Other genomic arrays Methylation ch IP on chip







- Slides: 21

Other genomic arrays: Methylation, ch. IP on chip… UBio Training Courses

SNP-arrays and copy number Genotyping arrays can detect CNVs

Copy numbers from SNP arrays

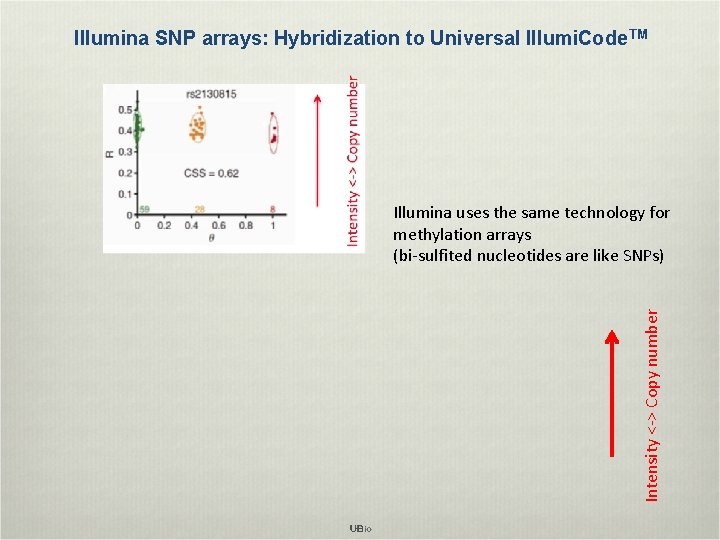
Illumina SNP arrays: Hybridization to Universal Illumi. Code TM Intensity <-> Copy number Illumina uses the same technology for methylation arrays (bi-sulfited nucleotides are like SNPs)

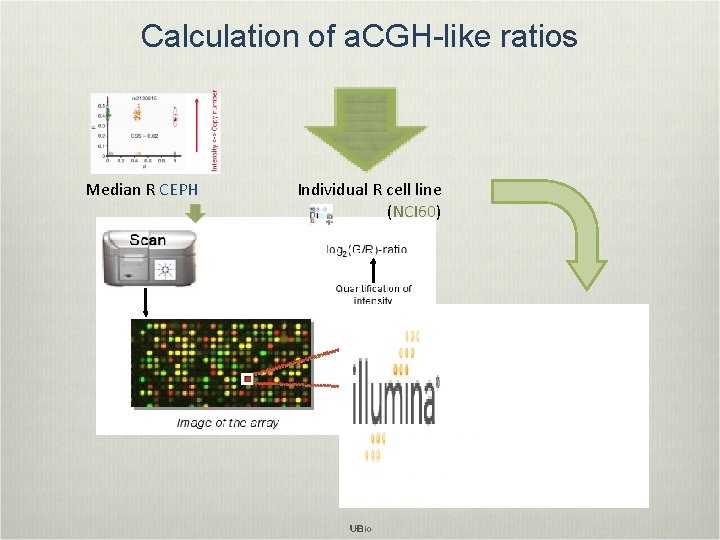
Calculation of a. CGH-like ratios Median R CEPH Individual R cell line (NCI 60)

Methylation arrays

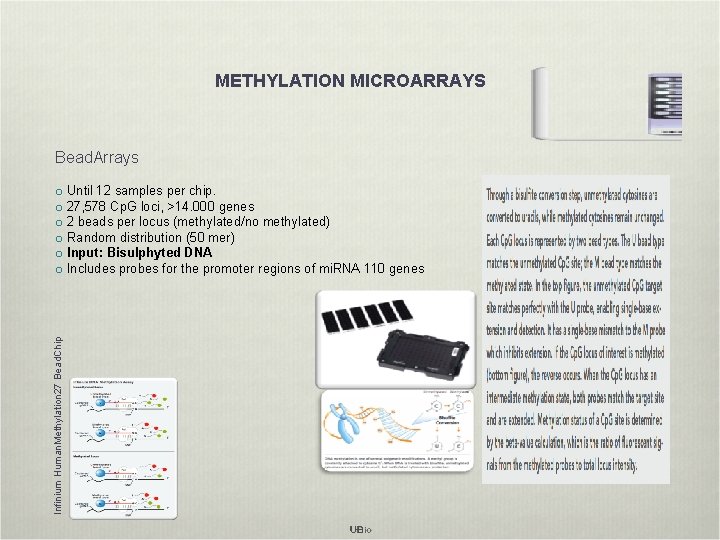
METHYLATION MICROARRAYS Bead. Arrays Infinium Human. Methylation 27 Bead. Chip o Until 12 samples per chip. o 27, 578 Cp. G loci, >14. 000 genes o 2 beads per locus (methylated/no methylated) o Random distribution (50 mer) o Input: Bisulphyted DNA o Includes probes for the promoter regions of mi. RNA 110 genes

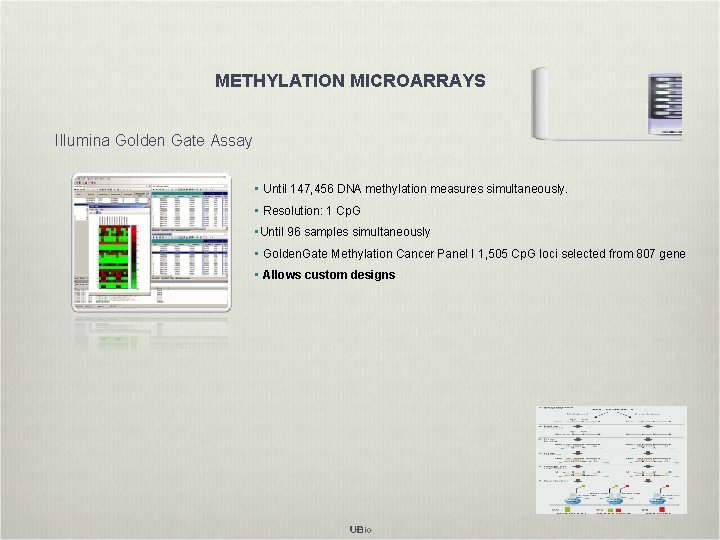
METHYLATION MICROARRAYS Illumina Golden Gate Assay • Until 147, 456 DNA methylation measures simultaneously. • Resolution: 1 Cp. G • Until 96 samples simultaneously • Golden. Gate Methylation Cancer Panel I 1, 505 Cp. G loci selected from 807 gene • Allows custom designs

METHYLATION MICROARRAYS SOFTWARE Bead Studio Genome Studio Methylation module http: //www. illumina. com/pages. ilmn? ID=196 Lumi package (Import, background correction, normalization) Beadarray package (Import, QC) Methylumi (Import, QC , normalization, differential meth. )

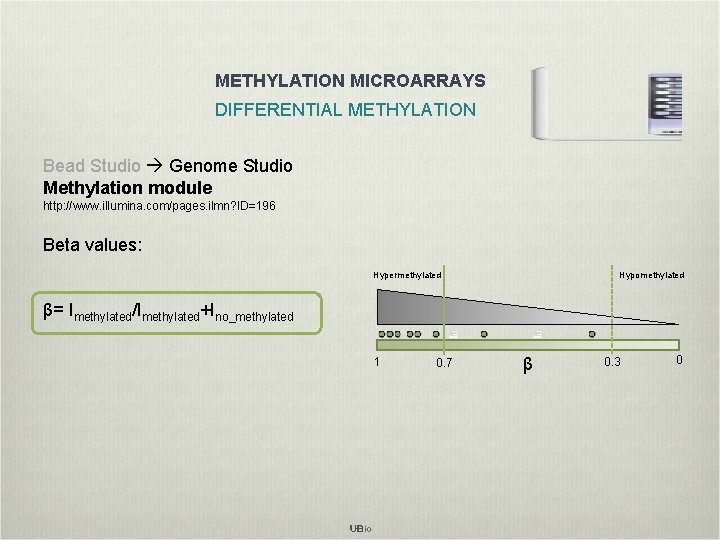
METHYLATION MICROARRAYS DIFFERENTIAL METHYLATION Bead Studio Genome Studio Methylation module http: //www. illumina. com/pages. ilmn? ID=196 Beta values: Hypermethylated Hypomethylated β= Imethylated/Imethylated+Ino_methylated 1 0. 7 β 0. 3 0

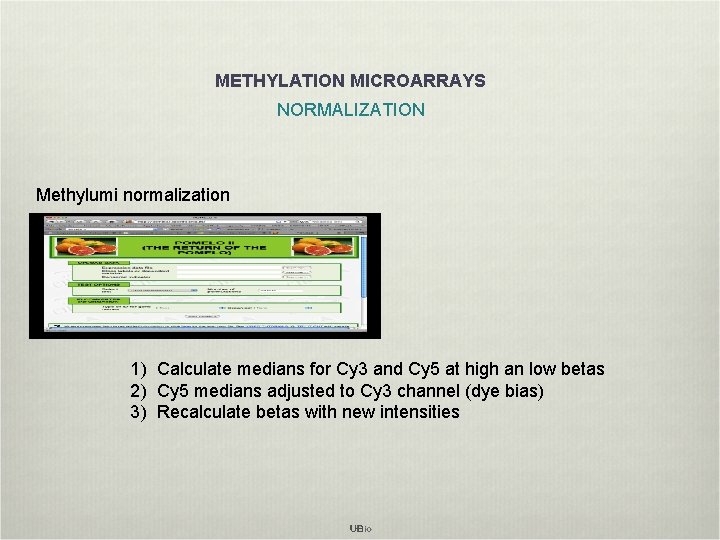
METHYLATION MICROARRAYS NORMALIZATION Methylumi normalization 1) Calculate medians for Cy 3 and Cy 5 at high an low betas 2) Cy 5 medians adjusted to Cy 3 channel (dye bias) 3) Recalculate betas with new intensities

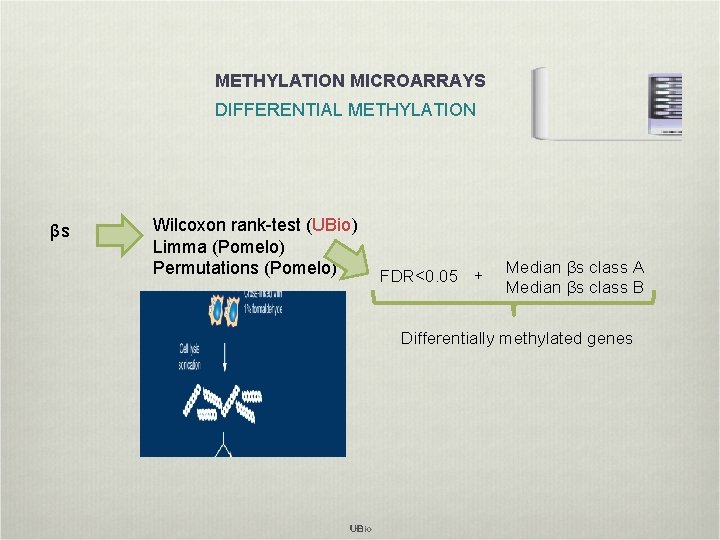
METHYLATION MICROARRAYS DIFFERENTIAL METHYLATION βs Wilcoxon rank-test (UBio) Limma (Pomelo) Permutations (Pomelo) FDR<0. 05 + Median βs class A Median βs class B Differentially methylated genes

Ch. IP on chip

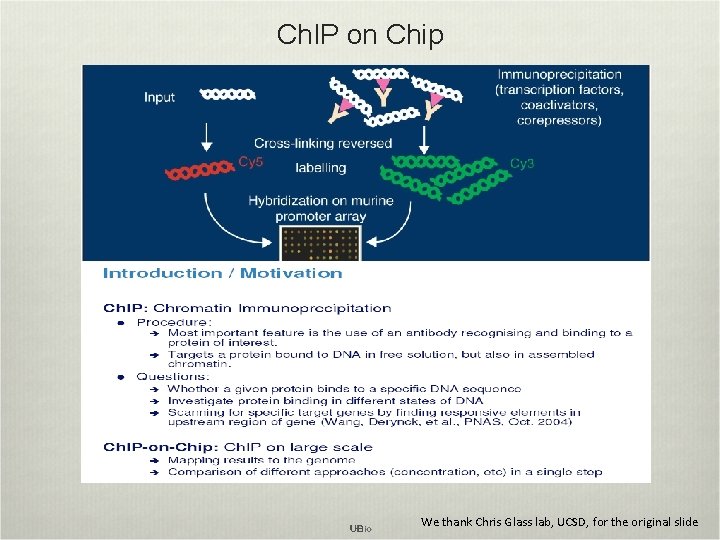
Ch. IP on Chip We thank Chris Glass lab, UCSD, for the original slide

Ch. IP on Chip Discover protein/DNA interactions!!

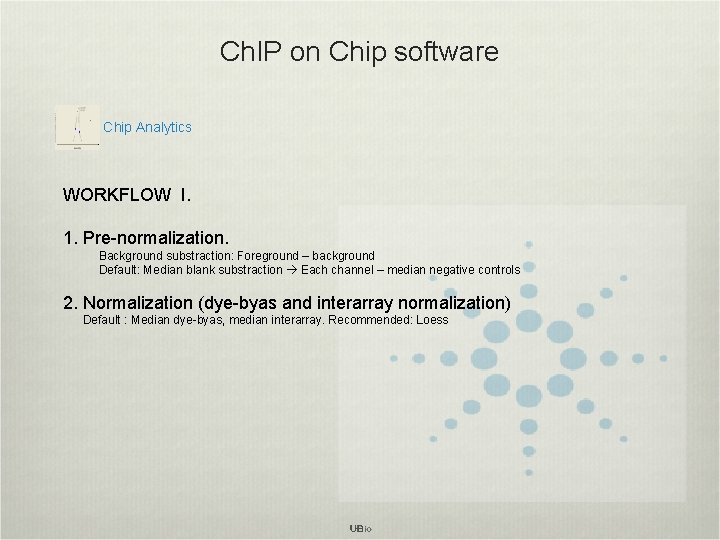
Ch. IP on Chip software Chip Analytics WORKFLOW I. 1. Pre-normalization. Background substraction: Foreground – background Default: Median blank substraction Each channel – median negative controls 2. Normalization (dye-byas and interarray normalization) Default : Median dye-byas, median interarray. Recommended: Loess

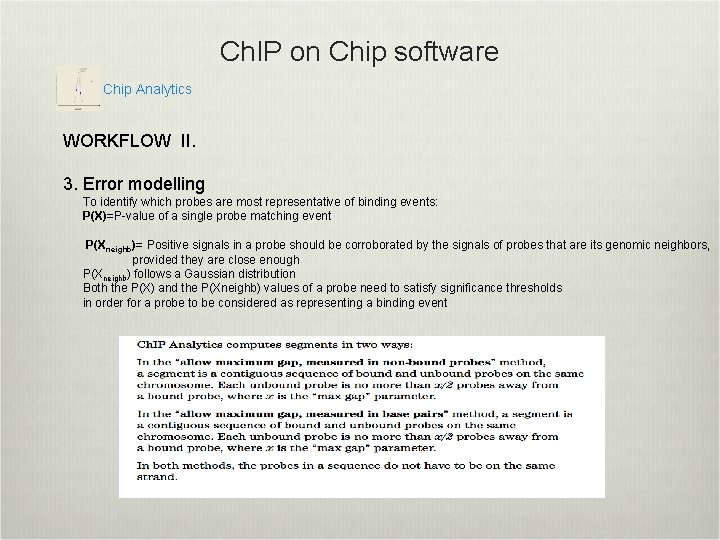
Ch. IP on Chip software Chip Analytics WORKFLOW II. 3. Error modelling To identify which probes are most representative of binding events: P(X)=P-value of a single probe matching event P(X neighb)= Positive signals in a probe should be corroborated by the signals of probes that are its genomic neighbors, provided they are close enough P(Xneighb) follows a Gaussian distribution Both the P(X) and the P(Xneighb) values of a probe need to satisfy significance thresholds in order for a probe to be considered as representing a binding event

Ch. IP on Chip software Chip Analytics WORKFLOW III. 4. Segment identification (clusters of enriched probes) bp 5. Gene identification -Segment, Gene or Probe report (Gene or probe ID, Chr, Start, End, p(X)…)

Co. Cas http: //www. ciml. univ-mrs. fr/software/cocas/index. html Agilent platform Normalization QC Report Genome Visualization Peak Finder Benoukraf et al. Bioinformatics 2009.

Weeder: Motif discovery in sequences from co-regulated genes (single specie). Weeder. H: Motif discovery in sequences from homologous genes. Pscan: Motif discovery in sequences from co-regulated genes (JASPAR, TRANSFAC matrices) UBio training courses: See “Course on Introduction to Sequence Analysis”

Thanks ! Visit UBio web ! http: //bioinfo. cnio. es/
 Methylation & chip-on-chip microarray platform
Methylation & chip-on-chip microarray platform Methylation vs acetylation
Methylation vs acetylation Hofmann exhaustive methylation
Hofmann exhaustive methylation Adenine methylation
Adenine methylation Genomic signal processing
Genomic signal processing Genomic equivalence
Genomic equivalence Genomic instability
Genomic instability Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic england
Genomic england Genomic
Genomic Genomic equivalence definition
Genomic equivalence definition Genomic england
Genomic england Genomic imprinting definition
Genomic imprinting definition Genomic england
Genomic england Arrays
Arrays Pascal 2d array
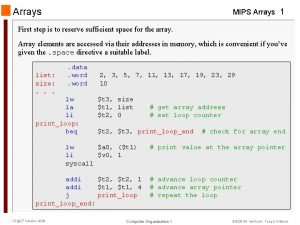
Pascal 2d array Mips array example
Mips array example Ragged array
Ragged array Facts about arrays
Facts about arrays Advantages of dynamic memory allocation
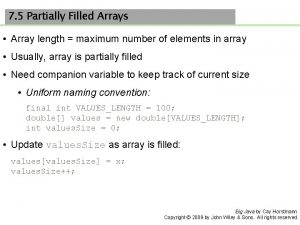
Advantages of dynamic memory allocation Java partially filled array
Java partially filled array Computer science arrays
Computer science arrays