DNA RNA Parameters total DNA RNA content nucleic

- Slides: 22

DNA & RNA Parameters • • • total DNA & RNA content nucleic acid sequence cell cycle analysis chromosome analysis reticulocyte analysis live/dead – membrane integrity • identify microorganisms Page 1 5: 33 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

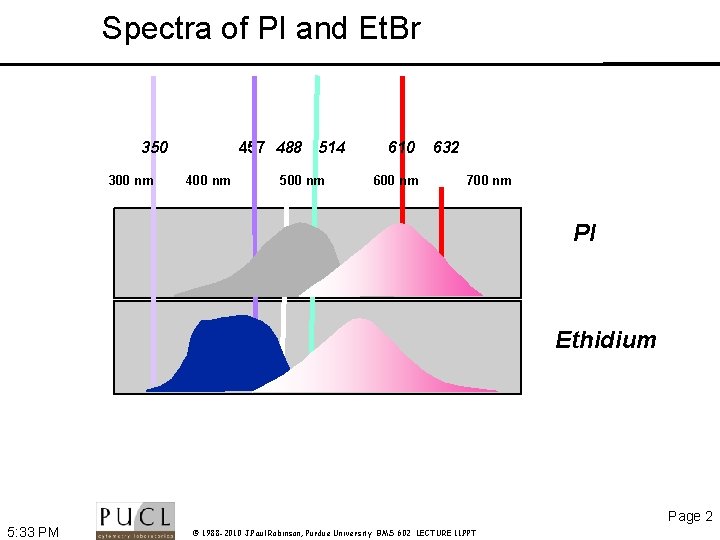
Spectra of PI and Et. Br 350 300 nm 457 488 514 400 nm 500 nm 610 600 nm 632 700 nm PI Ethidium Page 2 5: 33 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

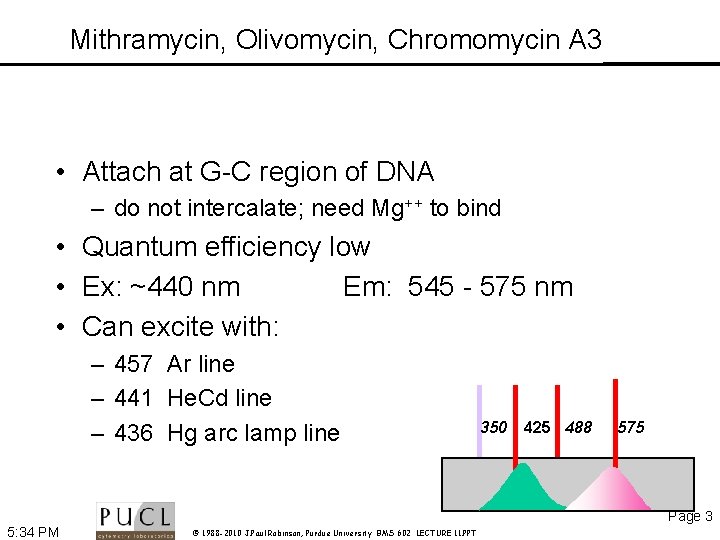
Mithramycin, Olivomycin, Chromomycin A 3 • Attach at G-C region of DNA – do not intercalate; need Mg++ to bind • Quantum efficiency low • Ex: ~440 nm Em: 545 - 575 nm • Can excite with: – 457 Ar line – 441 He. Cd line – 436 Hg arc lamp line 350 425 488 575 Page 3 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

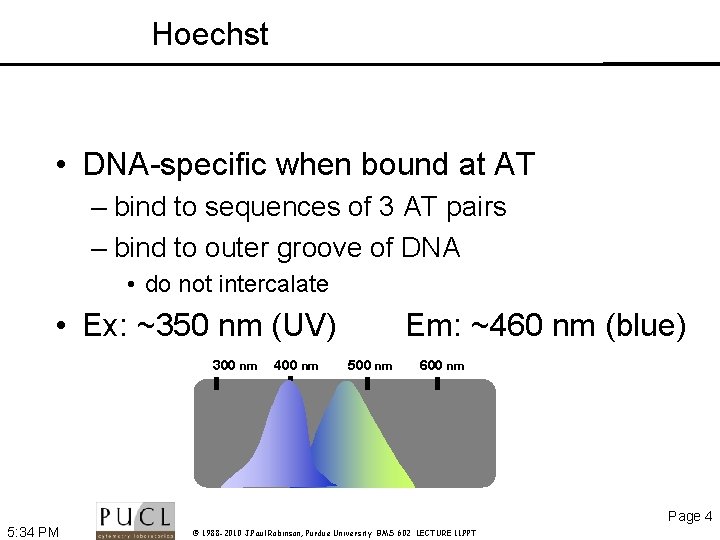
Hoechst • DNA-specific when bound at AT – bind to sequences of 3 AT pairs – bind to outer groove of DNA • do not intercalate • Ex: ~350 nm (UV) 300 nm 400 nm Em: ~460 nm (blue) 500 nm 600 nm Page 4 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

Cyanine Dyes • Thiazole orange, thiazole blue, thioflavin T and others • Stain both RNA and DNA • Quantum efficiency greatly increased when bound to NA – very low when unbound • Cross membranes of intact cells – will also enter mitochondria Page 5 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

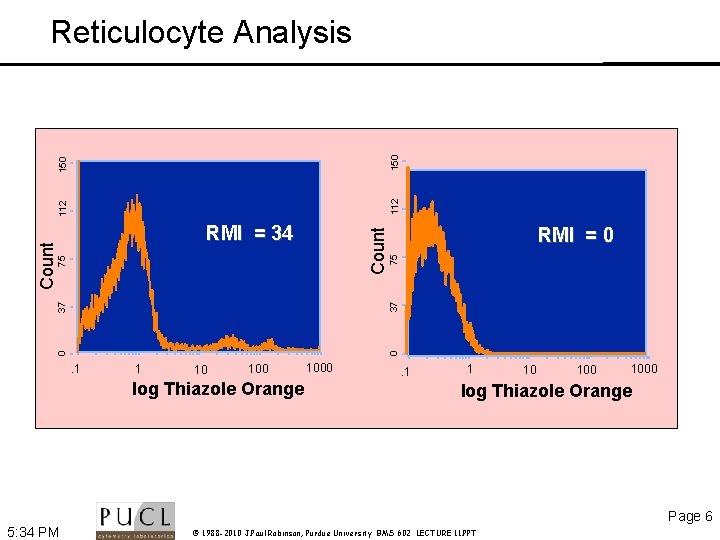
75 RMI = 0 37 75 Count RMI = 34 0 0 37 Count 112 150 Reticulocyte Analysis . 1 1 10 100 log Thiazole Orange 1000 . 1 1 10 1000 log Thiazole Orange Page 6 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

Cyanine Dyes • TOTO-1 , YOYO-1, TOTO-3 – developed to have high binding affinity and high quantum efficiency – homodimers of • thiazole orange, oxazole yellow, thiazole blue • positively charged side chains – do not penetrate intact membranes – not DNA-specific • treat cells with RNase Page 7 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

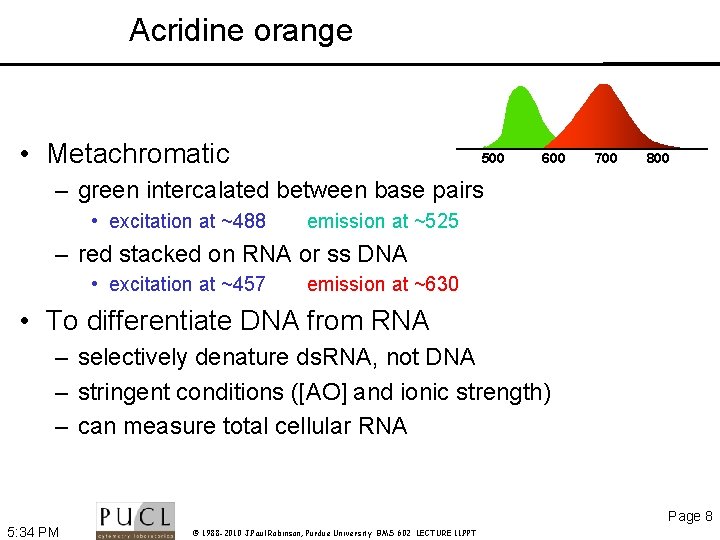
Acridine orange • Metachromatic 500 600 700 800 – green intercalated between base pairs • excitation at ~488 emission at ~525 – red stacked on RNA or ss DNA • excitation at ~457 emission at ~630 • To differentiate DNA from RNA – selectively denature ds. RNA, not DNA – stringent conditions ([AO] and ionic strength) – can measure total cellular RNA Page 8 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

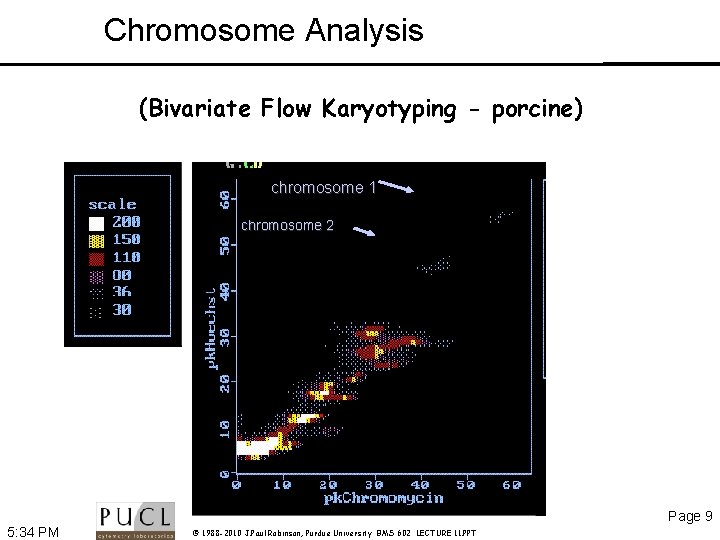
Chromosome Analysis (Bivariate Flow Karyotyping - porcine) chromosome 1 chromosome 2 Page 9 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

Dual Staining of Cells • • • Nuclear probes c-myc, c-fos, p 53 monoclonal Ab Cytoplasmic protooncogene probes ‘ras’, ‘neu’ monoclonal Ab Cell surface antigens p-glycoprotein Breast carcinoma • identify ploidy with PI • identify epithelial origin with cytokeratin antibody Page 10 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

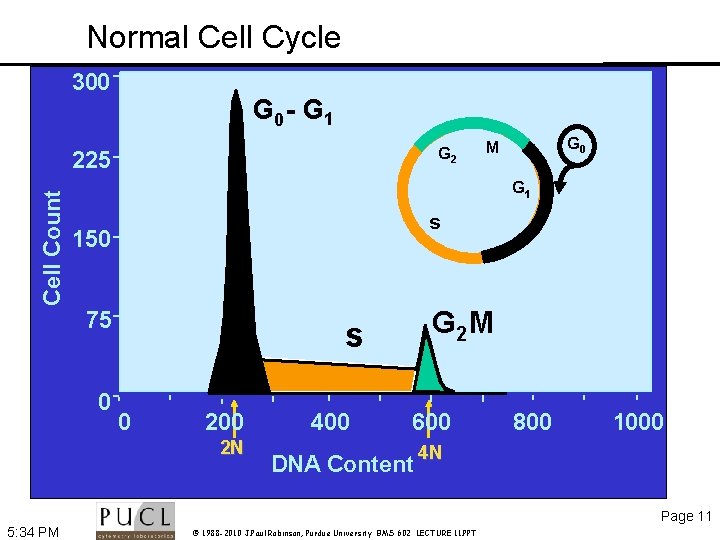
Normal Cell Cycle 300 G 0 - G 1 G 0 G 2 Cell Count 225 G 0 M G 1 s 150 75 0 G 2 M s 0 200 2 N 400 600 DNA Content 800 1000 4 N Page 11 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

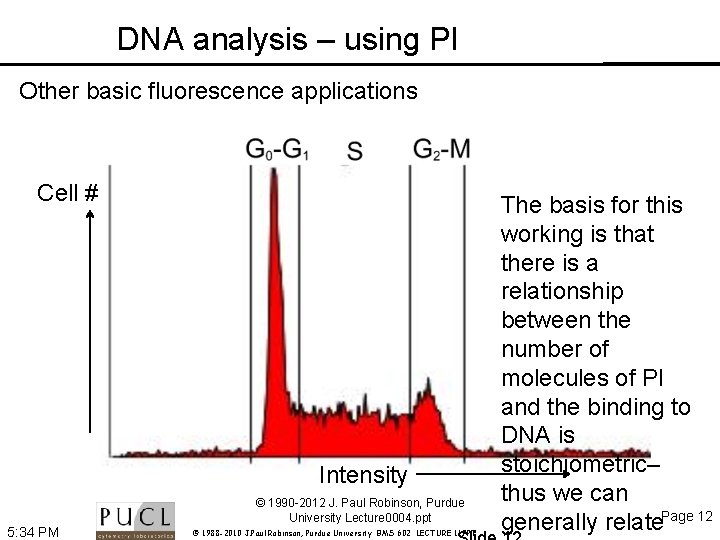
DNA analysis – using PI Other basic fluorescence applications Cell # Intensity © 1990 -2012 J. Paul Robinson, Purdue University Lecture 0004. ppt 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT The basis for this working is that there is a relationship between the number of molecules of PI and the binding to DNA is stoichiometric– thus we can generally relate. Page 12

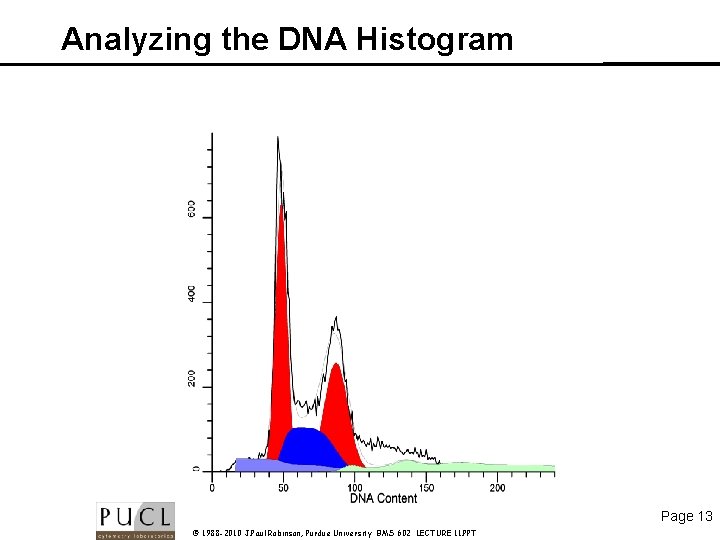
Analyzing the DNA Histogram Page 13 © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

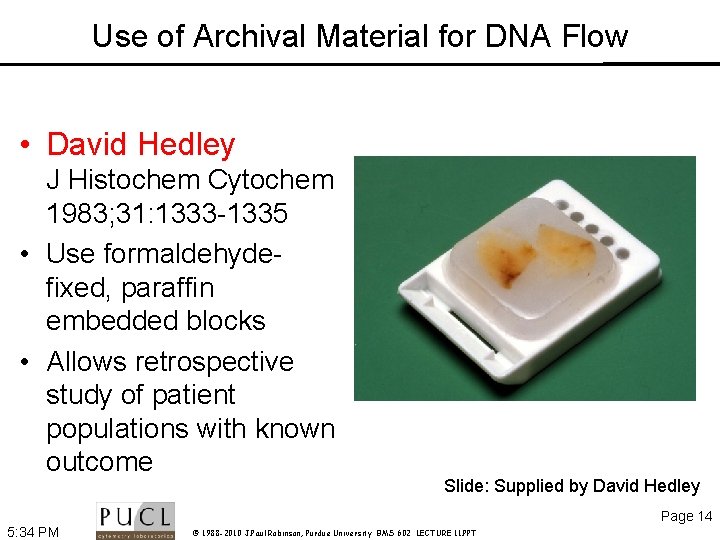
Use of Archival Material for DNA Flow • David Hedley J Histochem Cytochem 1983; 31: 1333 -1335 • Use formaldehydefixed, paraffin embedded blocks • Allows retrospective study of patient populations with known outcome Slide: Supplied by David Hedley Page 14 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

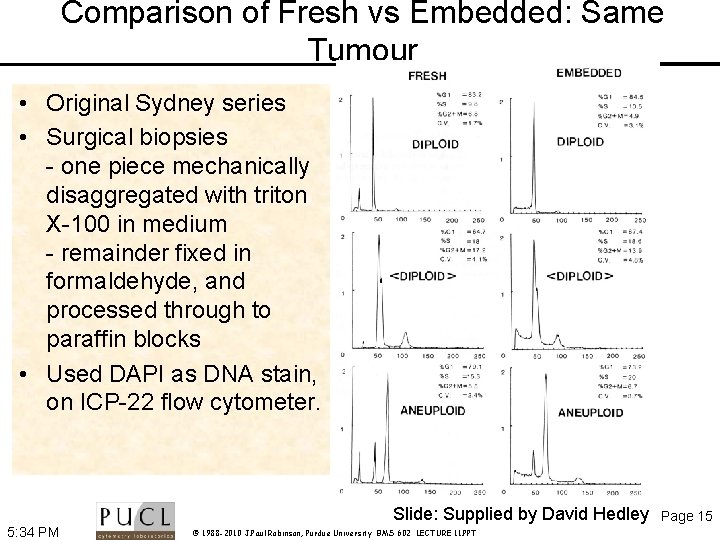
Comparison of Fresh vs Embedded: Same Tumour • Original Sydney series • Surgical biopsies - one piece mechanically disaggregated with triton X-100 in medium - remainder fixed in formaldehyde, and processed through to paraffin blocks • Used DAPI as DNA stain, on ICP-22 flow cytometer. Slide: Supplied by David Hedley 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT Page 15

Functional Assays • intracellular p. H • intracellular calcium • intracellular glutathione • oxidative burst • phagocytosis Page 16 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

Phagocytosis FITC-Labeled Bacteria Page 17 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

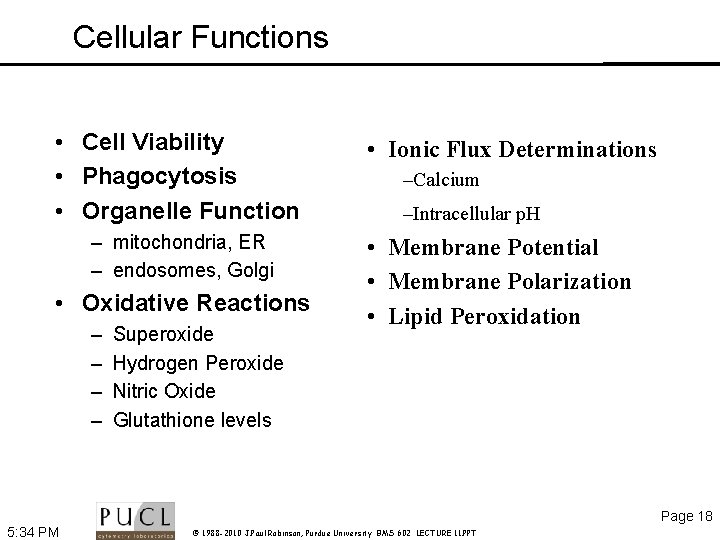
Cellular Functions • Cell Viability • Phagocytosis • Organelle Function – mitochondria, ER – endosomes, Golgi • Oxidative Reactions – – Superoxide Hydrogen Peroxide Nitric Oxide Glutathione levels • Ionic Flux Determinations –Calcium –Intracellular p. H • Membrane Potential • Membrane Polarization • Lipid Peroxidation Page 18 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

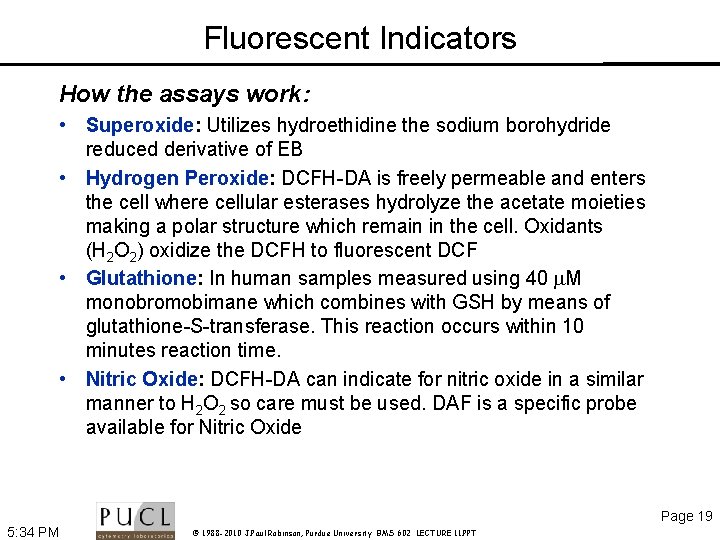
Fluorescent Indicators How the assays work: • Superoxide: Utilizes hydroethidine the sodium borohydride reduced derivative of EB • Hydrogen Peroxide: DCFH-DA is freely permeable and enters the cell where cellular esterases hydrolyze the acetate moieties making a polar structure which remain in the cell. Oxidants (H 2 O 2) oxidize the DCFH to fluorescent DCF • Glutathione: In human samples measured using 40 M monobromobimane which combines with GSH by means of glutathione-S-transferase. This reaction occurs within 10 minutes reaction time. • Nitric Oxide: DCFH-DA can indicate for nitric oxide in a similar manner to H 2 O 2 so care must be used. DAF is a specific probe available for Nitric Oxide Page 19 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

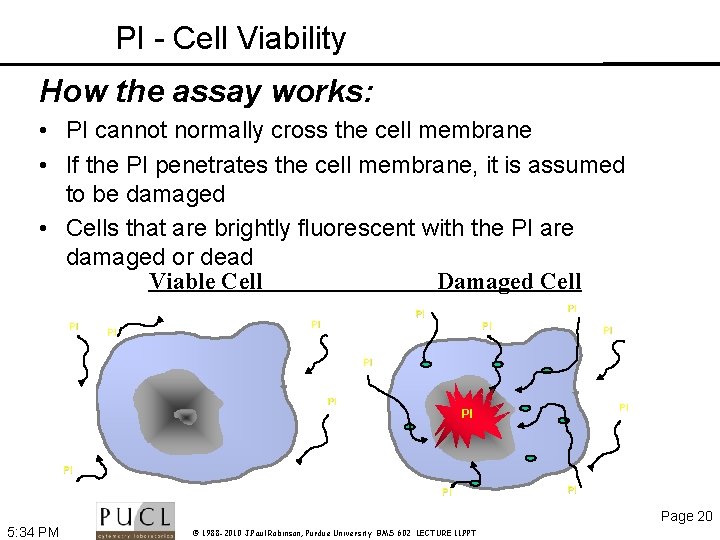
PI - Cell Viability How the assay works: • PI cannot normally cross the cell membrane • If the PI penetrates the cell membrane, it is assumed to be damaged • Cells that are brightly fluorescent with the PI are damaged or dead Viable Cell Damaged Cell PI PI PI PI Page 20 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

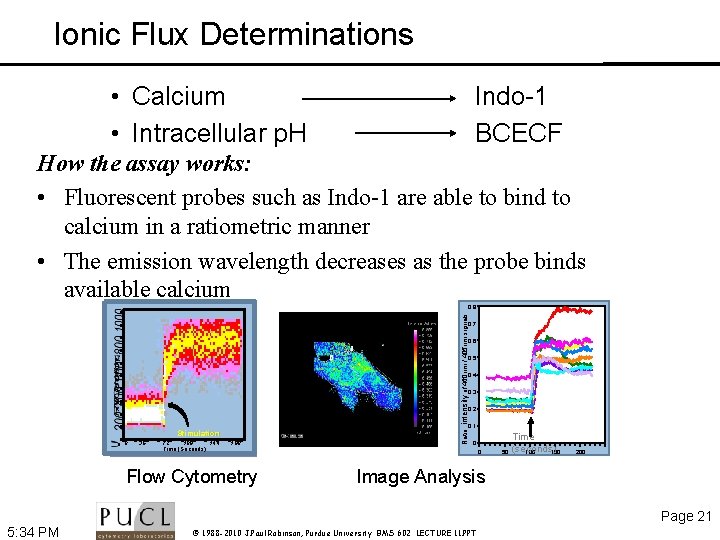
Ionic Flux Determinations • Calcium • Intracellular p. H Indo-1 BCECF How the assay works: • Fluorescent probes such as Indo-1 are able to bind to calcium in a ratiometric manner • The emission wavelength decreases as the probe binds available calcium Ratio: intensity of 460 nm / 405 nm signals 0. 8 0. 7 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 Stimulation 0 36 72 108 144 Time (Seconds) 180 Flow Cytometry 0 0 50 Time (seconds) 100 150 200 Image Analysis Page 21 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT

Phosphorylation assays • Phospho antibodies • Measure the phosphorylation state of intracellular proteins at the single cell level Page 22 5: 34 PM © 1988 -2010 J. Paul Robinson, Purdue University BMS 602 LECTURE 11. PPT