Clustering analysis of microarray gene expression data Ping








![Transition Profiles indicator[n] = (A[n-1] – A[n]) / (A[n] – A[n+1]) A[k] is the Transition Profiles indicator[n] = (A[n-1] – A[n]) / (A[n] – A[n+1]) A[k] is the](https://slidetodoc.com/presentation_image_h/0cc2498af9462e029963a4b8148d10f3/image-42.jpg)
![Reference l [1] Ying Xu, Victor Olman, and Dong Xu. Clustering Gene Expression Data Reference l [1] Ying Xu, Victor Olman, and Dong Xu. Clustering Gene Expression Data](https://slidetodoc.com/presentation_image_h/0cc2498af9462e029963a4b8148d10f3/image-43.jpg)

- Slides: 44

Clustering analysis of microarray gene expression data Ping Zhang November 19 th, 2008

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

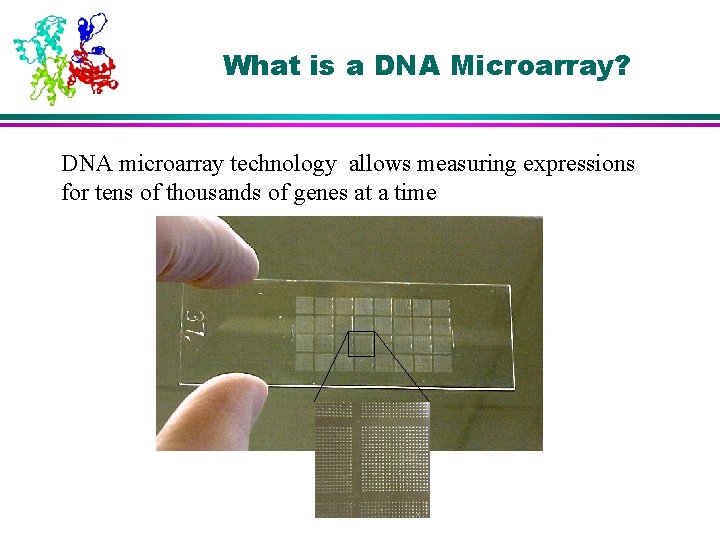
What is a DNA Microarray? DNA microarray technology allows measuring expressions for tens of thousands of genes at a time

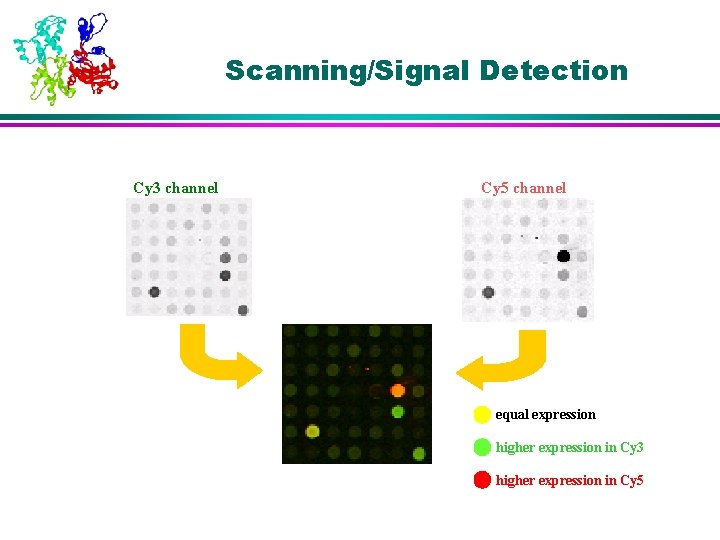
Scanning/Signal Detection Cy 3 channel Cy 5 channel equal expression higher expression in Cy 3 higher expression in Cy 5

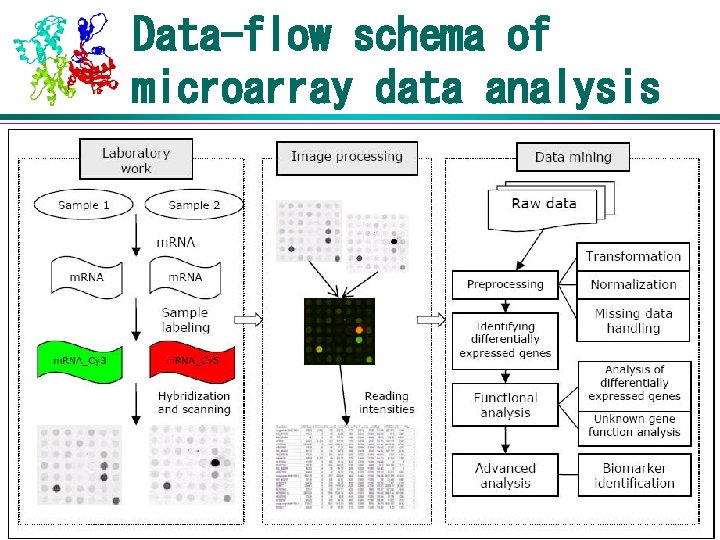
Data-flow schema of microarray data analysis

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

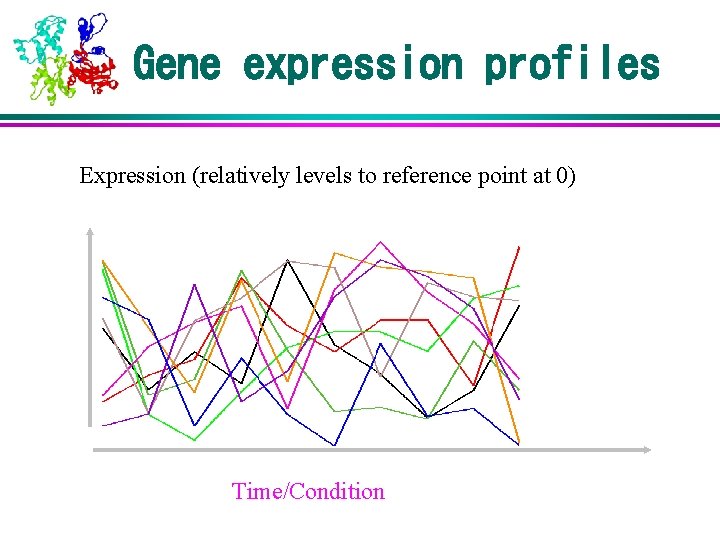
Gene expression profiles Expression (relatively levels to reference point at 0) Time/Condition

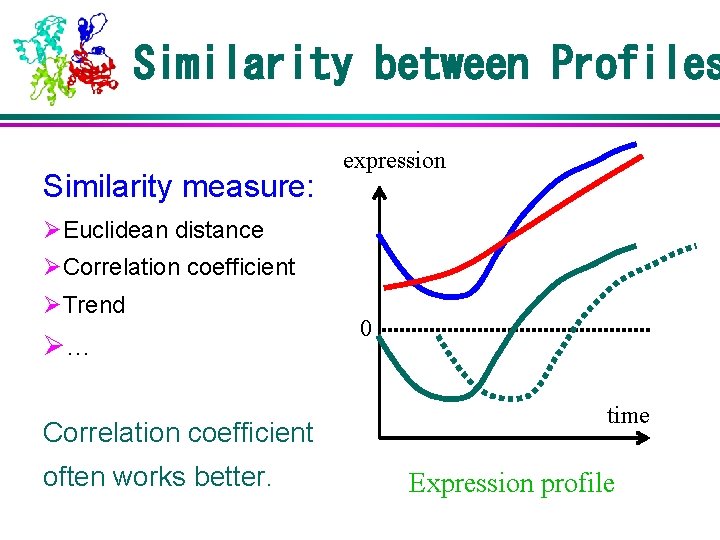
Similarity between Profiles Similarity measure: expression ØEuclidean distance ØCorrelation coefficient ØTrend Ø… Correlation coefficient often works better. 0 time Expression profile

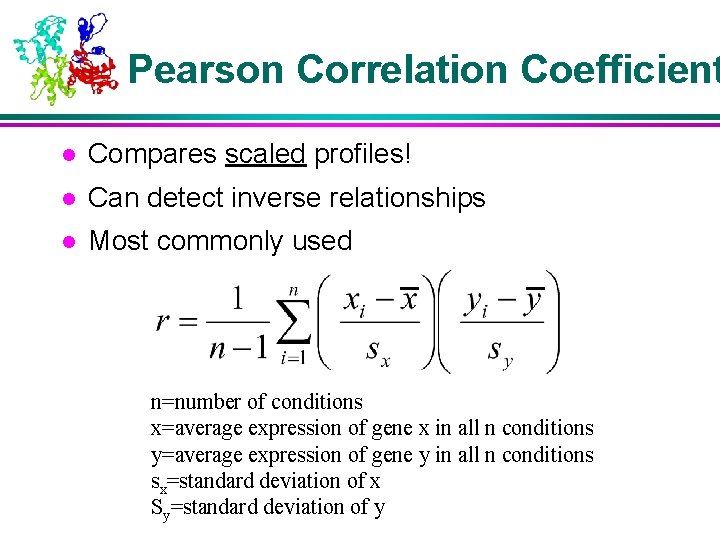
Pearson Correlation Coefficient l Compares scaled profiles! l Can detect inverse relationships l Most commonly used n=number of conditions x=average expression of gene x in all n conditions y=average expression of gene y in all n conditions sx=standard deviation of x Sy=standard deviation of y

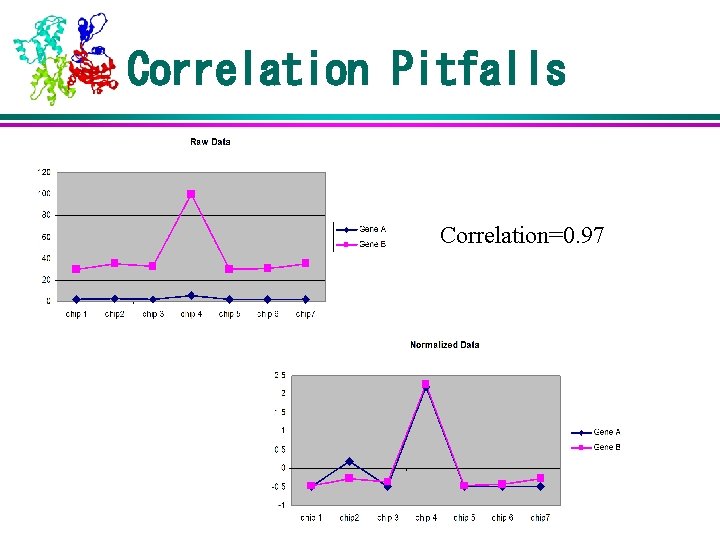
Correlation Pitfalls Correlation=0. 97

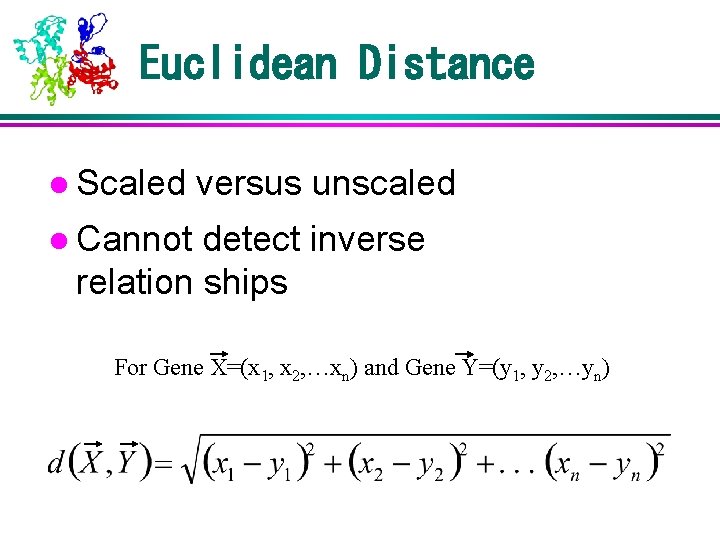
Euclidean Distance l Scaled versus unscaled l Cannot detect inverse relation ships For Gene X=(x 1, x 2, …xn) and Gene Y=(y 1, y 2, …yn)

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

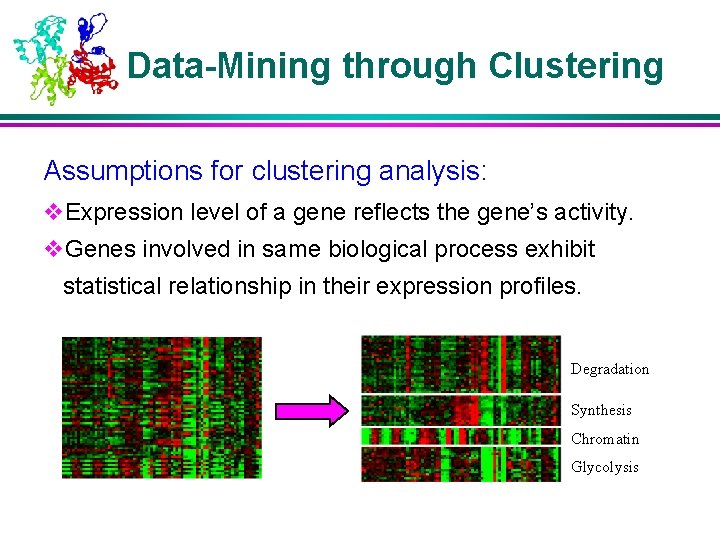
Data-Mining through Clustering Assumptions for clustering analysis: v. Expression level of a gene reflects the gene’s activity. v. Genes involved in same biological process exhibit statistical relationship in their expression profiles. Degradation Synthesis Chromatin Glycolysis

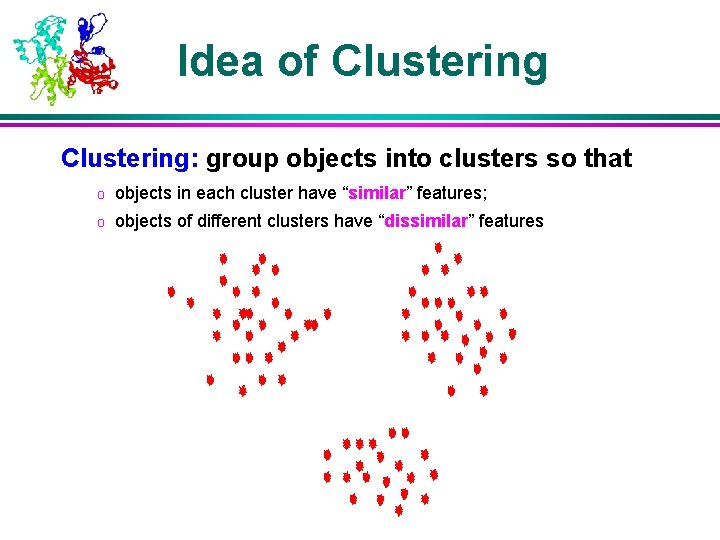
Idea of Clustering: group objects into clusters so that o objects in each cluster have “similar” features; o objects of different clusters have “dissimilar” features

Methods of Clustering • discriminant analysis (Fisher, 1931) • K-means (Lloyd, 1948) • hierarchical clustering • self-organizing maps (Kohonen, 1980) • support vector machines (Vapnik, 1985)

Issues in Cluster Analysis l. A lot of clustering algorithms l A lot of distance/similarity metrics l Which clustering algorithm runs faster and uses less memory? l How many clusters after all? l Are the clusters stable? l Are the clusters meaningful?

Which Clustering Method Should I Use? l What is the biological question? l Do I have a preconceived notion of how many clusters there should be? l How strict do I want to be? Spilt or Join? l Can a gene be in multiple clusters? l Hard or soft boundaries between clusters

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

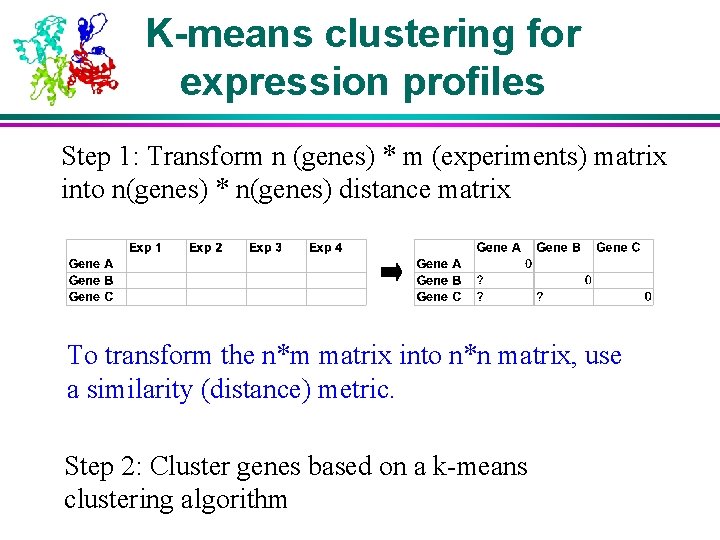
K-means clustering for expression profiles Step 1: Transform n (genes) * m (experiments) matrix into n(genes) * n(genes) distance matrix To transform the n*m matrix into n*n matrix, use a similarity (distance) metric. Step 2: Cluster genes based on a k-means clustering algorithm

K-means algorithm The most popular algorithm for clustering What is so attractive? • Simple • Fast • Mathematically correct • Invariant to dimension • Easy to implement

K-Means Clustering l Basic Ideas : using cluster centre (means) to represent cluster l Assigning data elements to the closet cluster (centre). l Goal: Minimize square error (intra-class dissimilarity) : 2 = l There is no hierarchy. l Must supply the number of clusters (k) into which the data are to be grouped.

K-means Clustering : Procedure (1) Initialization 1 Specify the number of cluster k -- for example, k = 4 Expression matrix conditions gene Each point is called “gene”

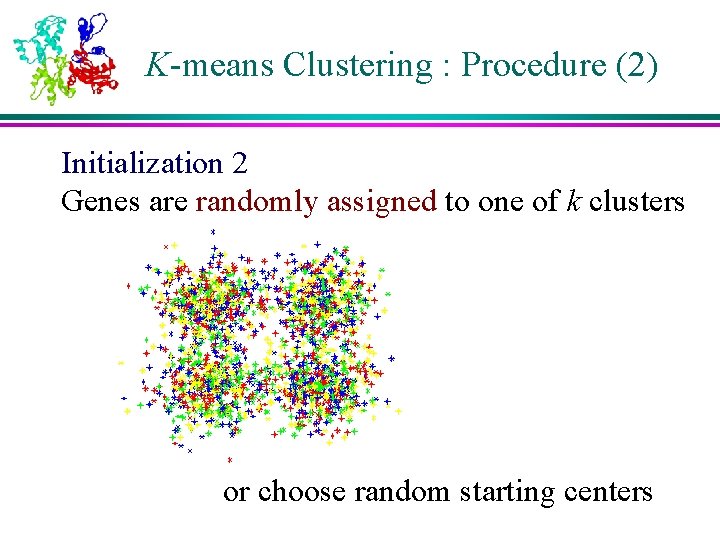
K-means Clustering : Procedure (2) Initialization 2 Genes are randomly assigned to one of k clusters or choose random starting centers

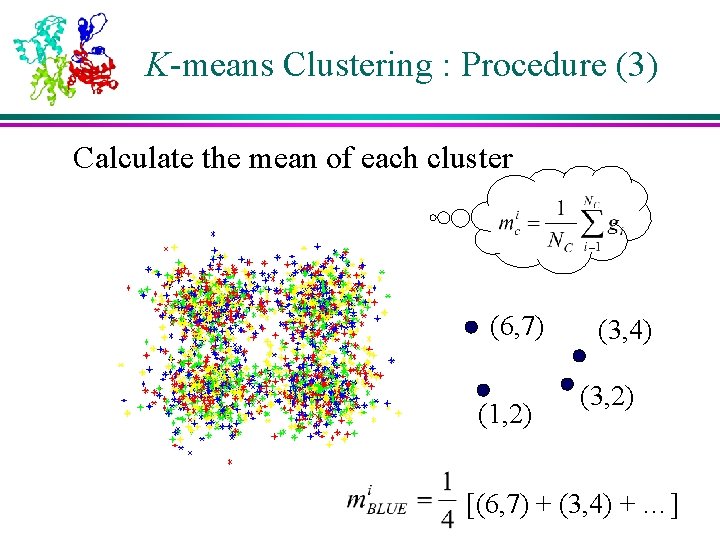
K-means Clustering : Procedure (3) Calculate the mean of each cluster (6, 7) (1, 2) (3, 4) (3, 2) [(6, 7) + (3, 4) + …]

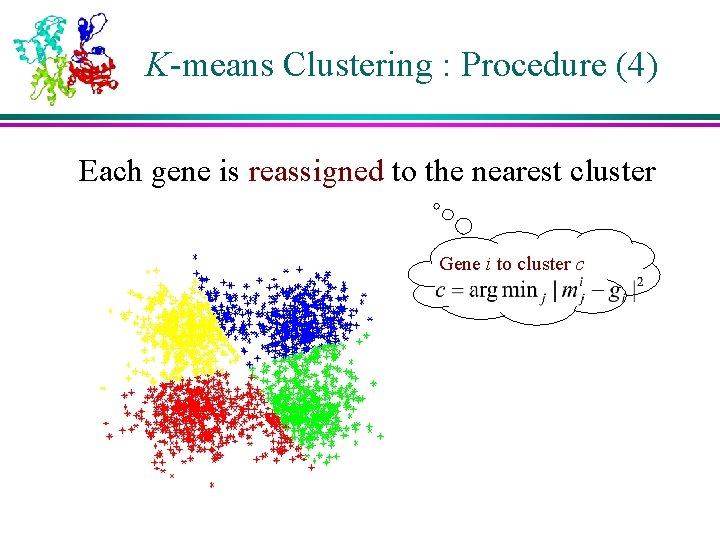
K-means Clustering : Procedure (4) Each gene is reassigned to the nearest cluster Gene i to cluster c

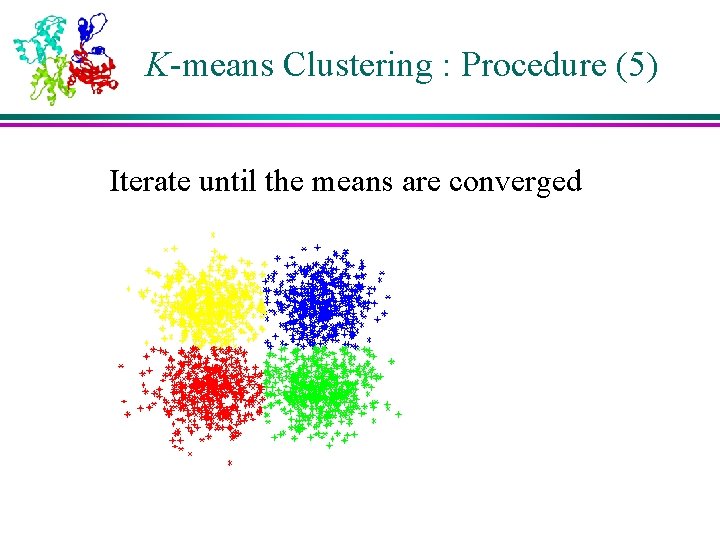
K-means Clustering : Procedure (5) Iterate until the means are converged

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

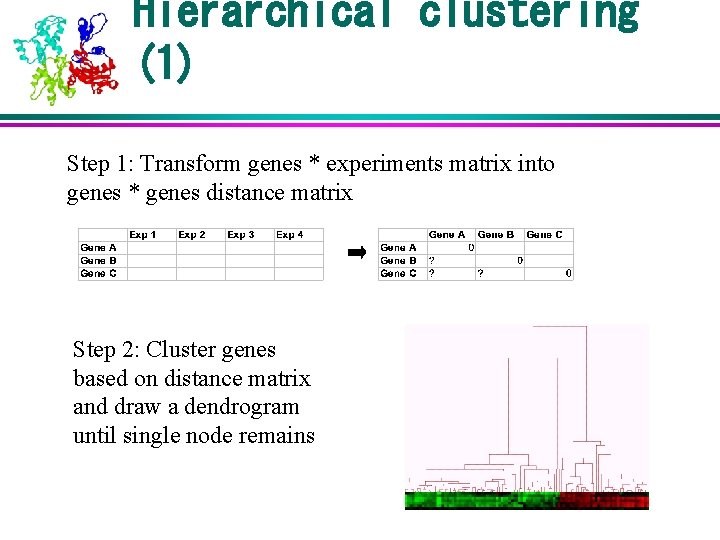
Hierarchical clustering (1) Step 1: Transform genes * experiments matrix into genes * genes distance matrix Step 2: Cluster genes based on distance matrix and draw a dendrogram until single node remains

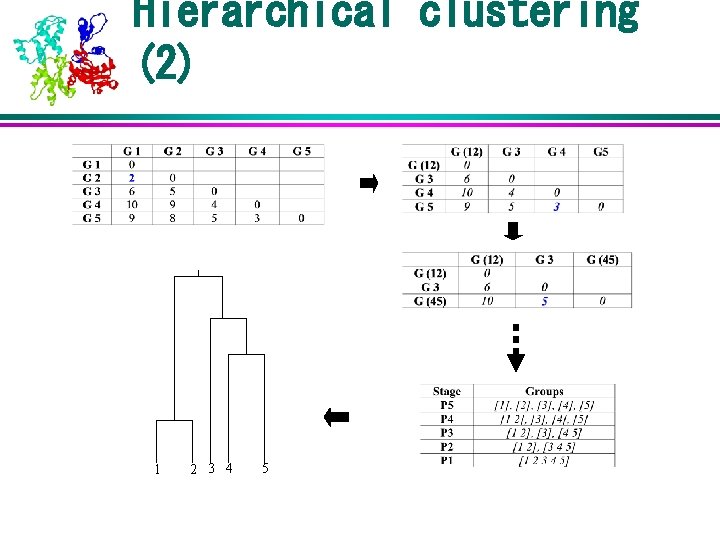
Hierarchical clustering (2) 1 2 3 4 5

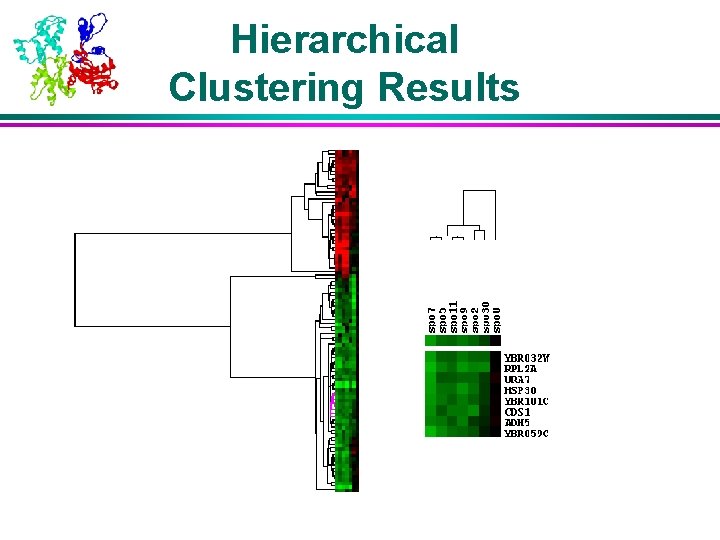
Hierarchical Clustering Results

Outline l Gene expression l Similarity between gene expression profiles l Concept of clustering l K-Means clustering l Hierarchical clustering l Minimum spanning tree-based clustering

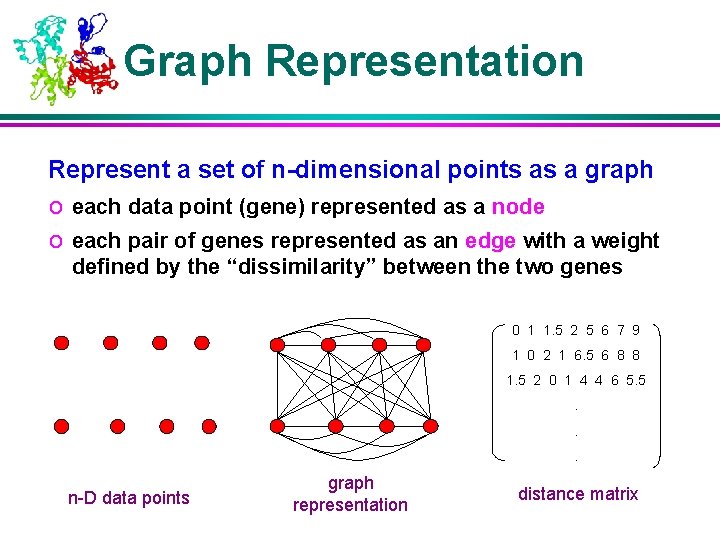
Graph Representation Represent a set of n-dimensional points as a graph o each data point (gene) represented as a node o each pair of genes represented as an edge with a weight defined by the “dissimilarity” between the two genes 0 1 1. 5 2 5 6 7 9 1 0 2 1 6. 5 6 8 8 1. 5 2 0 1 4 4 6 5. 5. . . n-D data points graph representation distance matrix

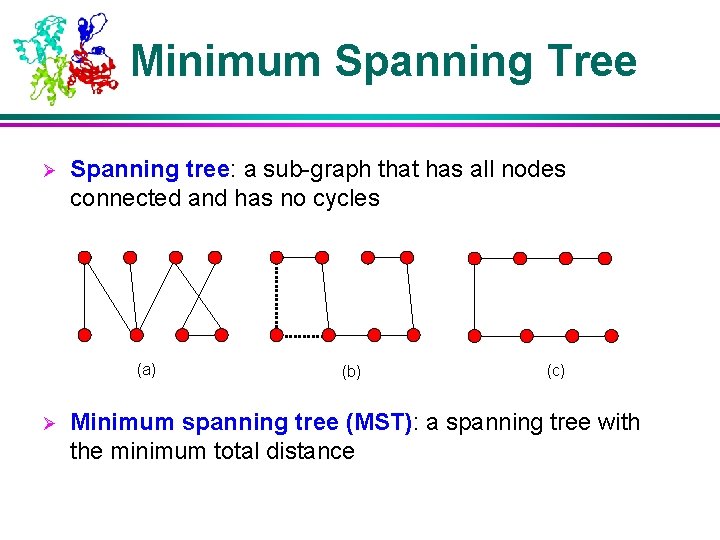
Minimum Spanning Tree Ø Spanning tree: a sub-graph that has all nodes connected and has no cycles (a) Ø (b) (c) Minimum spanning tree (MST): a spanning tree with the minimum total distance

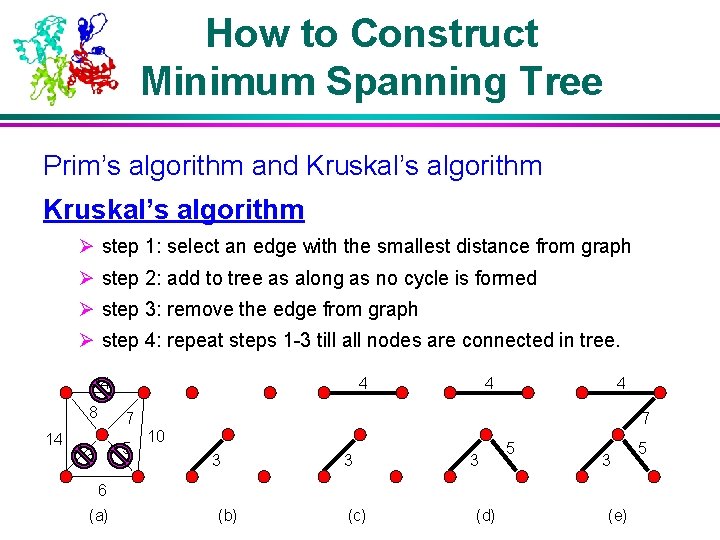
How to Construct Minimum Spanning Tree Prim’s algorithm and Kruskal’s algorithm Ø step 1: select an edge with the smallest distance from graph Ø step 2: add to tree as along as no cycle is formed Ø step 3: remove the edge from graph Ø step 4: repeat steps 1 -3 till all nodes are connected in tree. 4 8 4 4 4 7 14 5 3 7 10 3 3 3 5 3 6 (a) (b) (c) (d) (e) 5

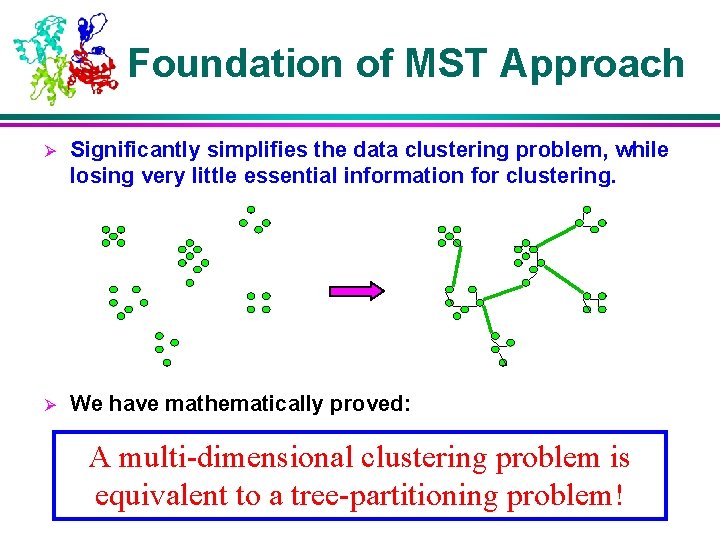
Foundation of MST Approach Ø Significantly simplifies the data clustering problem, while losing very little essential information for clustering. Ø We have mathematically proved: A multi-dimensional clustering problem is equivalent to a tree-partitioning problem!

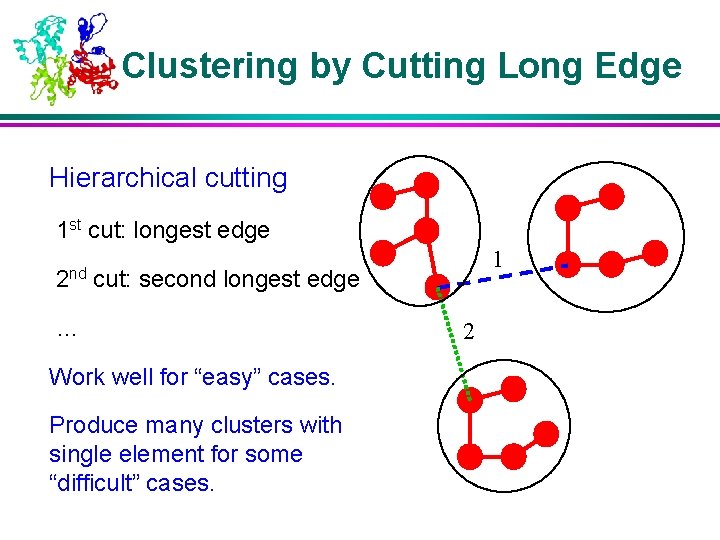
Clustering by Cutting Long Edge Hierarchical cutting 1 st cut: longest edge 2 nd 1 cut: second longest edge … Work well for “easy” cases. Produce many clusters with single element for some “difficult” cases. 2

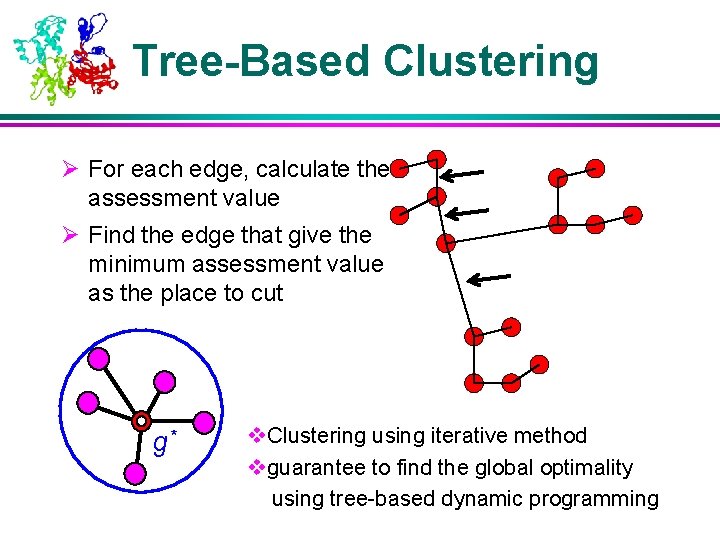
Tree-Based Clustering Ø For each edge, calculate the assessment value Ø Find the edge that give the minimum assessment value as the place to cut g* v. Clustering using iterative method vguarantee to find the global optimality using tree-based dynamic programming

Clustering through Removing Long MST-Edges l Objective: partition an MST into K subtrees so that the total edge-distance of all the K subtrees in minimized l Finding K-1 longest MST-edges and cutting them => we get K clusters l This works as long as the inter-cluster edgedistances are clearly larger than the intra-cluster edge-distances

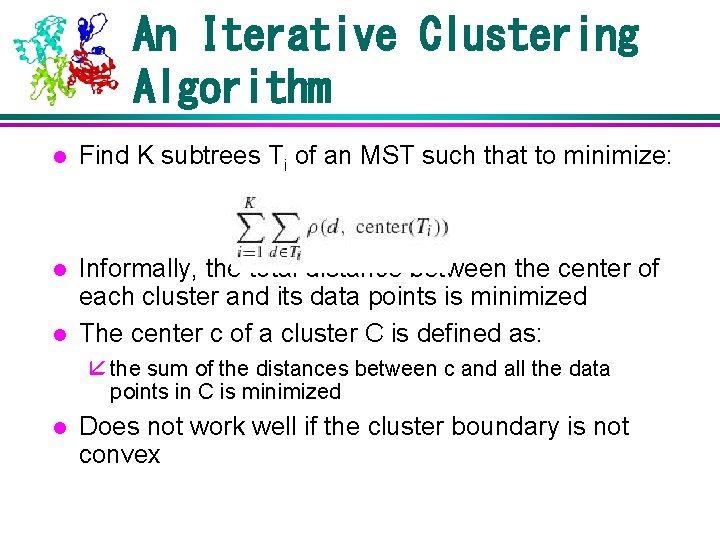
An Iterative Clustering Algorithm l Find K subtrees Ti of an MST such that to minimize: l Informally, the total distance between the center of each cluster and its data points is minimized The center c of a cluster C is defined as: å the sum of the distances between c and all the data l points in C is minimized l Does not work well if the cluster boundary is not convex

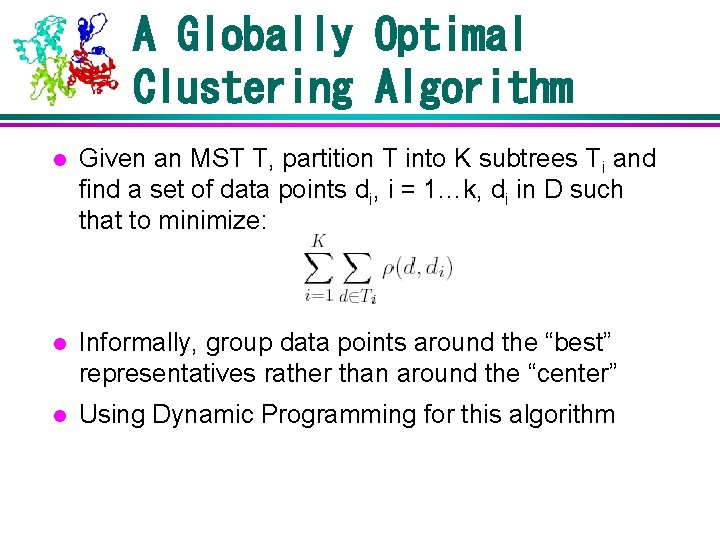
A Globally Optimal Clustering Algorithm l Given an MST T, partition T into K subtrees Ti and find a set of data points di, i = 1…k, di in D such that to minimize: l Informally, group data points around the “best” representatives rather than around the “center” l Using Dynamic Programming for this algorithm

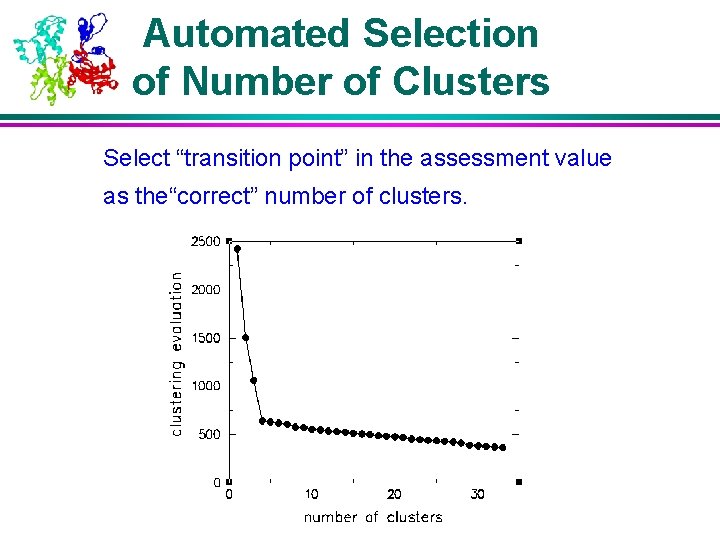
Automated Selection of Number of Clusters Select “transition point” in the assessment value as the“correct” number of clusters.
![Transition Profiles indicatorn An1 An An An1 Ak is the Transition Profiles indicator[n] = (A[n-1] – A[n]) / (A[n] – A[n+1]) A[k] is the](https://slidetodoc.com/presentation_image_h/0cc2498af9462e029963a4b8148d10f3/image-42.jpg)
Transition Profiles indicator[n] = (A[n-1] – A[n]) / (A[n] – A[n+1]) A[k] is the assessment value for partition with k clusters Our clustering of yeast data
![Reference l 1 Ying Xu Victor Olman and Dong Xu Clustering Gene Expression Data Reference l [1] Ying Xu, Victor Olman, and Dong Xu. Clustering Gene Expression Data](https://slidetodoc.com/presentation_image_h/0cc2498af9462e029963a4b8148d10f3/image-43.jpg)
Reference l [1] Ying Xu, Victor Olman, and Dong Xu. Clustering Gene Expression Data Using a Graph-Theoretic Approach: An Application of Minimum Spanning Trees. Bioinformatics. 18: 526 -535, 2002. l [2] Dong Xu, Victor Olman, Li Wang, and Ying Xu. EXCAVATOR: a computer program for gene expression data analysis. Nucleic Acid Research. 31: 5582 -5589. 2003. l Using slides from: Michael Hongbo Xie, Temple University (in 2006) Vipin Kumar, University of Minnesota Dong Xu, University of Missouri

Acknowledgement