SmallAngle Scattering Validation Task Force Virtual Meeting 4








- Slides: 16

Small-Angle Scattering Validation Task Force Virtual Meeting 4 May, 2017

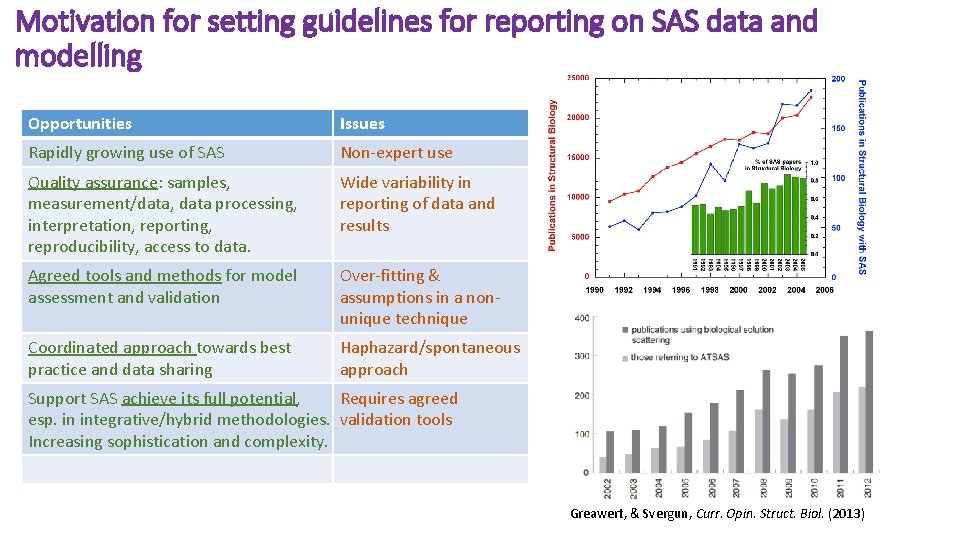
Motivation for setting guidelines for reporting on SAS data and modelling Opportunities Issues Rapidly growing use of SAS Non-expert use Quality assurance: samples, measurement/data, data processing, interpretation, reporting, reproducibility, access to data. Wide variability in reporting of data and results Agreed tools and methods for model assessment and validation Over-fitting & assumptions in a nonunique technique Coordinated approach towards best practice and data sharing Haphazard/spontaneous approach Support SAS achieve its full potential, Requires agreed esp. in integrative/hybrid methodologies. validation tools Increasing sophistication and complexity. Greawert, & Svergun, Curr. Opin. Struct. Biol. (2013)

Experience so far in developing standards • Community concerns : • could standards restrict opportunities for publishing? • potential for unreasonable/misguided standards? • is there ample opportunity for community input? • The process requires: • • a commitment to two way communication; structured planning reporting of progress in open access articles; leadership from experts across the international community • including providers/developers of instrumentation and analysis tools; • time to embed new practices and obtain the resources required. • International forums are major means for communication: • • • SAS and Journals Commissions of the International Union of Crystallography Triennial SAS meetings www. PDB SAS validation task force (SASvtf). Journal articles Commentary pieces in journals

The process so far • 2008 – IUCr Congress in Osaka, Japan • IUCr SAS Commission held discussions for establishing publication standards for the results of biomolecular SAS experiments. • 2012 – IUCr Journal Commission accepts SAS Commission draft publication guidelines for biomolecular SAS • http: //journals. iucr. org/services/sas/ • rationale for draft guidelines published: Jacques et al. (2012) Acta D 68, 620 • July 2012 – first meeting of the ww. PDB SAS validation task force (SASvtf) • report published: Trewhella et al. (2013) Structure 21, 875. • May 2014 – second meeting SASvtf • SAS 2012 and SAS 2015 • open sessions held to report on work of IUCr Commissions and SASvtf and to receive community input • Nov. 2016 – NIH R 24 grant submitted • to support ongoing community engagement with the objectives of the IHMvtf for archiving integrative/hybrid models. Unsuccessful

SASvtf Report Recommendations (2013) 1. Create a global repository • standard format X-ray and neutron SAS data • is searchable and freely accessible for download 2. A standard dictionary for data • defining terms, data format, data collection, samples, data processing and supporting information and for managing the SAS data repository; 3. Options for including models • SAS-derived shape and atomistic models • specific information regarding the uniqueness of the model 4. Criteria for quality assessment of data and models, • (sample & data quality, basic SAS parameters, data range & statisics, accuracy of model fit, uniqueness 5. There would be value to archive models derived from a diversity of hybrid data 6. Thought leaders from different disciplines in the field of structural biology should define • what should be archived in the PDB and • what complementary archives might be recommended. Trewhella, J. , Hendrickson, W. A. , Kleywegt, G. J. , Sali, A. , Sato, M. , Schwede, T. , Svergun, D. , Tainer, J. A. , Westbrook, J. , and Berman, H. M. “Report of the ww. PDB Small-Angle Scattering Task Force: Data Requirements for Biomolecular Modeling and the PDB, ” Structure 21, 875 -881, 2013.

I/HMvtf Report Recommendations (2014) 1. Models with all relevant experimental data and metadata, as well as experimental and computational protocols should be archived; inclusivity is key. 2. A flexible model representation needs to be developed, allowing for multi-scale models, multistate models, ensembles of models, and models related by time or other order. 3. Procedures for estimating the uncertainty of integrative models should be developed, validated, and adopted. 4. A federated system of model and data archives should be created. 5. Publication standards for integrative models should be established. Sali, A. , Berman, H. M. , Schwede, T. , Trewhella, J. , Kleywegt, G. , Burley, S. K. , Markley, J. , Nakamura, H. , Adams, P. , Bonvin, A. M. J. J. , Chiu, W. , Dal Peraro, M. , Di Maio, F. , Ferrin, T. E. , Grünewald, K. , Gutmanas, A. , Henderson, R. , Hummer, G. , Iwasaki, K. , Johnson, G. , Lawson, C. L. , Meiler, J. , Marti. Renom, M. A. , Montelione, G. T. , Nilges, M. , Nussinov, R. , Patwardhan, A. , Rappsilber, J. , Read, R. J. , Saibil, H. , Schröder, G. F. , Schwieters, C. D. , Seidel, C. A. M. , Svergun, D. , Topf, M. , Ulrich, E. L. , Velankar, S. , Westbrook, J. D. “Outcome of the First ww. PDB Hybrid/Integrative Methods Task Force Workshop”, Structure 23, 1156 -1167, 2015.

Progress on recommendations of the SASvtf and I/Mvtf • sas. CIF developed and extended as a standard dictionary • with definitions of terms for SAS data collection and for managing SAS data • Kachala et al. (2016) J Appl Crystallogr. 49, 302 • sas. CIF is an extension of the IUCr Crystallographic Information Framework • enables SAS databases to inter-operate with the ww. PDB • future support for integrative/hybrid biomolecular structures and assemblies. • SASBDB established as a SAS data and model repository • (Valentini et al. (2015) NAR 43, D 357) • BIOISIS –similarities and differences to SASBDB, agreement at 12 May 2014 SASvtf meeting to data exchange via SAScif • Integrative/Hybrid Methods workshop held • (Hinxton, Oct. 2014) and recommendations published (Sali et al. (2015) Structure 23, 1156) • SAS/NMR hybrid data project incorporate SAS and NMR data supporting hybrid modelling into the ww. PDB One-Dep system

Current activities/plans for the next period • Jan-July 2017 • Schedule SASvtf meeting with expanded membership for broader SAS expertise: • Review and update preliminary publication guidelines • take the high level recommendations of the SASvft report to more specific, implementable recommendations for both data and model validation; • Aug. 2017: IUCr Congress Hyderabad • Present the updated publication guidelines at an open meeting. • 2017 -2018 • Approach structural biology journals to propose adoption of publication guidelines for biomolecular SAS to include deposition of SAS data in a freely accessible and searchable archive; • Continue to support SAS inclusion in the work of the Integrative/Hybrid Methods validation task force (IHMvtf)

Questions for Today • Outstanding issues on reporting on and deposition of SAS data and models • Where to publish our recommendations once finalised • Potential sub-group activities for unresolved matters • Validation of models based on combined analysis of NMR and SAXS data. • Other business

Reporting on/deposition of SAS data and models •

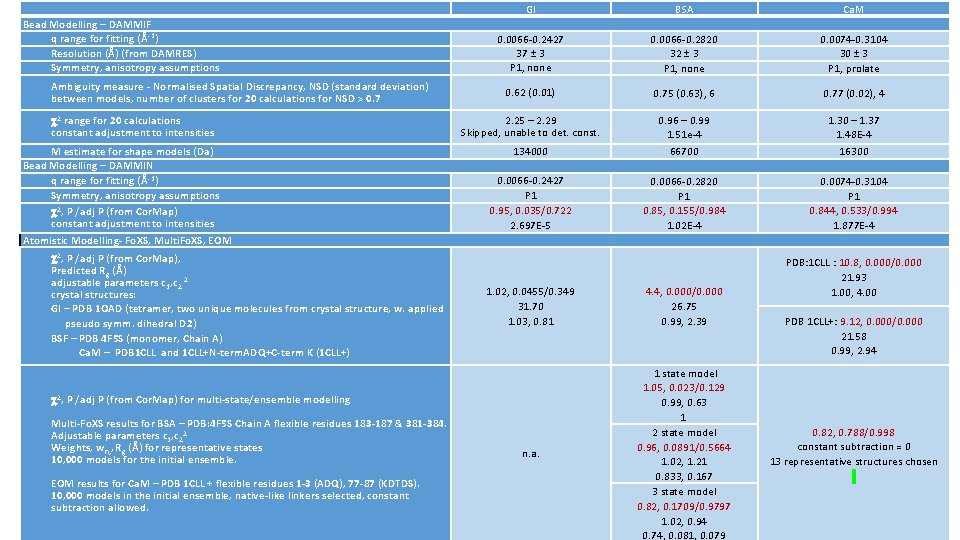
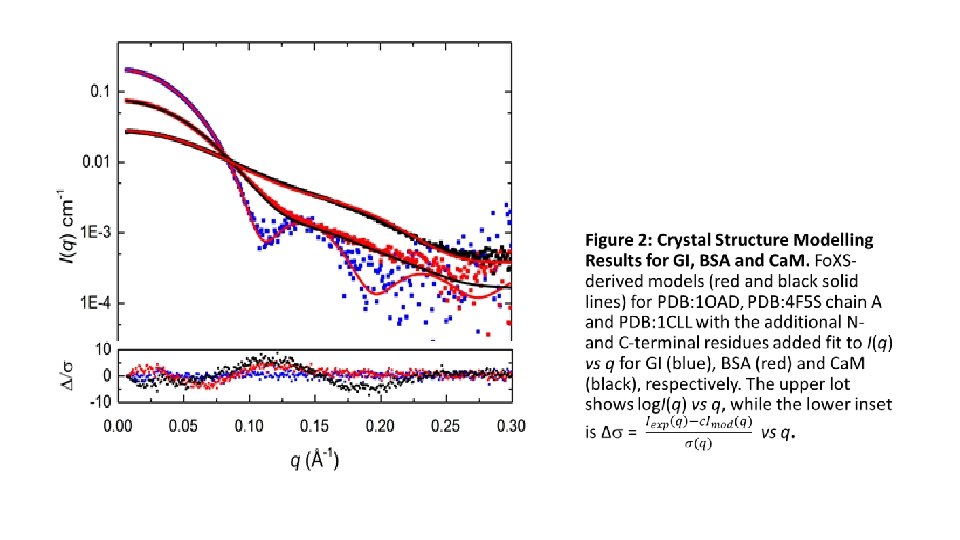
Bead Modelling – DAMMIF q range for fitting (Å-1) Resolution (Å) (from DAMRES) Symmetry, anisotropy assumptions Ambiguity measure - Normalised Spatial Discrepancy, NSD (standard deviation) between models, number of clusters for 20 calculations for NSD > 0. 7 2 range for 20 calculations constant adjustment to intensities M estimate for shape models (Da) Bead Modelling – DAMMIN q range for fitting (Å-1) Symmetry, anisotropy assumptions 2, P /adj P (from Cor. Map) constant adjustment to intensities Atomistic Modelling- Fo. XS, Multi. Fo. XS, EOM 2, P /adj P (from Cor. Map), Predicted Rg (Å) adjustable parameters c 1, c 2 2 crystal structures: GI – PDB 1 OAD (tetramer, two unique molecules from crystal structure, w. applied pseudo symm. dihedral D 2) BSF – PDB 4 F 5 S (monomer, Chain A) Ca. M – PDB 1 CLL and 1 CLL+N-term. ADQ+C-term K (1 CLL+) GI BSA Ca. M 0. 0066 -0. 2427 37 ± 3 P 1, none 0. 0066 -0. 2820 32 ± 3 P 1, none 0. 0074 -0. 3104 30 ± 3 P 1, prolate 0. 62 (0. 01) 0. 75 (0. 63), 6 0. 77 (0. 02), 4 2. 25 – 2. 29 Skipped, unable to det. const. 134000 0. 96 – 0. 99 1. 51 e-4 66700 1. 30 – 1. 37 1. 48 E-4 16300 0. 0066 -0. 2427 P 1 0. 95, 0. 035/0. 722 2. 697 E-5 0. 0066 -0. 2820 P 1 0. 85, 0. 155/0. 984 1. 02 E-4 0. 0074 -0. 3104 P 1 0. 844, 0. 533/0. 994 1. 877 E-4 1. 02, 0. 0455/0. 349 31. 70 1. 03, 0. 81 4. 4, 0. 000/0. 000 26. 75 0. 99, 2. 39 n. a. 1 state model 1. 05, 0. 023/0. 129 0. 99, 0. 63 1 2 state model 0. 96, 0. 0891/0. 5664 1. 02, 1. 21 0. 833, 0. 167 3 state model 0. 82, 0. 1709/0. 9797 1. 02, 0. 94 0. 74, 0. 081, 0. 079 2, P /adj P (from Cor. Map) for multi-state/ensemble modelling Multi-Fo. XS results for BSA – PDB: 4 F 5 S Chain A flexible residues 183 -187 & 381 -384. Adjustable parameters c 1, c 22 Weights, wn, , Rg (Å) for representative states 10, 000 models for the initial ensemble. EOM results for Ca. M – PDB 1 CLL + flexible residues 1 -3 (ADQ), 77 -87 (KDTDS). 10, 000 models in the initial ensemble, native-like linkers selected, constant subtraction allowed. PDB: 1 CLL : 10. 8, 0. 000/0. 000 21. 93 1. 00, 4. 00 PDB 1 CLL+: 9. 12, 0. 000/0. 000 21. 58 0. 99, 2. 94 0. 82, 0. 788/0. 998 constant subtraction = 0 13 representative structures chosen


Where to publish? • Acta D was proposed for the current manuscript based on the fact that it is a follow up to the preliminary guidelines from 2012, will have strong support from the editor (Ted Baker), is also of an appropriate length and structure, and likely would be open access by virtue of the subject matter. • The ww. PDB task forces traditionally have published their findings in Structure. If we agree to a, should we consider a short summary commentary for Structure?

Potential SASvtf sub-group activities Model scattering profiles from pdb coordinates: This area is a matter of continued research. An expert sub-group could look into this area in detail with a subset of well-known, robust, globular proteins with complete crystal structures and for which high quality SAXS data are available. It would be of significant value in understanding the uncertainties in the context of hybrid structural modelling and provides an important foundation for potential recommendations from this group.

Validation of hybrid NMR/SAXS data/models • Members of the ww. PDB NMR task force is meeting in New Hampshire Friday June 16, at the Grand Summit Hotel at Sunday River, Newby Maine (GRC on Computational Aspects of Biomolecular NMR) and have tentatively put on this topic on their agenda. Can anyone from this group attend? • Shall we agree to provide the draft manuscript to the NMR task force? • How shall we manage future interactions with the NMR task force as NMRSAXS hybrid modelling continues to grow and topic 4 a above continues to develop? • If the subgroup looking into modelling scattering profiles from pdb coordinates is established, should we recruit one or two NMR colleagues?

Other business?