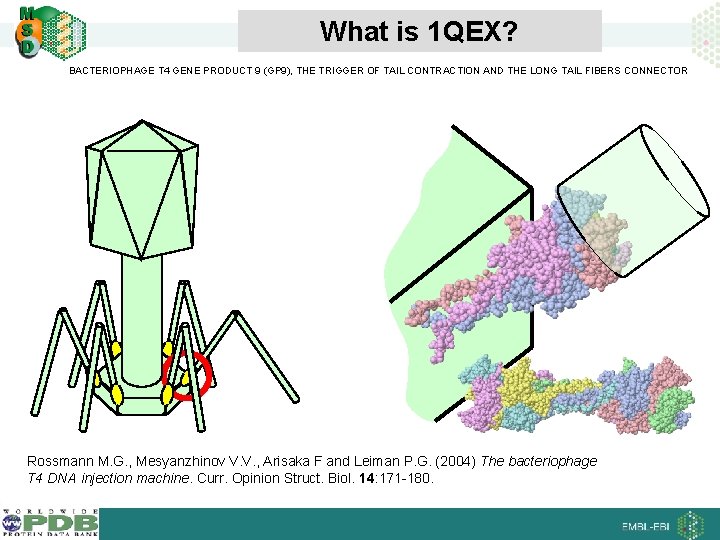
What is 1 QEX? BACTERIOPHAGE T 4 GENE PRODUCT 9 (GP 9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR Rossmann M. G. , Mesyanzhinov V. V. , Arisaka F and Leiman P. G. (2004) The bacteriophage T 4 DNA injection machine. Curr. Opinion Struct. Biol. 14: 171 -180.

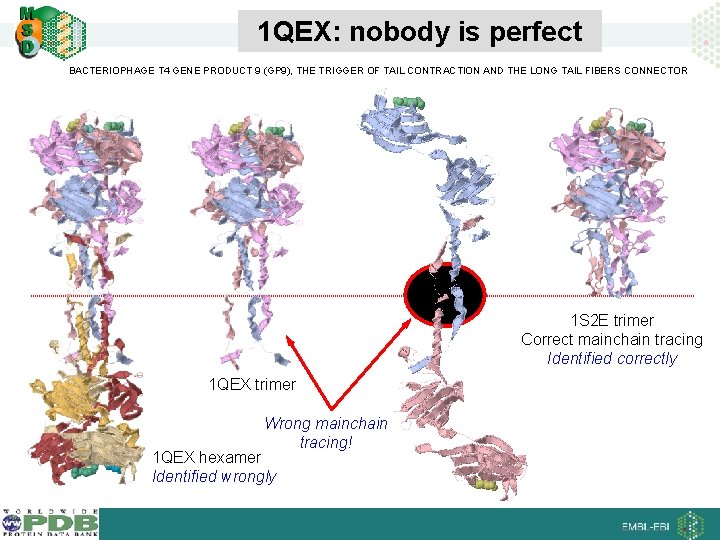
1 QEX: nobody is perfect BACTERIOPHAGE T 4 GENE PRODUCT 9 (GP 9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR 1 S 2 E trimer Correct mainchain tracing Identified correctly 1 QEX trimer Wrong mainchain tracing! 1 QEX hexamer Identified wrongly

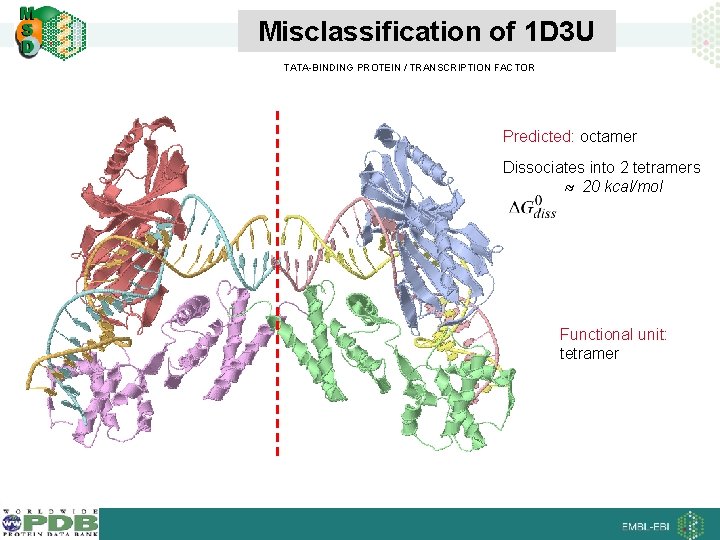
Misclassification of 1 D 3 U TATA-BINDING PROTEIN / TRANSCRIPTION FACTOR Predicted: octamer Dissociates into 2 tetramers 20 kcal/mol Functional unit: tetramer

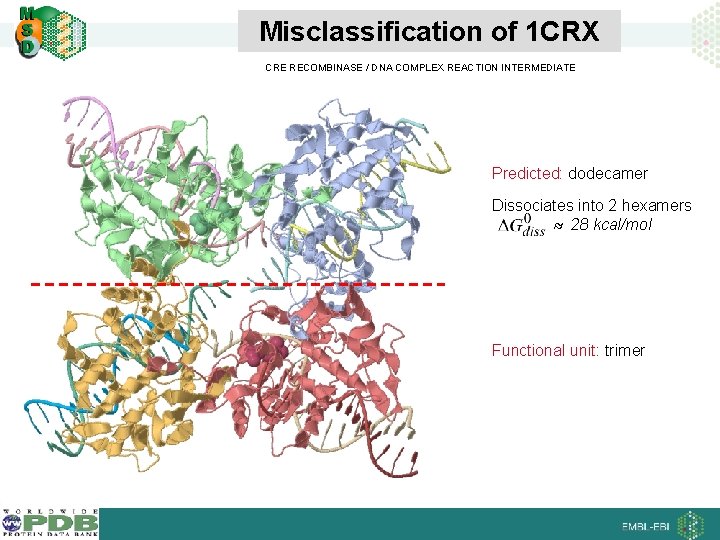
Misclassification of 1 CRX CRE RECOMBINASE / DNA COMPLEX REACTION INTERMEDIATE Predicted: dodecamer Dissociates into 2 hexamers 28 kcal/mol Functional unit: trimer

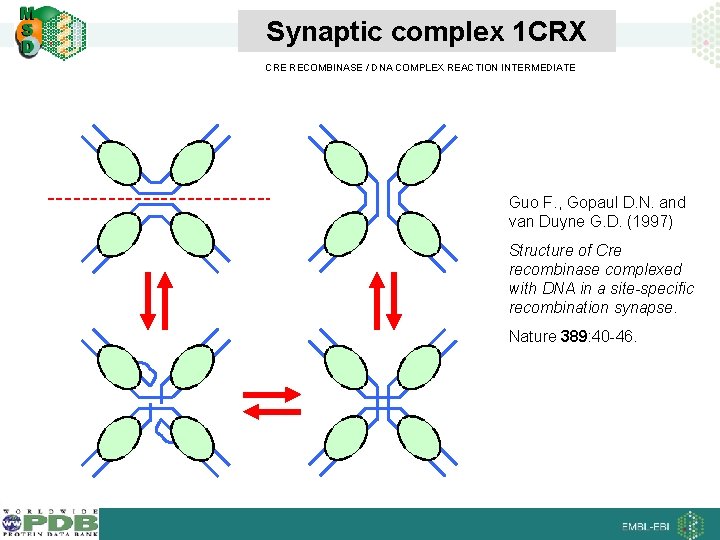
Synaptic complex 1 CRX CRE RECOMBINASE / DNA COMPLEX REACTION INTERMEDIATE Guo F. , Gopaul D. N. and van Duyne G. D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389: 40 -46.

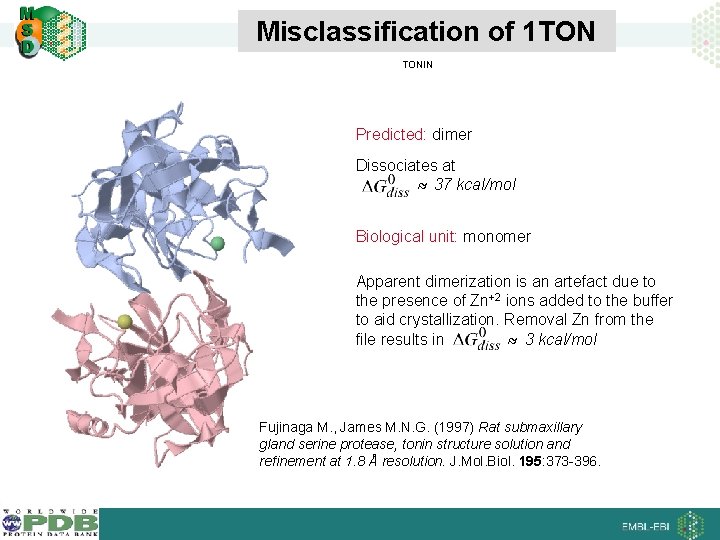
Misclassification of 1 TON TONIN Predicted: dimer Dissociates at 37 kcal/mol Biological unit: monomer Apparent dimerization is an artefact due to the presence of Zn+2 ions added to the buffer to aid crystallization. Removal Zn from the file results in 3 kcal/mol Fujinaga M. , James M. N. G. (1997) Rat submaxillary gland serine protease, tonin structure solution and refinement at 1. 8 Å resolution. J. Mol. Biol. 195: 373 -396.

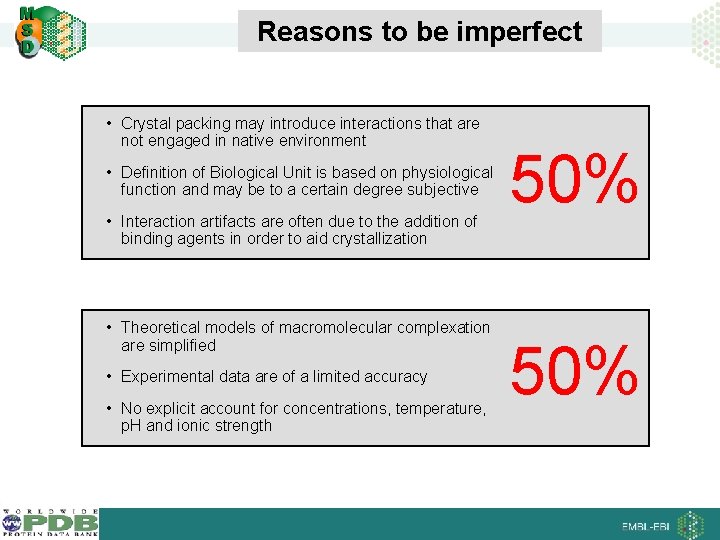
Reasons to be imperfect • Crystal packing may introduce interactions that are not engaged in native environment • Definition of Biological Unit is based on physiological function and may be to a certain degree subjective • Interaction artifacts are often due to the addition of binding agents in order to aid crystallization • Theoretical models of macromolecular complexation are simplified • Experimental data are of a limited accuracy • No explicit account for concentrations, temperature, p. H and ionic strength 50%

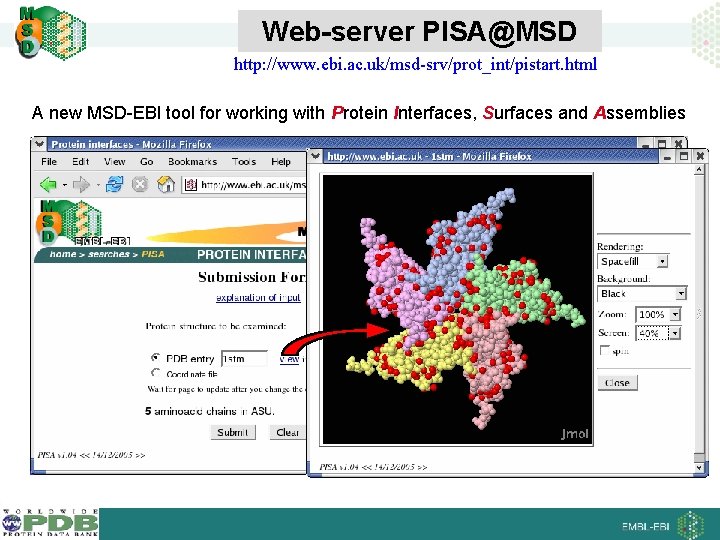
Web-server PISA@MSD http: //www. ebi. ac. uk/msd-srv/prot_int/pistart. html A new MSD-EBI tool for working with Protein Interfaces, Surfaces and Assemblies

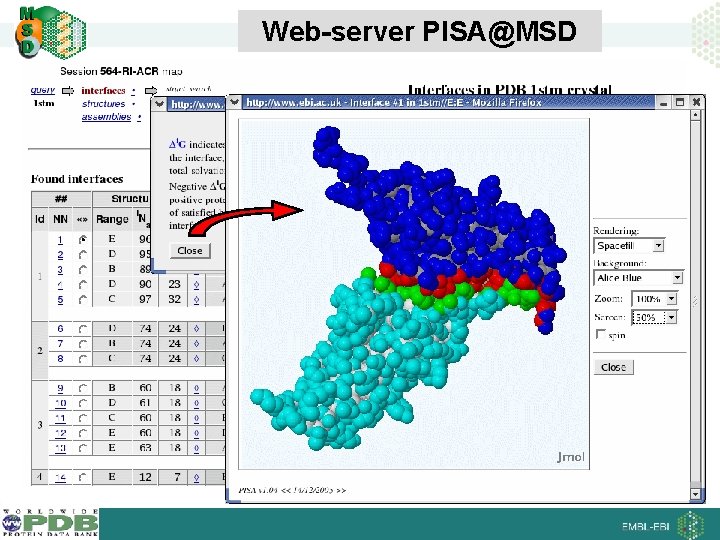
Web-server PISA@MSD

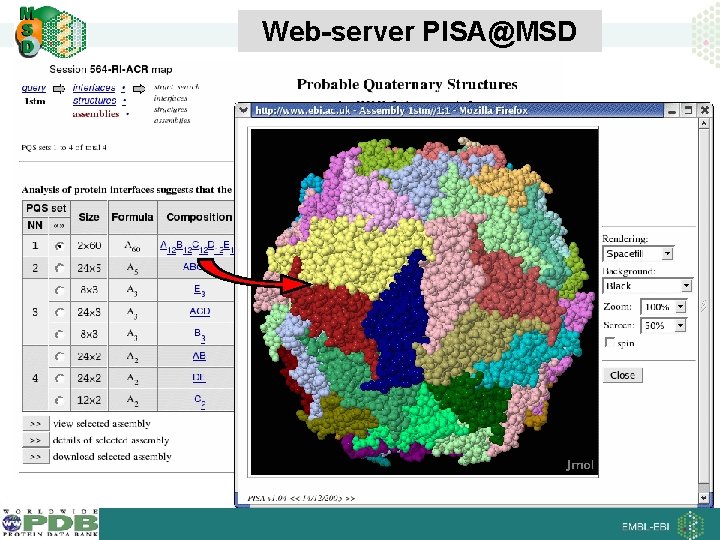
Web-server PISA@MSD

Conclusions Chemical-thermodynamical models for protein complex stability allow one to recover biological units from protein crystallography data at 80 -90% success rate Considerable part of misclassifications is due to the difference of experimental and native environments and artificial interactions induced by crystal packing Functional significance of protein interfaces cannot be reliably inferred only from their properties. Due to entropy contribution and entangled interactions, interface function is also subject to protein complex composition and geometry Protein interface and assembly analysis software (PISA) is available for public use as a web-service from MSD@EBI. Acknowledgement. This work has been supported by research grant No. 721/B 19544 from the Biotechnology and Biological Sciences Research Council (BBSRC) UK.
 Central pocket vs plain whorl
Central pocket vs plain whorl Severity hirarc
Severity hirarc Interactions between ais and internal and external parties
Interactions between ais and internal and external parties Livestock breed identification
Livestock breed identification Qualitative and quantitative research
Qualitative and quantitative research Unit 5 macroeconomics lesson 2 activity 45
Unit 5 macroeconomics lesson 2 activity 45 The properties and interactions of magnets
The properties and interactions of magnets Sphere interactions examples
Sphere interactions examples 6.1 habitats, niches, and species interactions answer key
6.1 habitats, niches, and species interactions answer key Fundamental and incidental interactions
Fundamental and incidental interactions Factors that influence the communication process
Factors that influence the communication process Symbiosis and species interactions keystone webquest
Symbiosis and species interactions keystone webquest Naive bayes pays attention to complex interactions and
Naive bayes pays attention to complex interactions and Modular architecture vs integrated architecture
Modular architecture vs integrated architecture Niches and community interactions
Niches and community interactions Regional and transregional interactions
Regional and transregional interactions Fundamental particles and interactions
Fundamental particles and interactions Regional and transregional interactions
Regional and transregional interactions Noncovalent interactions
Noncovalent interactions Interactions among branches of government
Interactions among branches of government Predation symbiotic relationship
Predation symbiotic relationship Wikimedia commons
Wikimedia commons Sertraline interactions
Sertraline interactions Grapefruit-drug interactions chart
Grapefruit-drug interactions chart Interactions within ecosystems grade 7
Interactions within ecosystems grade 7 Examples of abiotic factors
Examples of abiotic factors Section 20-1 review species interactions
Section 20-1 review species interactions Photosynthesis is important to animals because flocabulary
Photosynthesis is important to animals because flocabulary Moa of h2 antagonist
Moa of h2 antagonist Types of interactions
Types of interactions Circulatory system interactions with other systems
Circulatory system interactions with other systems