DNA Extraction RNA Extraction The Different Types and


















- Slides: 44

DNA Extraction /RNA Extraction

The Different Types and Varieties of Nucleic Acid Target Molecules • must be readily accessible to primers and DNA polymerase and free from inhibitory concentrations of contaminating proteins, lipids, carbohydrates and salts • Blood samples • RNA • DNA • Serum samples

A Brief Description of DNA and RNA Targets A. Bacteria B. Viruses 1. Class I: Double stranded DNA viruses. human papilloma viruses (cervical cancer). Replicate in the host cell cytoplasm. 2. Class II: Single stranded DNA viruses One example is Parvovirus B 19 3. Class III: Double stranded RNA viruses example is rotaviruses 4. Class IV: single stranded RNA viruses example is Poliomyelitis virus 5. Class V: single stranded RNA viruses, Influenza viruses 6. Class VI: Retroviruses, HIV

Contaminations…. • Blood • Inhibitors • kits • RNase 1. gloves should be worn 2. use of disposable RNase free plastic……. 3. work surfaces are also …RNase free 4. inactivated by…. . DEPC (diethyl pyrocarbonate) or commercially available …… RNase AWAY, RNase ERASE, RNA Clean 5. DEPC in water being used to remove RNase 6. DEPC also hydrolyses the imidazole ring of purines and destroys nucleic acids. 7. 180°C to inactivate the DEPC 8. DEPC…. toxic carcinogen that………. safety precautions • DNase

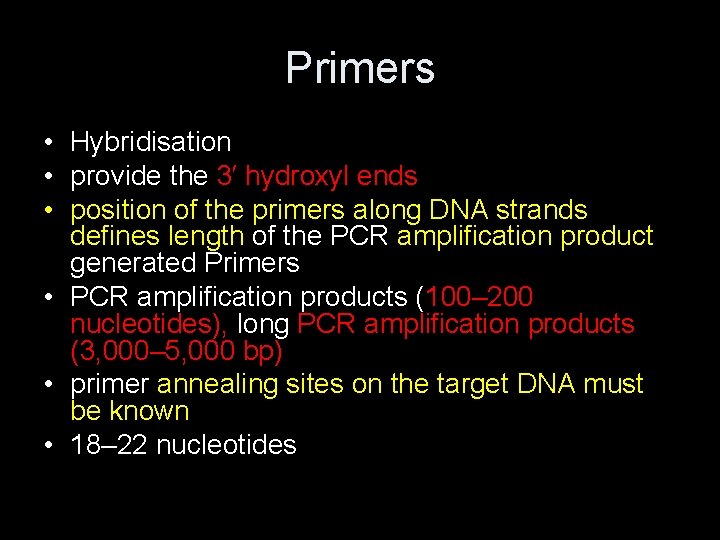
Primers

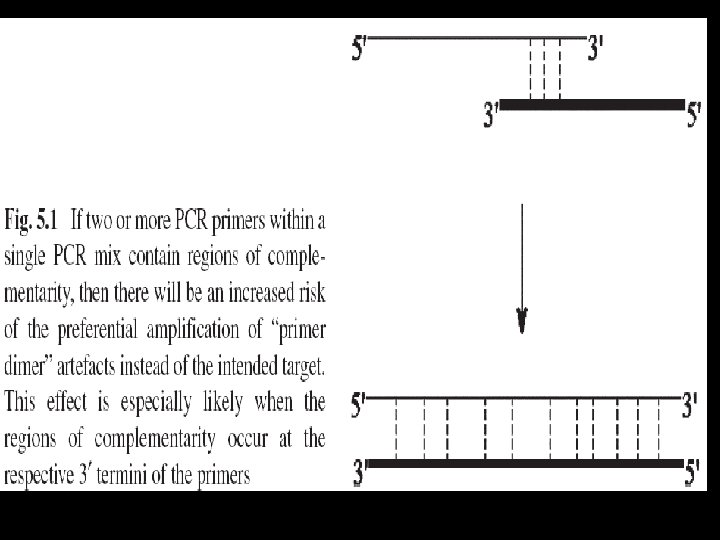
Primers • Hybridisation • provide the 3′ hydroxyl ends • position of the primers along DNA strands defines length of the PCR amplification product generated Primers • PCR amplification products (100– 200 nucleotides), long PCR amplification products (3, 000– 5, 000 bp) • primer annealing sites on the target DNA must be known • 18– 22 nucleotides

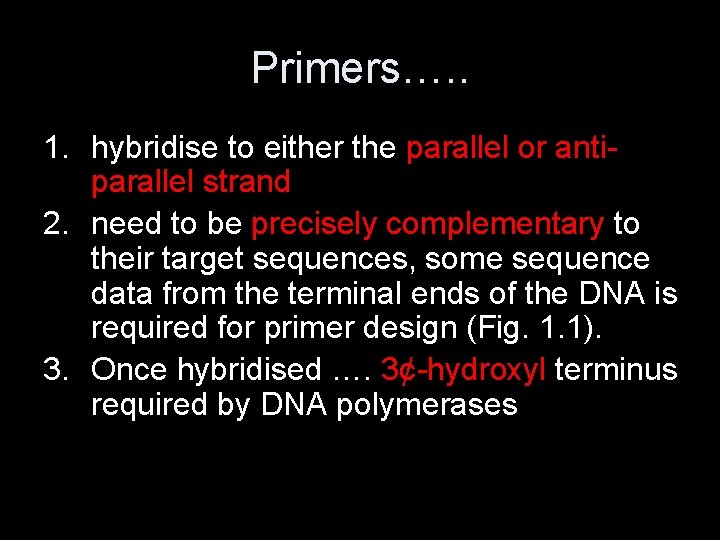
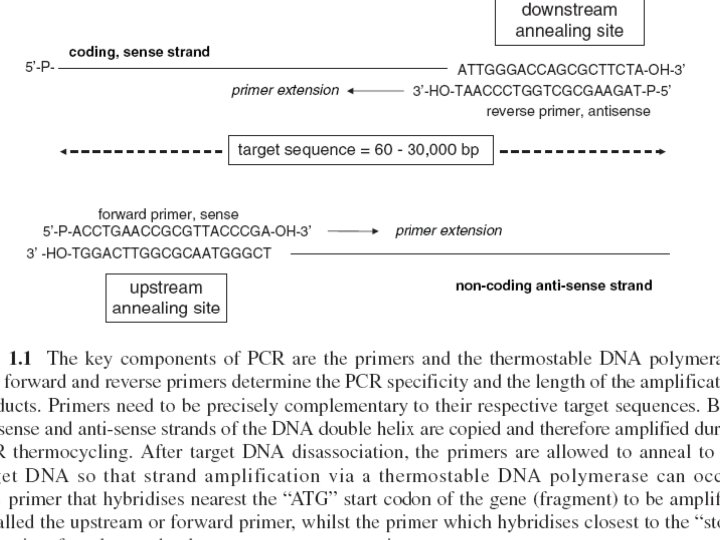
Primers…. . 1. hybridise to either the parallel or antiparallel strand 2. need to be precisely complementary to their target sequences, some sequence data from the terminal ends of the DNA is required for primer design (Fig. 1. 1). 3. Once hybridised …. 3¢-hydroxyl terminus required by DNA polymerases


PCR Primer Design and Quality Requirements 1. 2. 3. 4. 5. 6. base composition length of primer GC-content annealing temperatures, no internal complementary regions Complementary regions…. . primer/primer hybridisation…… “primer dimer” 7. secondary structure…. . primer dimers 8. primer-dimer formation…. . multiplex PCR protocols, 9. Y=G, z=C, x=T, and w=A


main rules for effective primer design • 18 and 30 nucleotides (bp) in length. • no internal complementary regions (e. g. multiple guanosine and cytosine repeats) • GC content 50% • genetically stable…. . Conserved sequences • inosine nucleotide • annealing temperatures… 65°C • annealing temperatures …. . ± 5°C

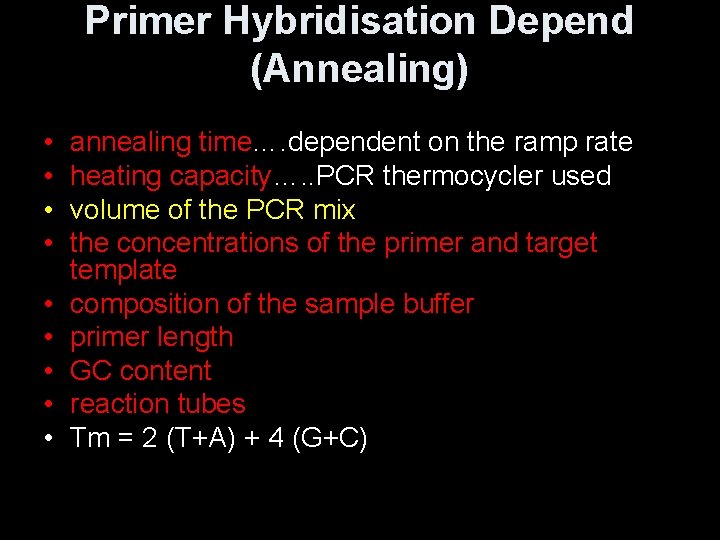
Primer Hybridisation Depend (Annealing) • • • annealing time…. dependent on the ramp rate heating capacity…. . PCR thermocycler used volume of the PCR mix the concentrations of the primer and target template composition of the sample buffer primer length GC content reaction tubes Tm = 2 (T+A) + 4 (G+C)

Primer Synthesis • Commercial companies • nucleic acid synthesising machines (e. g. the ABI 3400 DNA synthesizer, USA).

Effect of Mismatches Between PCR Primer and Target 1. Mutations in the target sequence or incorrectly designed PCR primers 2. resulting in incomplete base pairing between the two nucleic acid molecules…. . 3. lowers the annealing temperature (Tm)……reducing the yield of specific PCR amplification products

Primer Concentration • 0. 1– 1 μM concentration …. . (in 50 μl) • but excess of primer……. non-specific PCR products so

Deoxynucleotide Triphosphates (DNTPs)

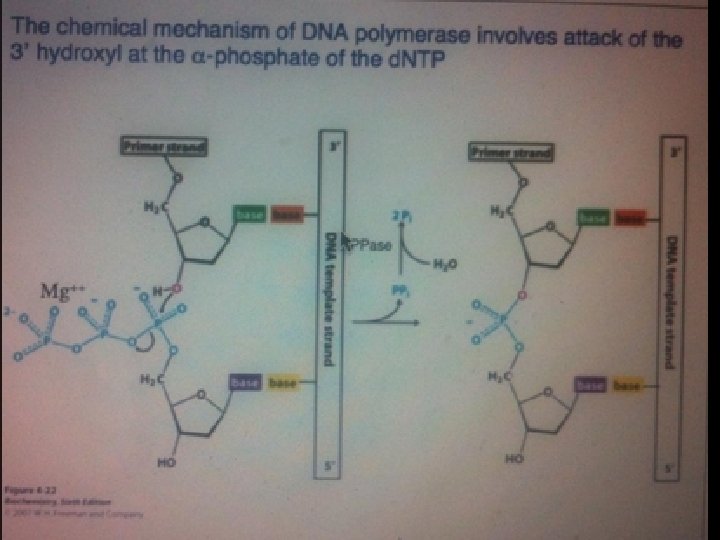
DNTPs…. 1. building blocks of nucleic acid molecules 2. necessary components of PCR mixes 3. The four individual deoxynucleotides…in PCR 1. 2. 3. 4. Deoxyadenosine triphosphate, d. ATP Deoxythymidine triphosphate, d. TTP Deoxycytosine triphosphate, d. CTP Deoxyguanosin triphosphate, d. GTP 4. purchased either individually or as an mix 5. stable when stored at − 20°C 6. Acidic in nature…so. . . working and stock solutions …. to be neutralized with alkaline compounds 7. 10 m. M stock solutions 8. d. NTPs may be lost due to non-specific heat inactivation…… during a 40 cycle PCR program…. . loss of d. NTPs may be in 50% of the original amount added to the PCR mix

DNTPs…. • d. NTPs…p. H 7. 2. . . negative charge…. ability to bind both mono- and divalent cations…. . mg ions • Mg ions …. bound by d. NTPs


Factors Affecting the Choice of d. NTP Concentration • use a concentration of d. NTPs …. only a fraction at the end of the PCR cycling • if the d. NTP limiting. . . rate of d. NTP incorporation will be reduced • 10μl……. . 0. 2 μl

Modified d. NTPs Typical PCR protocol…. all four d. NTPs 1. specially modified d. NTPs may also be added to the PCR 2. helix destabilizing nucleotide>>7 -deaza- 2′deoxyguanosine 3. Inosine is a naturally occurring nucleotide structural build in block t. RNA 4. helps to overcome amplification problems 5. solve problems related to sequence variation within the target DNA. 6. useful in amplifying Cp. G islands (GC rich sequence found in the promoter sequences of many higher eukaryotes)

Disadvantage of the use of Modified d. NTPs • 7 -deaza-2′-deoxyguanosine is very difficult to stain using ethidium bromide. • So d. GTP is also added to the PCR mix at a concentration 25% • their presence modifies restriction digest recognition sites…. these sites may no longer be recognized by restriction enzymes.

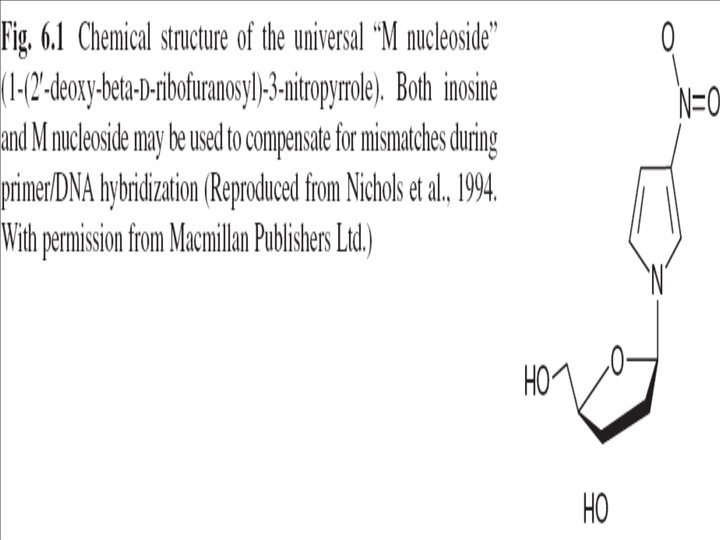
M-nucleoside…… • Alternatively, another universal nucleoside (1(2′-deoxy-β-D-ribofuranosyl)-3 -nitropyrrole) has been described in the literature • This nucleoside is known as “M-nucleoside” • maintains the ability …. four normal d. NTPs • its incorporation in PCR primers affects the Tm of primer annealing


The PCR Buffer

PCR Buffer • (10 X concentrated) PCR reaction buffer • DNA polymerase activity… p. H conditions should be maintained • ammonium sulphate help to remove any inhibitory products, such as pyrophosphate, …. . accumulate during PCR amplification.

Monovalent Ions

Monovalent Ions…… stimulate the activity of polymerases – Sodium (Na+), – potassium (K+) and – ammonium (NH+4) ions • Potassium ions have an optimum stimulatory effect on PCR DNA polymerases • NH+ 4 ions compete for the hydrogen bonds

Divalent

Magnesium Ions…… • important ingredient of PCR reaction • co-factor for thermostable DNA polymerase activity, stimulating the enzymes • optimization is frequently performed in combination with an experiment to determine the optimum d. NTP concentration • magnesium ion concentration range of between 0. 5 and 5 Mm • elevated magnesium ion concentrations inhibit PCR……double stranded DNA …actually stabilized …. • increasing the risk of nonspecific product amplification

Taq and Other Thermostable DNA Polymerases

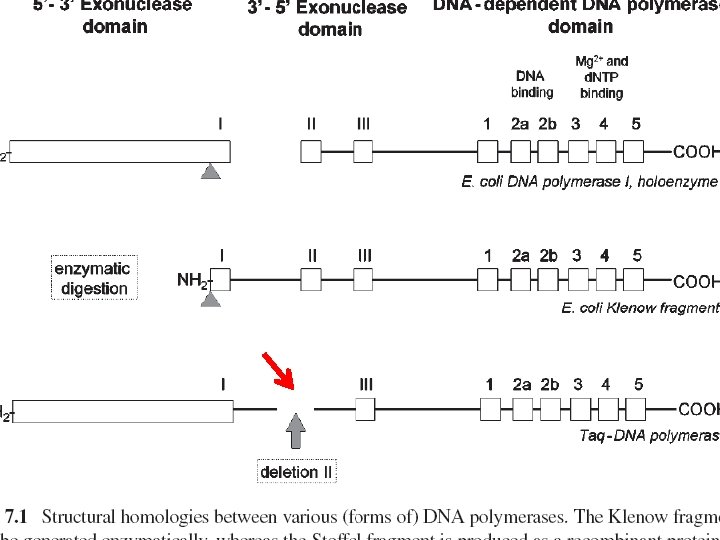
DNA Polymerases…… Ø DNA polymerase…. the success of PCR • Klenow fragment of DNA-dependent DNA polymerase I…. disadvantage • 1 lacks a 5¢-3¢ exonuclease activity. . • optimum reaction temperature at 37°C… • impossible to generate PCR…. longer than 400 bp

Taq polymerase “in 1976 Chien described a 94 k. D thermostable DNA-dependant DNA polymerase derived from a eubacterium called Thermus aquaticus” Ø 3¢-5¢ proofreading domain…. missing


The Advantages and Disadvantages of Taq over Klenow Fragment DNA Polymerase

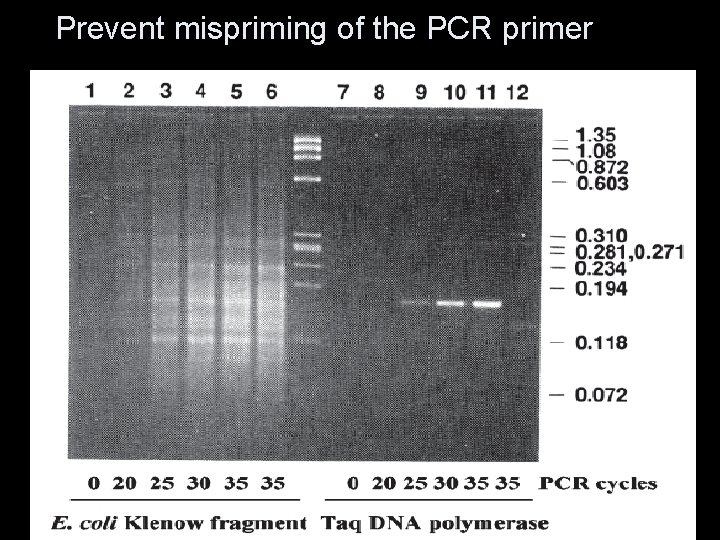
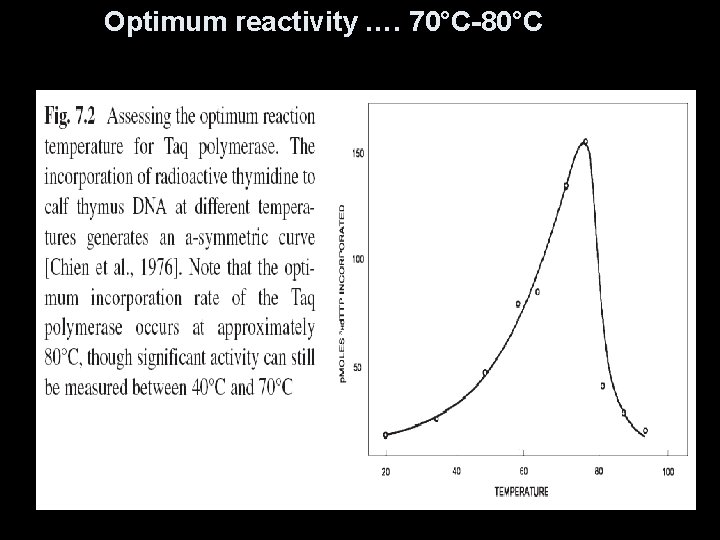
Advantages… 1. 2. 3. 4. 5. 6. Heat stable … half-life of 130 minutes at a temperature of 92. 5°C. Reaction vial not… opened in order to add fresh enzyme … reducing contamination optimum reactivity …. 70°C-80°C…helps to prevent any secondary or tertiary structure High optimum temperature …prevent mispriming of the PCR primer Has increased processivity at elevated incubation temperature. Generates significantly higher yields of amplification products (ten times higher) 60 nucleotides per second at 72°C, but – only 1. 5 nucleotides per second at 37°C 7. 8. Enzyme is not inhibited by chemical contaminants remaining after nucleic acid extraction, e. g. chloroform Taq enzyme frequently adds an overhanging nucleotide

Prevent mispriming of the PCR primer

Optimum reactivity …. 70°C-80°C

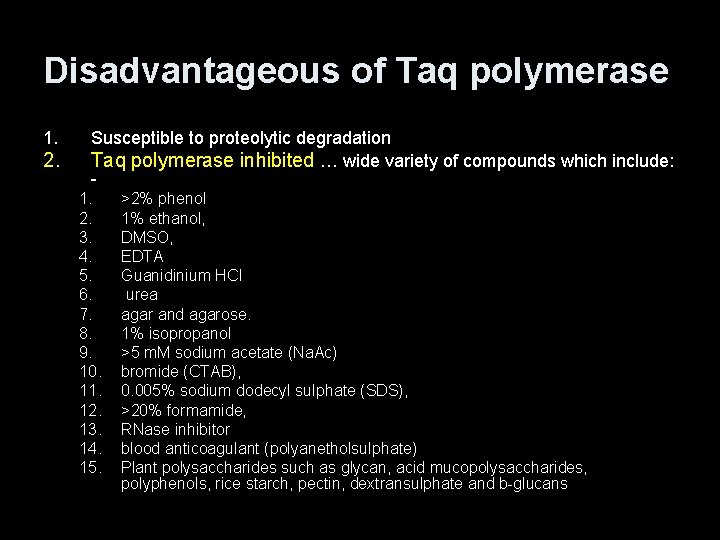
Disadvantageous of Taq polymerase 1. 2. Susceptible to proteolytic degradation Taq polymerase inhibited … wide variety of compounds which include: 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. >2% phenol 1% ethanol, DMSO, EDTA Guanidinium HCl urea agar and agarose. 1% isopropanol >5 m. M sodium acetate (Na. Ac) bromide (CTAB), 0. 005% sodium dodecyl sulphate (SDS), >20% formamide, RNase inhibitor blood anticoagulant (polyanetholsulphate) Plant polysaccharides such as glycan, acid mucopolysaccharides, polyphenols, rice starch, pectin, dextransulphate and b-glucans

Analysis of PCR Amplification Products

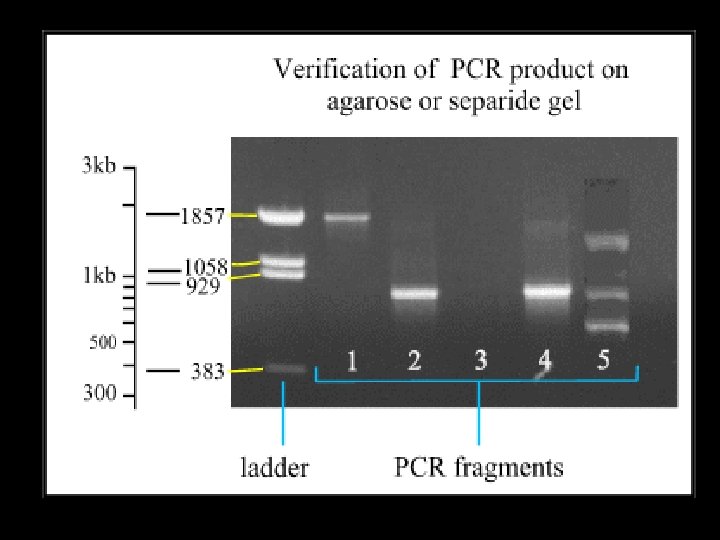
Gel Electrophoresis (i) Simple agarose gel electrophoresis (ii) Simple polyacrylamide gel electrophoresis (PAGE) (iii) Restriction Fragment Length Polymorphism (RFLP) analysis and oligomer restriction (iv) Single Stranded Conformation Polymorphism (SSCP) analysis (v) Denaturing Gradient Gel Electrophoresis (DGGE) (vi) Image analysis of PCR amplification products in gel electrophoresis systems (vii) Excising and cleaning PCR amplimers from electrophoresis gels

Simple agarose gel electrophoresis 1. polysaccharide agarose (poly d-galactose 3, 6 anhydro-l galactose 2. purified from marine algae 3. solid compound at room temperature…. powder


PCR Failures… Smear of amplification products…. may be caused by…. 1. 2. 3. 4. 5. Degraded PCR primers, Contamination from previous PCRs, Excessive quantity of Taq High magnesium ion concentration Number of cycles in the PCR Amount of template DNA 6. DNA added to the PCR mix may be impure or degraded………