CS 882 Fall 2006 Lecture 7 Computing Protein





- Slides: 41

CS 882, Fall 2006 Lecture 7. Computing Protein Structures • Current attempts: • Threading: RAPTOR • Consensus: ACE • Fragment assembly Can we compute the protein structures eventually? Your projects.

Homologous proteins have similar structure and functions n Being homologous means that they have evolved from a common ancestral gene. Hence at least in the past they had the same structure and function. n Caution: old genes can be recruited for new functions. Example: a structural protein in eye lens is homologous to an ancient glycolytic enzyme. n Homology search is done by BLAST, or Pattern. Hunter for more sensitivity. BLAST will work with over 30% sequence identity.

Conserving core regions n Homologous proteins usually have conserved core regions. n When we model one protein after a similar protein with known structure, the main problem becomes modeling loop regions. n Modeling loops can also depend on database to some degree. n Side chains: on a few side-chain conformations frequently occur – they are called rotamers, there is a such a database.

Primary, secondary, and tertiary n There are many secondary structure prediction programs. However, without considering tertiary structure, we will never be correct solely predicting secondary structures. n Most tertiary structure prediction programs today depend on good secondary predictions. This is also not good: you cannot get right tertiary structure with wrong starting information. n They must be done together.

There are not too many candidates! n There are only about 1000 topologically different n n n domain structures. There is no reason whatsoever that we cannot compute their structures accurately. Ab initio method – we have heard about it. Another promising method is threading (separate lecture). After threading, an important step is “refinement”, perhaps by fragment assembly. This will be a separate topic (Xin Gao). Folding membrane proteins is a quite different topic (Richard Jang). Now we go to threading.

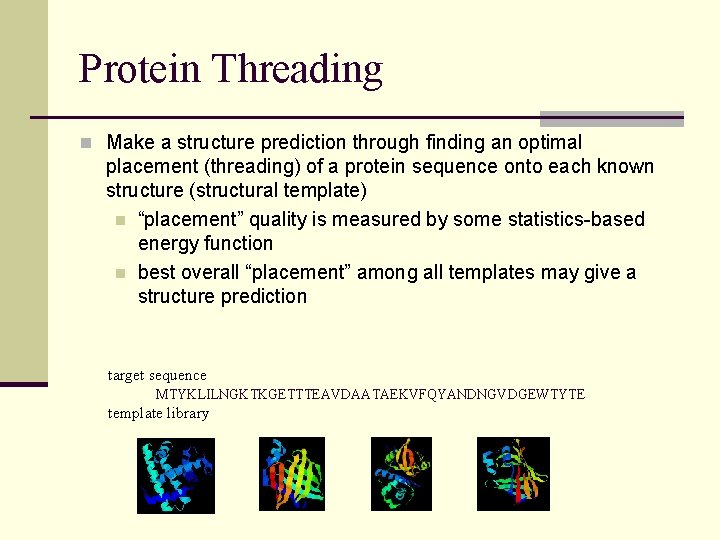
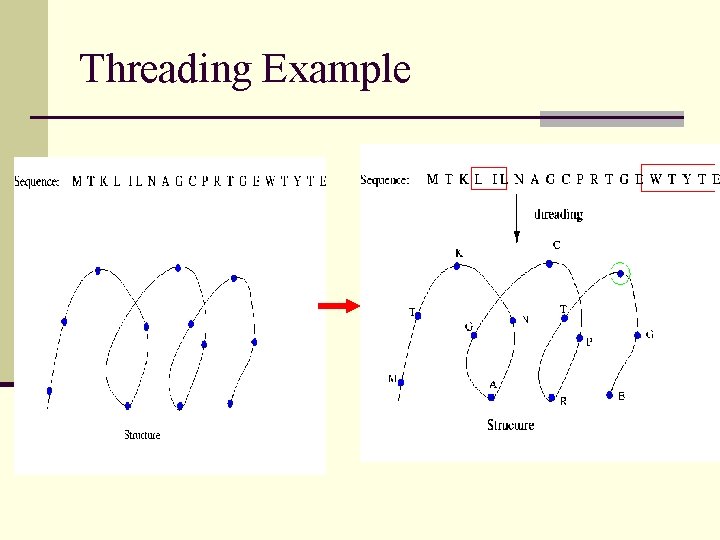
Protein Threading n Make a structure prediction through finding an optimal placement (threading) of a protein sequence onto each known structure (structural template) n “placement” quality is measured by some statistics-based energy function n best overall “placement” among all templates may give a structure prediction target sequence MTYKLILNGKTKGETTTEAVDAATAEKVFQYANDNGVDGEWTYTE template library

Threading Example

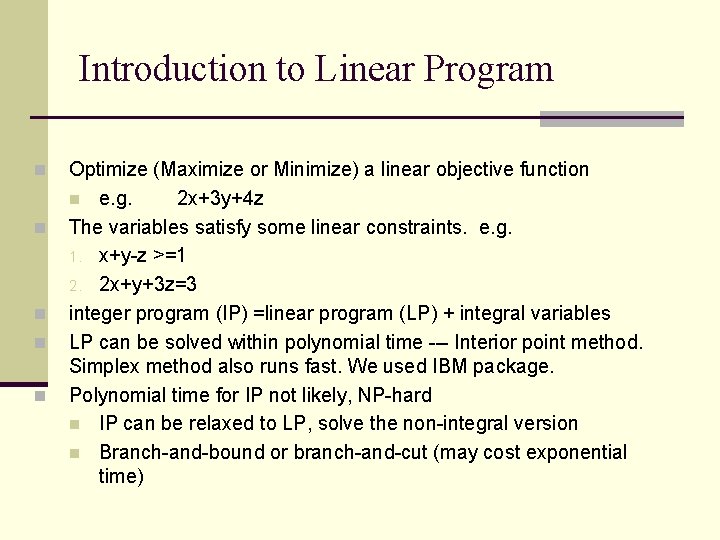
Introduction to Linear Program n n n Optimize (Maximize or Minimize) a linear objective function n e. g. 2 x+3 y+4 z The variables satisfy some linear constraints. e. g. 1. x+y-z >=1 2. 2 x+y+3 z=3 integer program (IP) =linear program (LP) + integral variables LP can be solved within polynomial time --- Interior point method. Simplex method also runs fast. We used IBM package. Polynomial time for IP not likely, NP-hard n IP can be relaxed to LP, solve the non-integral version n Branch-and-bound or branch-and-cut (may cost exponential time)

Why Integer Programming? n Treat pairwise potentials rigorously n critical for fold-level targets n Existing Exact algorithms for pairwise potentials High memory requirement, or n Expensive computational time n n Exploit correlations between various kinds of item scores in the energy function n 99% real data generate integral solutions directly, no branch-and-bound needed.

Different approaches n Approximation Algorithm n Interaction-Frozen Algorithm (A. Godzik et al. ) n Monte Carlo Sampling (T. Madej et al. ) n Double dynamic programming (D. Jones et al. ) n Recursive dynamic programming (R. Thiele et al. ) n Exact Algorithm n Branch-and-bound (R. H. Lathrop et al. ) n n Exploit the relationship among various scoring parameters, fast self-threading Divide-and-conquer (Y. Xu et al. ) n Exploit the topological structure of template contact graphs

Formulating Protein Threading by LP • Protein Threading Needs: 1. 2. 3. 4. Construction of Template Library Design of Energy Function Sequence-Structure Alignment Template Selection and Model Construction

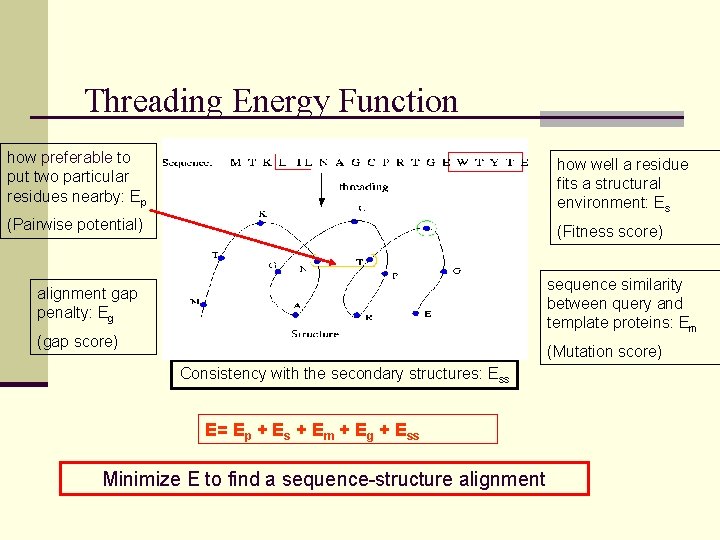
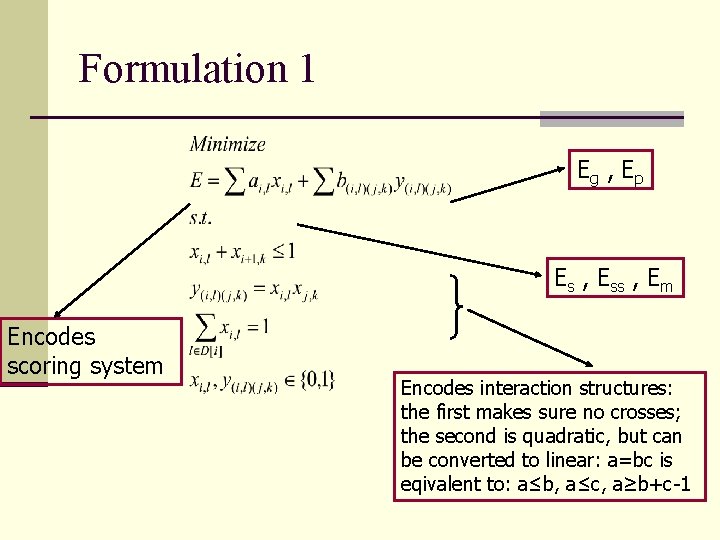
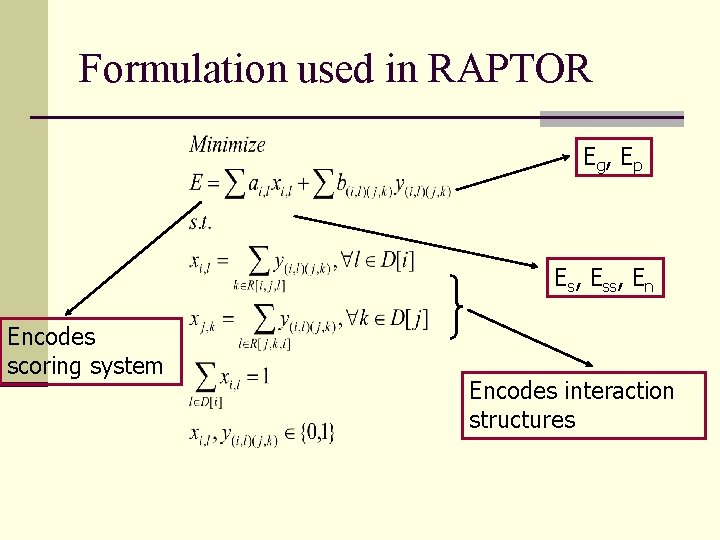
Threading Energy Function how preferable to put two particular residues nearby: Ep how well a residue fits a structural environment: Es (Pairwise potential) (Fitness score) sequence similarity between query and template proteins: Em alignment gap penalty: Eg (gap score) (Mutation score) Consistency with the secondary structures: Ess E= Ep + Es + Em + Eg + Ess Minimize E to find a sequence-structure alignment

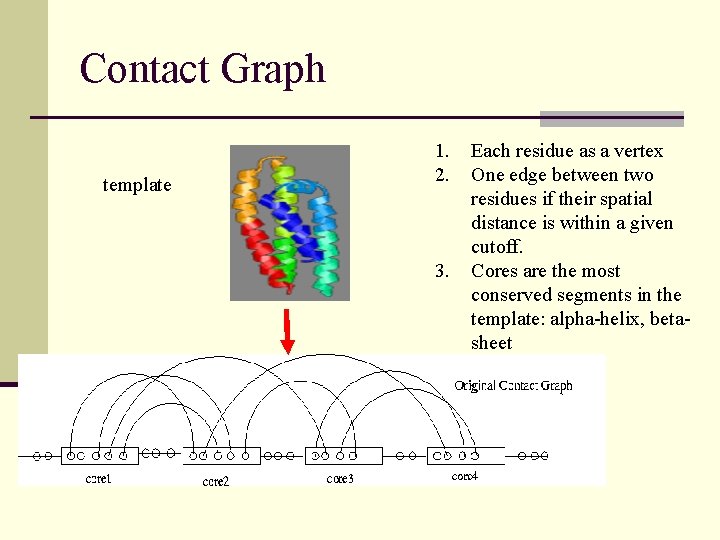
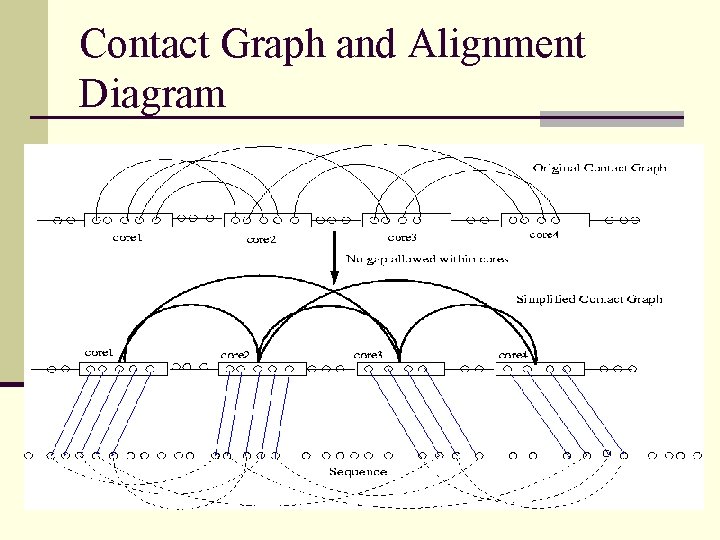
Contact Graph template 1. 2. 3. Each residue as a vertex One edge between two residues if their spatial distance is within a given cutoff. Cores are the most conserved segments in the template: alpha-helix, betasheet

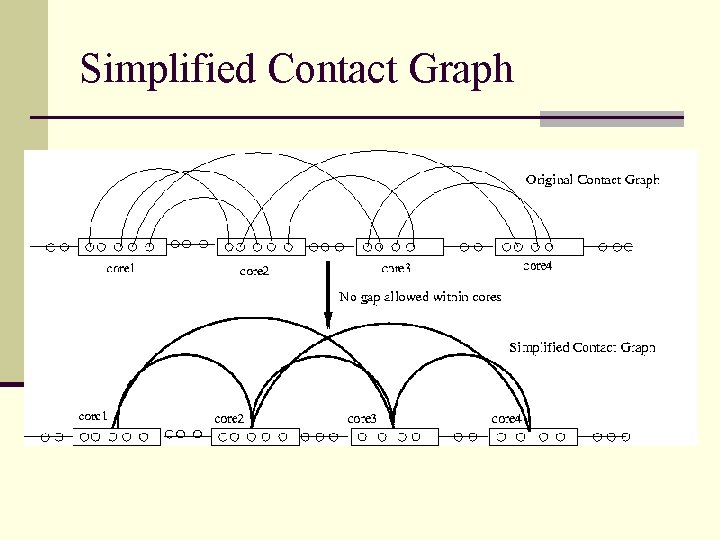
Simplified Contact Graph

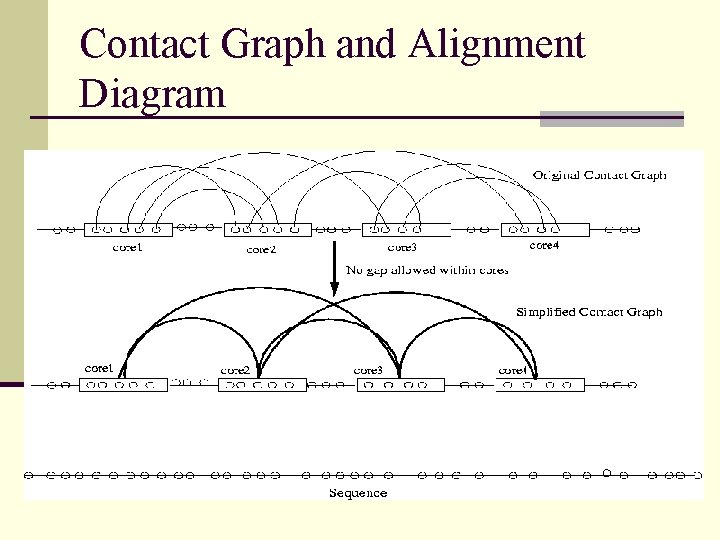
Contact Graph and Alignment Diagram

Contact Graph and Alignment Diagram

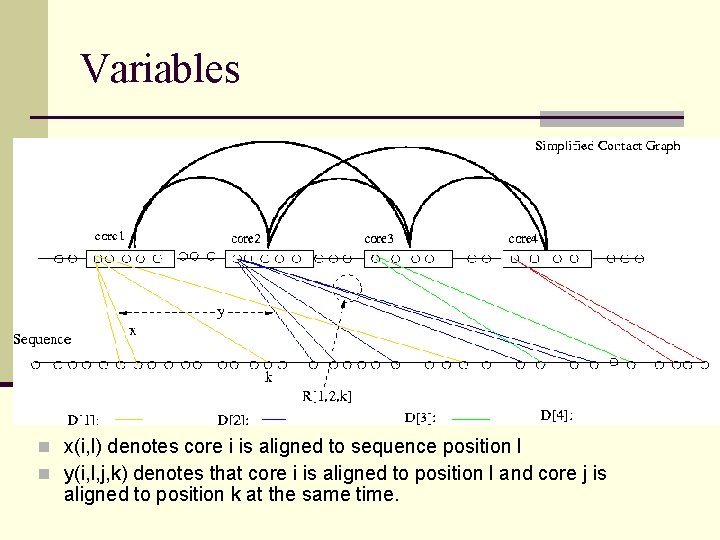
Variables n x(i, l) denotes core i is aligned to sequence position l n y(i, l, j, k) denotes that core i is aligned to position l and core j is aligned to position k at the same time.

Formulation 1 Eg , Ep Es , Ess , Em Encodes scoring system Encodes interaction structures: the first makes sure no crosses; the second is quadratic, but can be converted to linear: a=bc is eqivalent to: a≤b, a≤c, a≥b+c-1

Formulation used in RAPTOR Eg , Ep Es, Ess, En Encodes scoring system Encodes interaction structures

Solving the Problem Practically 1. More than 99% threading instances can be solved directly by linear programming, the rest can be solved by branch-and-bound with only several branch nodes 2. Less memory consumption 3. Less computational time 4. Easy to extend to incorporate other constraints

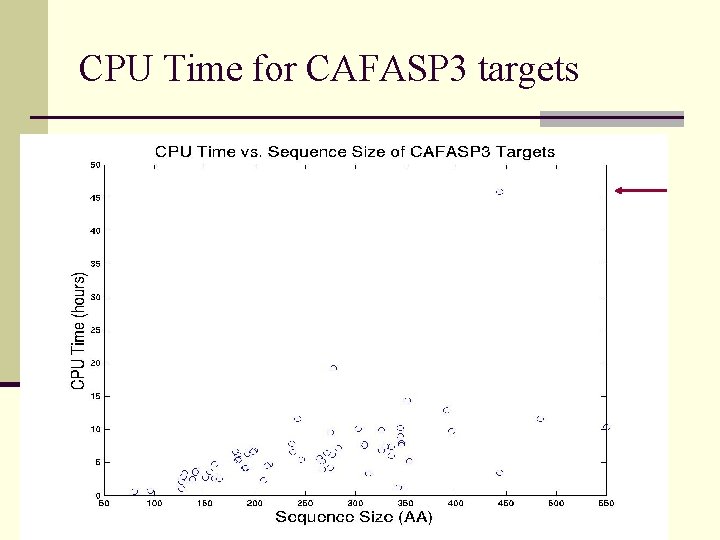
CPU Time for CAFASP 3 targets

Fold Recognition n Support Vector Machines (SVM) Approach n Features are extracted from the alignments n A threading pair is treated as a positive pattern only if they are in at least fold-level similarity n 60, 000 threading pairs are employed to train SVM model. n 5% more targets are recognized by SVM approach than the traditional z-Score

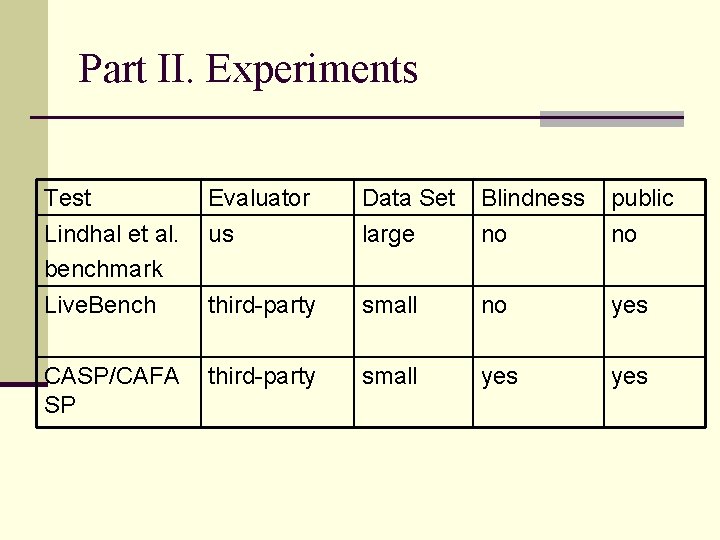
Part II. Experiments Test Lindhal et al. benchmark Evaluator us Data Set large Blindness no public no Live. Bench third-party small no yes CASP/CAFA SP third-party small yes

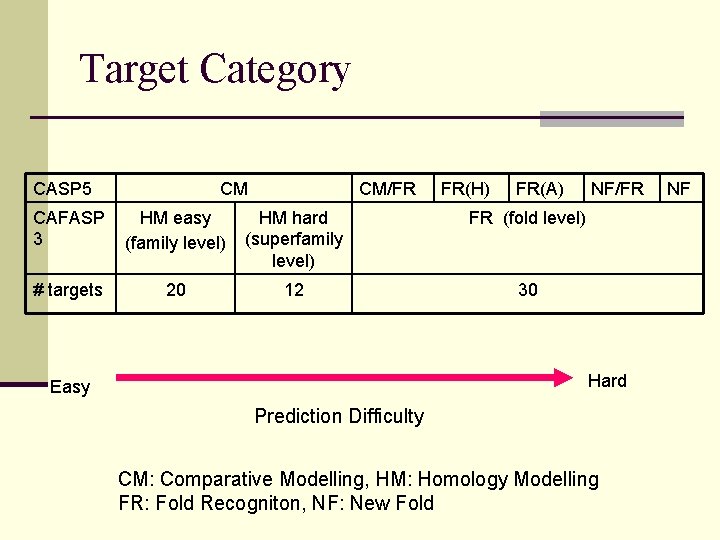
Target Category CASP 5 CM CM/FR FR(H) FR(A) CAFASP 3 HM easy (family level) HM hard (superfamily level) FR (fold level) # targets 20 12 30 NF/FR Hard Easy Prediction Difficulty CM: Comparative Modelling, HM: Homology Modelling FR: Fold Recogniton, NF: New Fold NF

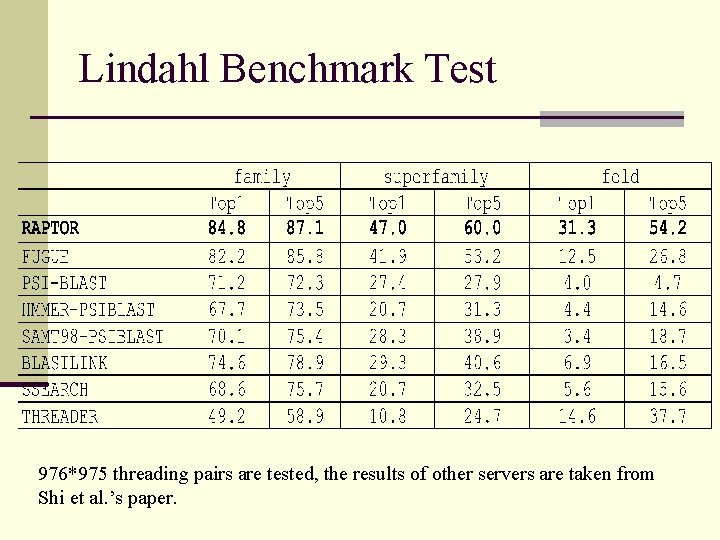
Lindahl Benchmark Test 976*975 threading pairs are tested, the results of other servers are taken from Shi et al. ’s paper.

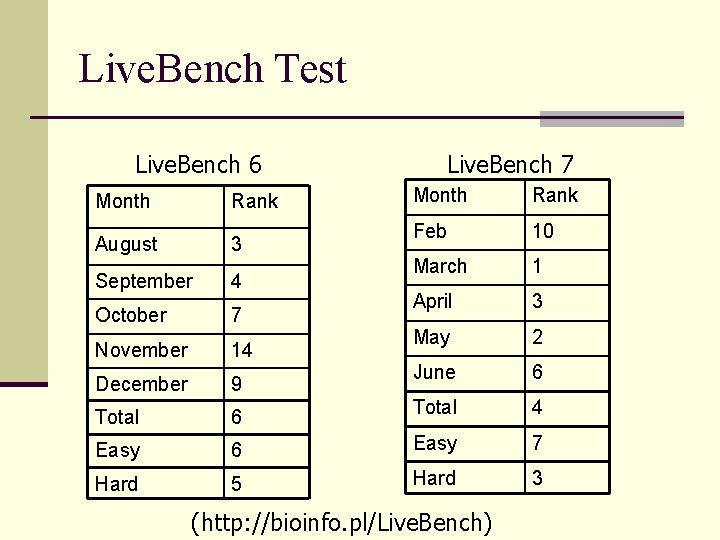
Live. Bench Test Live. Bench 6 Live. Bench 7 Month Rank Feb 10 March 1 April 3 May 2 June 6 6 Total 4 Easy 6 Easy 7 Hard 5 Hard 3 Month Rank August 3 September 4 October 7 November 14 December 9 Total (http: //bioinfo. pl/Live. Bench)

CASP 5/CAFASP 3 n 62 targets n Time allowed for each target: n Individual Servers: 48 hours n Meta Servers: 48 hours n Predictors: computer program, no manual intervention (CAFASP 3) n Evaluated by computer program n RAPTOR was voted by CASP 5 attendees as the most novel approach, at http: //forcasp. org CAFASP 3: The Third Critical Assessment of Fully Automated Structure Prediction

CAFASP 3 Evaluation Criteria n Model n n Only the first submission considered for each target, each server can submit 10 models for each target, n Max. Sub (evaluation program) n n n Superimpose the predicted structure with the experimental structure Calculate the length of maximum superimposable subsegment within 5Å RMSD one prediction is regarded as correct only if the length is above a given value.

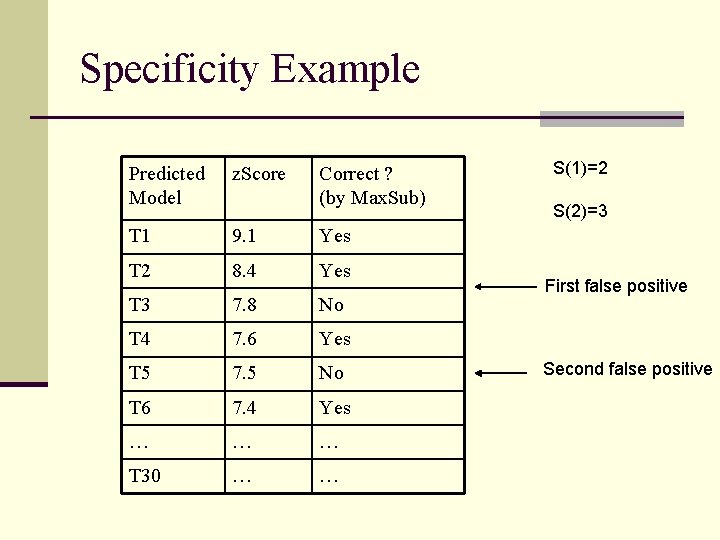
CAFASP 3 Evaluation Criteria n Sensitivity (N-1 Rule) n One miss allowed for each server, i. e. , the first models of N-1 out of N targets ranked n Specificity n Rank the first models of all targets according to their z. Scores n S(M): # Correct before the first M false positives n Average of S(1), S(2), …, S(5)

Specificity Example Predicted Model z. Score Correct ? (by Max. Sub) T 1 9. 1 Yes T 2 8. 4 Yes T 3 7. 8 No T 4 7. 6 Yes T 5 7. 5 No T 6 7. 4 Yes … … … T 30 … … S(1)=2 S(2)=3 First false positive Second false positive

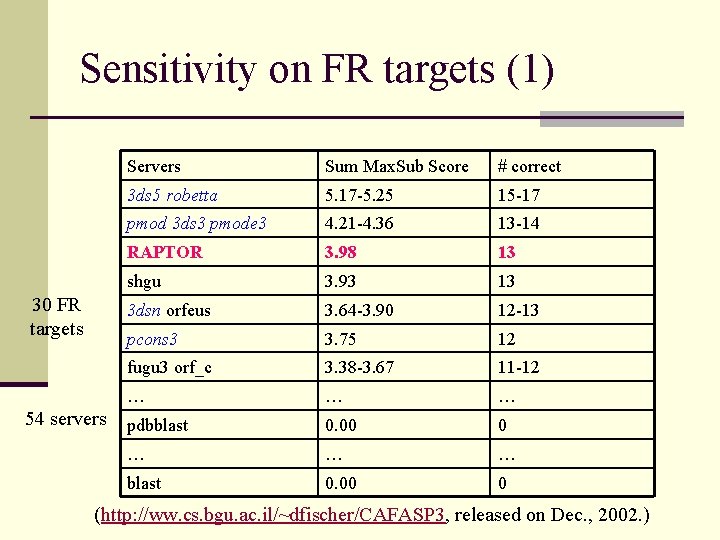
Sensitivity on FR targets (1) 30 FR targets 54 servers Sum Max. Sub Score # correct 3 ds 5 robetta 5. 17 -5. 25 15 -17 pmod 3 ds 3 pmode 3 4. 21 -4. 36 13 -14 RAPTOR 3. 98 13 shgu 3. 93 13 3 dsn orfeus 3. 64 -3. 90 12 -13 pcons 3 3. 75 12 fugu 3 orf_c 3. 38 -3. 67 11 -12 … … … pdbblast 0. 00 0 … … … blast 0. 00 0 (http: //ww. cs. bgu. ac. il/~dfischer/CAFASP 3, released on Dec. , 2002. )

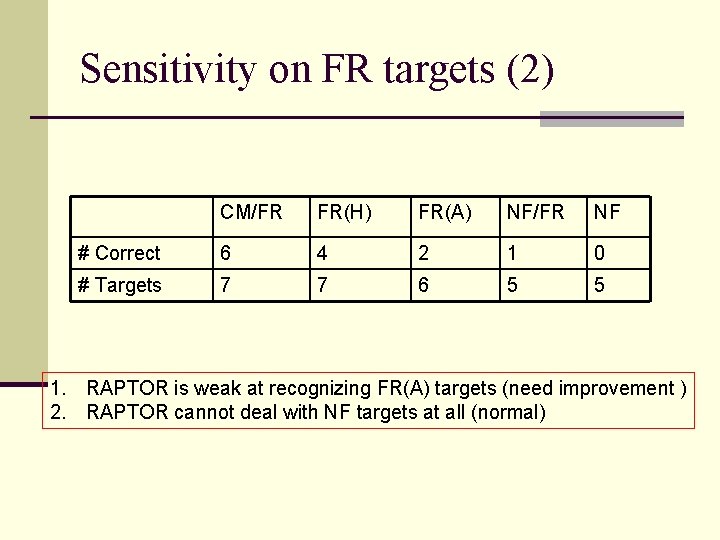
Sensitivity on FR targets (2) CM/FR FR(H) FR(A) NF/FR NF # Correct 6 4 2 1 0 # Targets 7 7 6 5 5 1. RAPTOR is weak at recognizing FR(A) targets (need improvement ) 2. RAPTOR cannot deal with NF targets at all (normal)

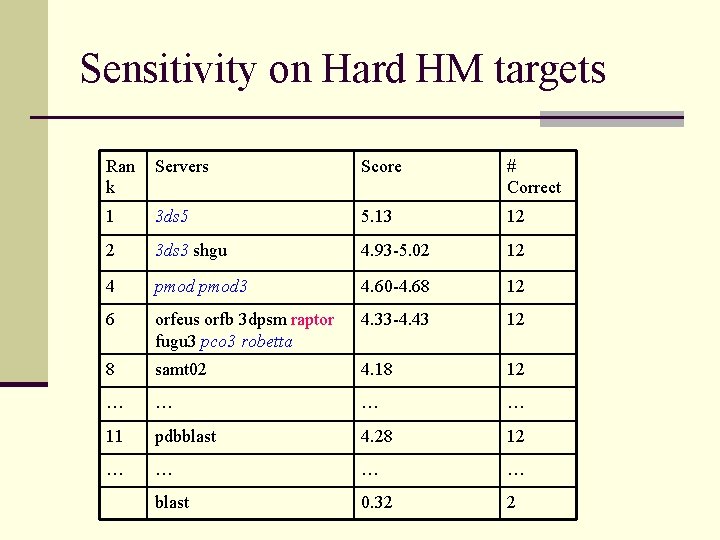
Sensitivity on Hard HM targets Ran k Servers Score # Correct 1 3 ds 5 5. 13 12 2 3 ds 3 shgu 4. 93 -5. 02 12 4 pmod 3 4. 60 -4. 68 12 6 orfeus orfb 3 dpsm raptor fugu 3 pco 3 robetta 4. 33 -4. 43 12 8 samt 02 4. 18 12 … … 11 pdbblast 4. 28 12 … … blast 0. 32 2

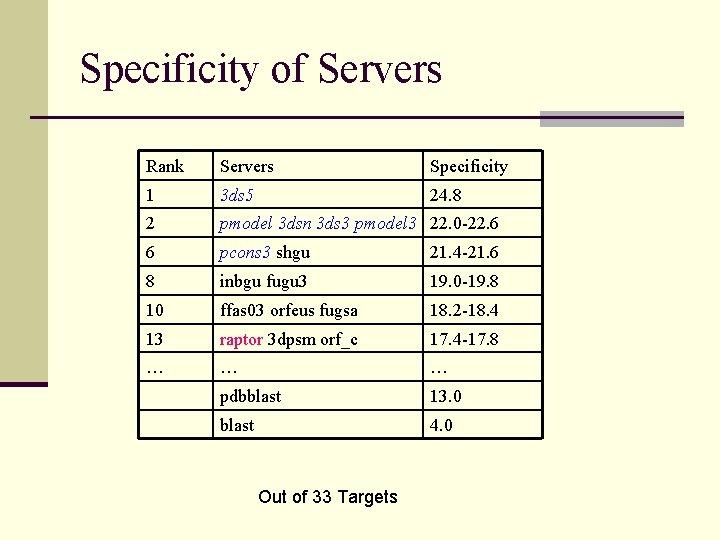
Specificity of Servers Rank Servers Specificity 1 3 ds 5 24. 8 2 pmodel 3 dsn 3 ds 3 pmodel 3 22. 0 -22. 6 6 pcons 3 shgu 21. 4 -21. 6 8 inbgu fugu 3 19. 0 -19. 8 10 ffas 03 orfeus fugsa 18. 2 -18. 4 13 raptor 3 dpsm orf_c 17. 4 -17. 8 … … … pdbblast 13. 0 blast 4. 0 Out of 33 Targets

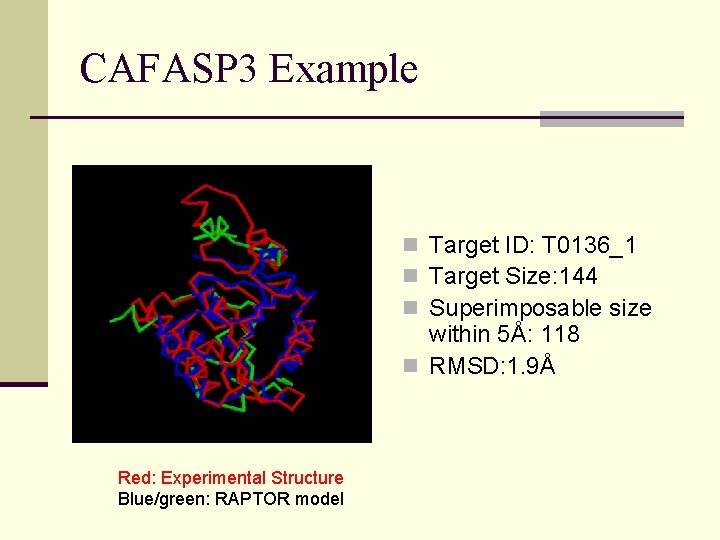
CAFASP 3 Example n Target ID: T 0136_1 n Target Size: 144 n Superimposable size within 5Å: 118 n RMSD: 1. 9Å Red: Experimental Structure Blue/green: RAPTOR model

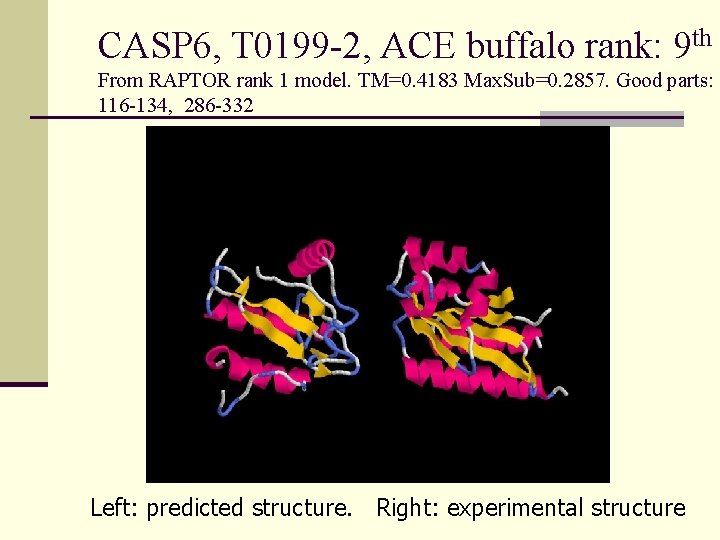
CASP 6, T 0199 -2, ACE buffalo rank: 9 th From RAPTOR rank 1 model. TM=0. 4183 Max. Sub=0. 2857. Good parts: 116 -134, 286 -332 Left: predicted structure. Right: experimental structure

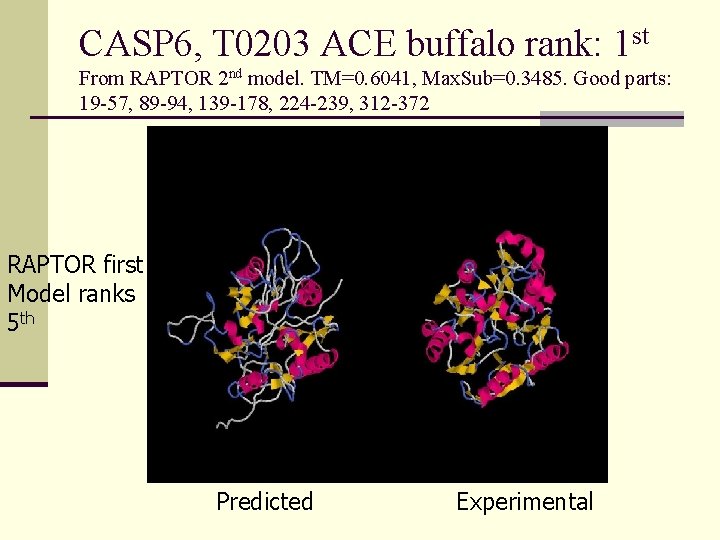
CASP 6, T 0203 ACE buffalo rank: 1 st From RAPTOR 2 nd model. TM=0. 6041, Max. Sub=0. 3485. Good parts: 19 -57, 89 -94, 139 -178, 224 -239, 312 -372 RAPTOR first Model ranks 5 th Predicted Experimental

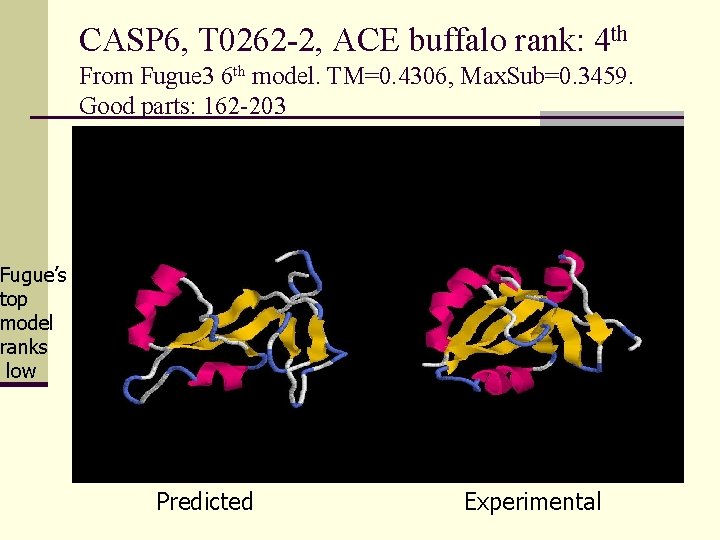
CASP 6, T 0262 -2, ACE buffalo rank: 4 th From Fugue 3 6 th model. TM=0. 4306, Max. Sub=0. 3459. Good parts: 162 -203 Fugue’s top model ranks low Predicted Experimental

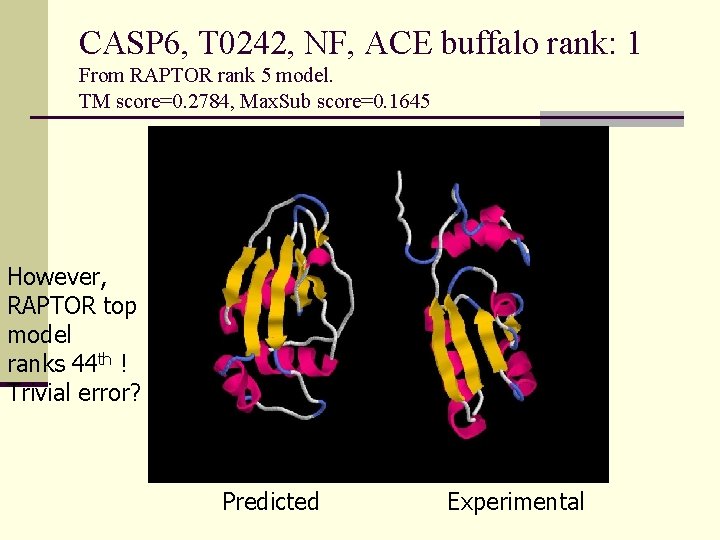
CASP 6, T 0242, NF, ACE buffalo rank: 1 From RAPTOR rank 5 model. TM score=0. 2784, Max. Sub score=0. 1645 However, RAPTOR top model ranks 44 th ! Trivial error? Predicted Experimental

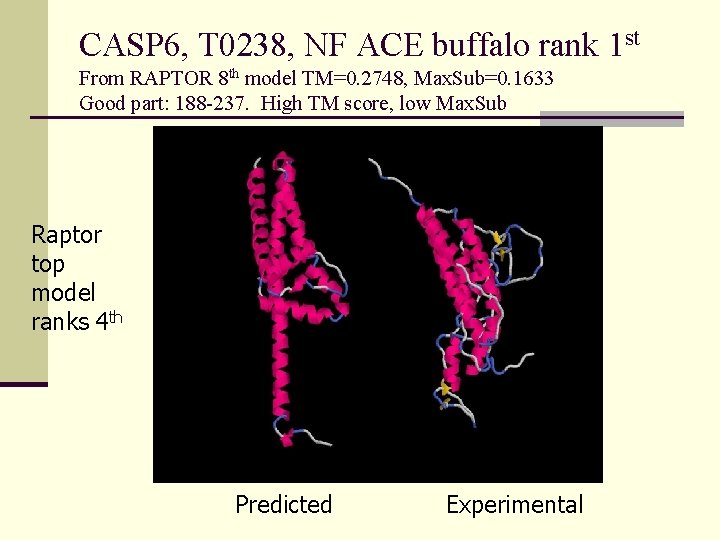
CASP 6, T 0238, NF ACE buffalo rank 1 st From RAPTOR 8 th model TM=0. 2748, Max. Sub=0. 1633 Good part: 188 -237. High TM score, low Max. Sub Raptor top model ranks 4 th Predicted Experimental

About RAPTOR n Jinbo Xu’s Ph. D. thesis work. n The RAPTOR system has benefited significantly from PROSPECT (Ying Xu, Dong Xu, et al). n Currently distributed by BSI. n References: J. Xu, M. Li, D. Kim, Y. Xu, Journal of Bioinformatics and Computational Biology, 1: 1(2003), 95 -118. J. Xu, M. Li, PROTEINS: Structure, Function, and Genetics, CASP 5 special issue.