MIAME and Array Express a standard for microarray








- Slides: 36

MIAME and Array. Express – a standard for microarray gene expression data and the public database at EBI Susanna-Assunta Sansone (Toxicogenomics project coordinator) Microarray Informatics Team EMBL- EBI (European Bioinformatics Institute) Transcriptome Symposium, April 2002 CHU Pitié-Salpêtrière, Université Paris VI

Why have a public database? § EMBL- EBI centre for research and services in bioinformatics that makes and maintains public db: • EMBL Nucleotide Sequence, SWISS-PROT, Ensembl, MSD, etc. § Practical reasons: • Easy data access • Resolves local storage issues • Common data exchange formats can be developed § Scientific reasons: • • Curation can be applied Annotation can be controlled Additional info can be stored that is missing in publications Improve data comparison ! § Public standard can be applied

Talk structure § MIAME standard § MIAME annotation challenge: • MGED Bio. Material Ontology § Uses of MIAME concepts: • Array. Express: a public repository for gene expression data • MIAMExpress submission and annotation tool

Talk structure § MIAME standard

Standard for microarray data Why? § § § § Size of dataset Different platforms - nylon, glass Different technologies - oligos, spotted References to external db not stable! Array annotation Sample annotation Data sharing needs standardized way to annotate and record the information!

Standard for microarray data MGED Group § Microarray Gene Expression Data Group: EBI + world’s largest microarray labs and companies (Sanger, Stanford, TIGR, Universite D'Aix-Marseille II, Affymetrics, Agilent, NCBI, DDBJ, etc. ) § MGED Group aims to • Facilitate adoption of standards for: – Experiment annotation – Data representation • Introduce standard for: – Experimental controls – Data normalization methods

General MIAME principles § Minimum information about a microarray experiment § NOT a formal specification BUT a set of guidelines § Sufficient information must be recorded to: • Correctly interpret and verify the results • Replicate the experiments § Structured information must be recorded to: • Query and correctly retrieve the data • Analyse the data § MIAME- Brazma et al. , Nature Genetics, 2001

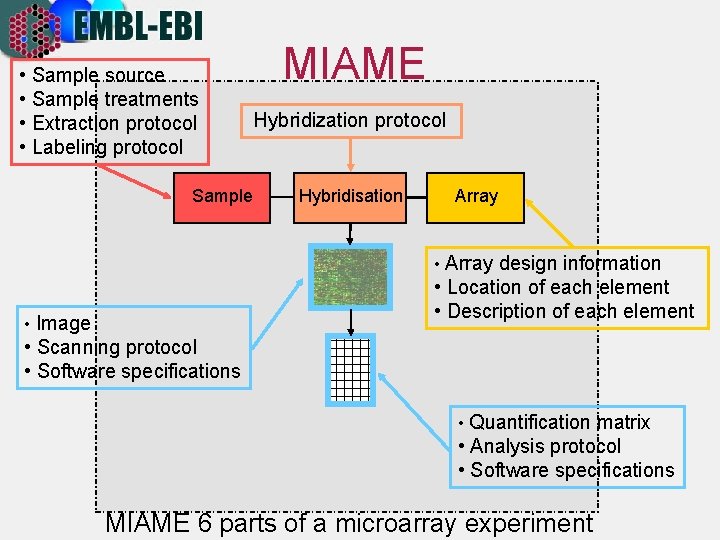
• Sample source • Sample treatments • Extraction protocol • Labeling protocol Sample MIAME Hybridization protocol Hybridisation Array • Array design information • Location of each element • Description of each element • Image • Scanning protocol • Software specifications • Quantification matrix • Analysis protocol • Software specifications MIAME 6 parts of a microarray experiment

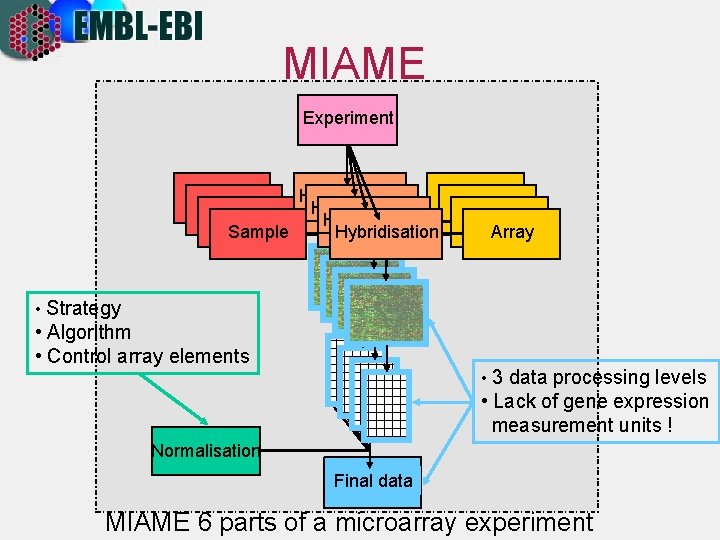
MIAME Experiment Hybridisation Array Sample • Strategy • Algorithm • Control array elements • 3 data processing levels • Lack of gene expression measurement units ! Normalisation Final data MIAME 6 parts of a microarray experiment

MIAME – Annotation challenge § Annotation implementations are required ! • Avoid/reduce free text descriptions • Use of controlled terms • Definitions and sources for each term • Remove of synonyms, or use of synonym mappings • Data curation at source (LIMS) • Integration of controlled terms in query interfaces § Facilitate data queries-analysis…….

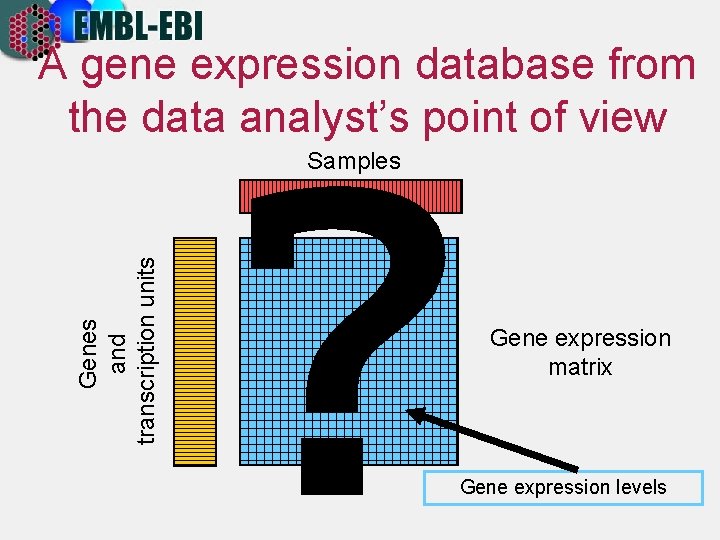
A gene expression database from the data analyst’s point of view Genes and transcription units Samples ? Gene expression matrix Gene expression levels

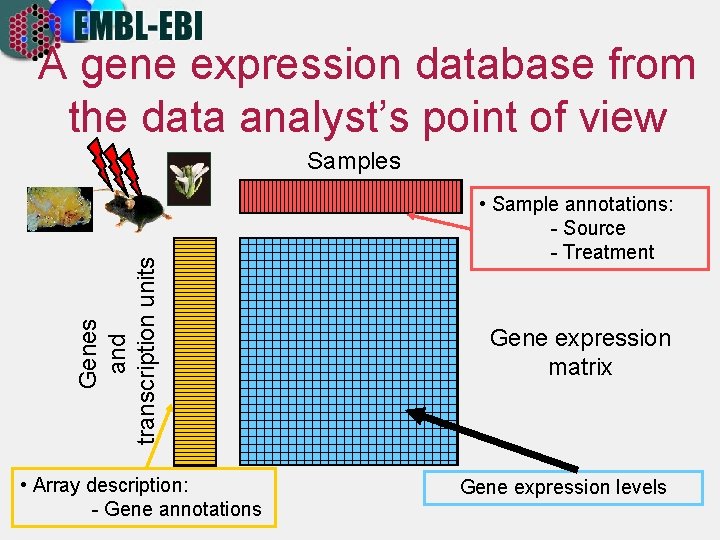
A gene expression database from the data analyst’s point of view Genes and transcription units Samples • Array description: - Gene annotations • Sample annotations: - Source - Treatment Gene expression matrix Gene expression levels

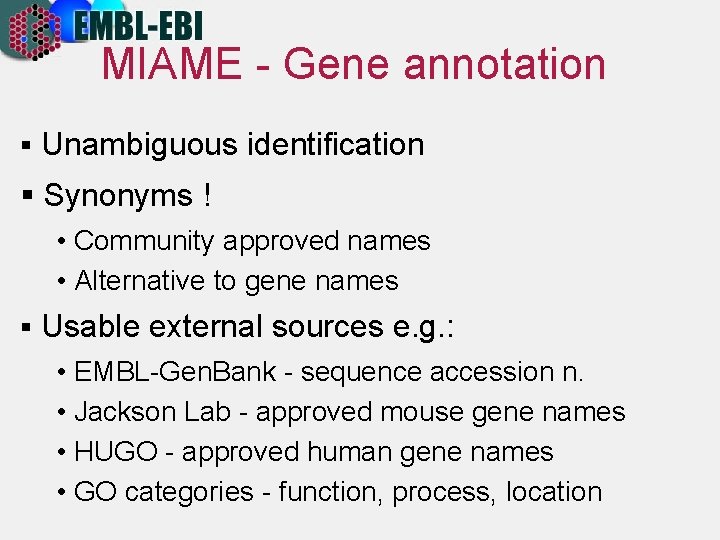
MIAME - Gene annotation § Unambiguous identification § Synonyms ! • Community approved names • Alternative to gene names § Usable external sources e. g. : • EMBL-Gen. Bank - sequence accession n. • Jackson Lab - approved mouse gene names • HUGO - approved human gene names • GO categories - function, process, location

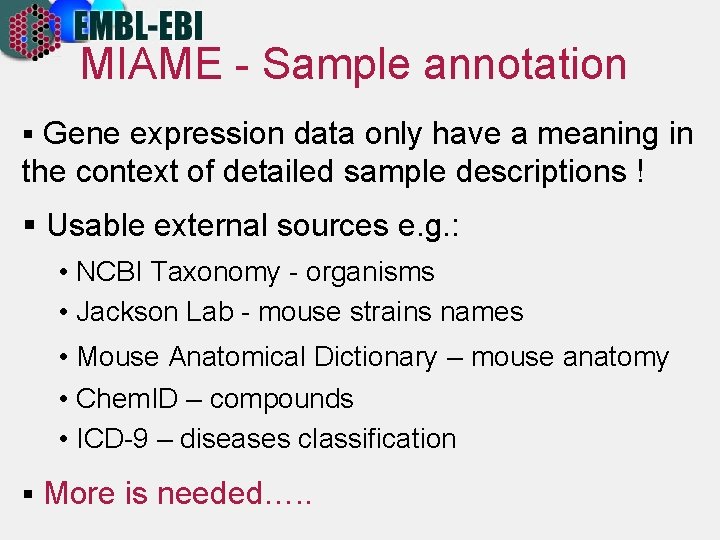
MIAME - Sample annotation § Gene expression data only have a meaning in the context of detailed sample descriptions ! § Usable external sources e. g. : • NCBI Taxonomy - organisms • Jackson Lab - mouse strains names • Mouse Anatomical Dictionary – mouse anatomy • Chem. ID – compounds • ICD-9 – diseases classification § More is needed…. .

Annotation – implementations required! § Need an ontology to describe the sample: • Defining controlled vocabularies and…… • …. Using existing external ontologies § Integrate the ontology in LIMS and databases: • Develop browser or interface for the ontology • Develop internal editing tools for the ontology § However some free text description is unavoidable

Talk structure § MIAME standard § MIAME annotation challenge: • MGED Bio. Material Ontology

What CV and ontology are? § Controlled Vocabulary (CV): • Set of restrictive terms used to describe something, in the simplest case it could be a list § Ontology is more then a CV: • Describes the relationship between the terms in a structured way, provides semantics and constraints • Capture knowledge and make it machine processable

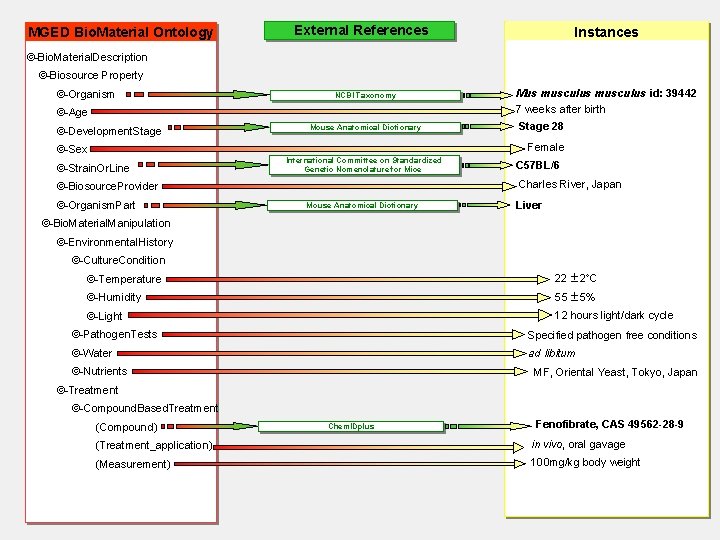
Sample annotation – MGED Bio. Material Ontology § Under construction by Chris Stoeckert (Univ. of Penn. ) and MGED members § Use OILed (rdf, daml and html files available) § Motivated by MIAME and guided by ‘case scenarios’ § Defines terms, provides constraints, develops CVs for sample annotation § Links also to external CVs and ontologies § Will be extended to other part of a microarray experiment that need to be described

Sample annotation – MGED Bio. Material Ontology an example Sample source and treatment description, and its correct annotation using the MGED Bio. Material Ontology classes and correspondent external references: “Seven week old C 57 BL/6 N mice were treated with fenofibrate. Liver was dissected out, RNA prepared………”

MGED Bio. Material Ontology External References Instances ©-Bio. Material. Description ©-Biosource Property ©-Organism NCBI Taxonomy ©-Age ©-Development. Stage Mouse Anatomical Dictionary Mus musculus id: 39442 7 weeks after birth Stage 28 Female ©-Sex ©-Strain. Or. Line International Committee on Standardized Genetic Nomenclature for Mice Charles River, Japan ©-Biosource. Provider ©-Organism. Part C 57 BL/6 Mouse Anatomical Dictionary Liver ©-Bio. Material. Manipulation ©-Environmental. History ©-Culture. Condition ©-Temperature 22 2 C ©-Humidity 55 5% ©-Light 12 hours light/dark cycle ©-Pathogen. Tests Specified pathogen free conditions ©-Water ad libitum ©-Nutrients MF, Oriental Yeast, Tokyo, Japan ©-Treatment ©-Compound. Based. Treatment (Compound) Chem. IDplus Fenofibrate, CAS 49562 -28 -9 (Treatment_application) in vivo, oral gavage (Measurement) 100 mg/kg body weight

Talk structure § MIAME standard § Sample annotation: • MGED Bio. Material Ontology § Uses of MIAME concepts: • Array. Express a public repository for gene expression data • MIAMEpress submission and annotation tool

Uses of MIAME concepts § Specifies the content of the information: • Sufficient • Structured § Uses: • Creation of MIAME-compliant LIMS or databases e. g: Array. Express • Development of submission/annotation tool for generating MIAME-compliant information e. g. : MIAMExpress

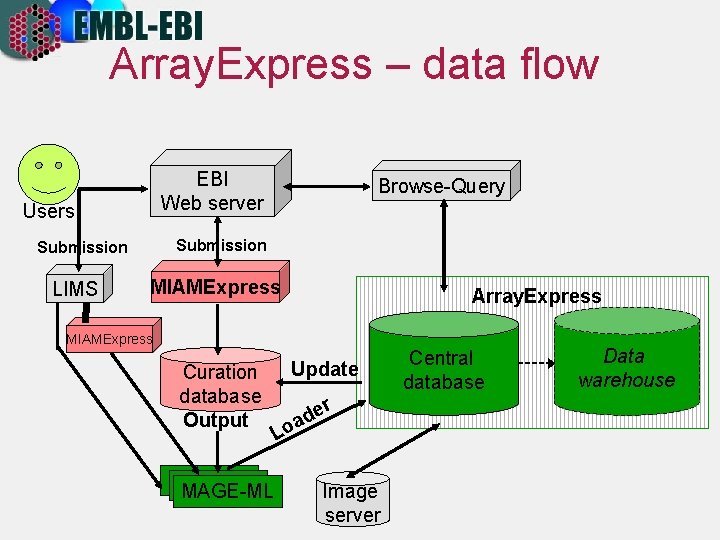
Array. Express – data flow EBI Web server Users Submission LIMS Browse-Query MIAMExpress Array. Express MIAMExpress Curation database Output MAGE-ML Update r Lo e ad Image server Central database Data warehouse

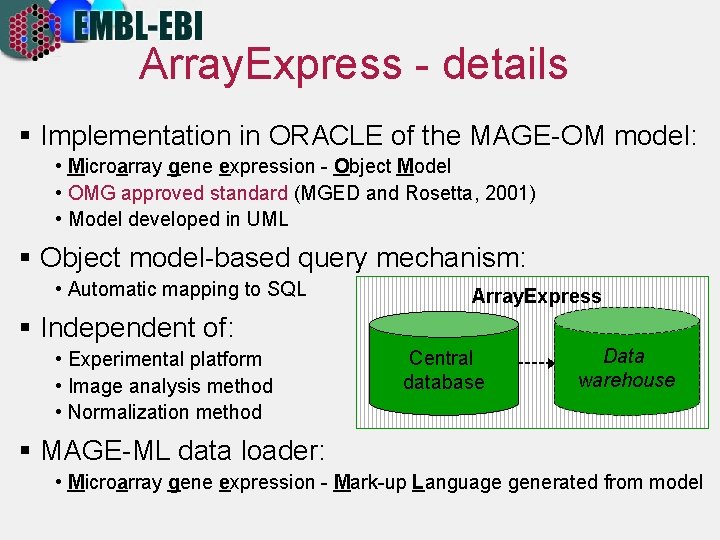
Array. Express - details § Implementation in ORACLE of the MAGE-OM model: • Microarray gene expression - Object Model • OMG approved standard (MGED and Rosetta, 2001) • Model developed in UML § Object model-based query mechanism: • Automatic mapping to SQL Array. Express § Independent of: • Experimental platform • Image analysis method • Normalization method Central database Data warehouse § MAGE-ML data loader: • Microarray gene expression - Mark-up Language generated from model

Array. Express – conceptual model Experiment Hybridisation Array Sample Normalisation Final data MIAME 6 parts of a microarray experiment

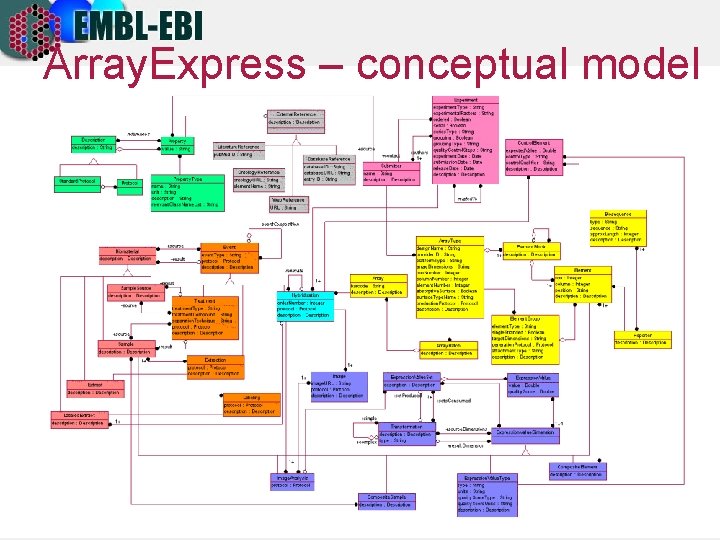
Array. Express – simplified model • Classes are represented by boxes • Classes describe objects • Related classes are grouped together in packages • MAGE-OM has 16 packages, ~ 150 tables

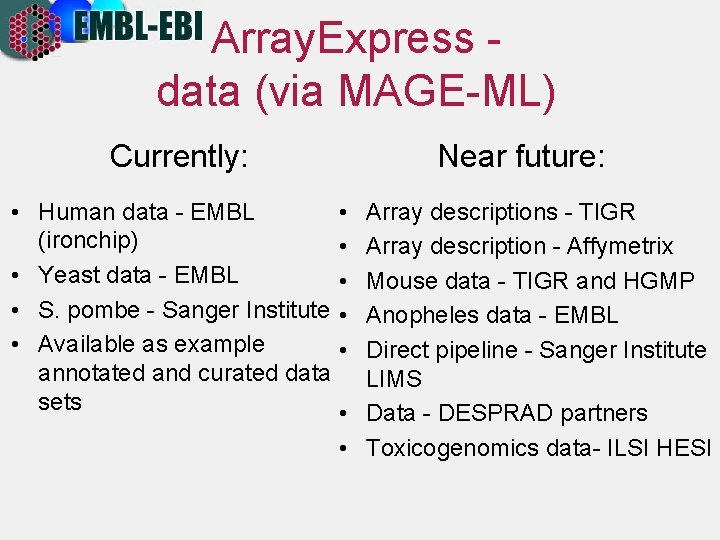
Array. Express data (via MAGE-ML) Currently: Near future: • Human data - EMBL • (ironchip) • • Yeast data - EMBL • • S. pombe - Sanger Institute • • Available as example • annotated and curated data sets • Array descriptions - TIGR Array description - Affymetrix Mouse data - TIGR and HGMP Anopheles data - EMBL Direct pipeline - Sanger Institute LIMS Data - DESPRAD partners • Toxicogenomics data- ILSI HESI

Array. Express – query interface First release 12 Januray 2002

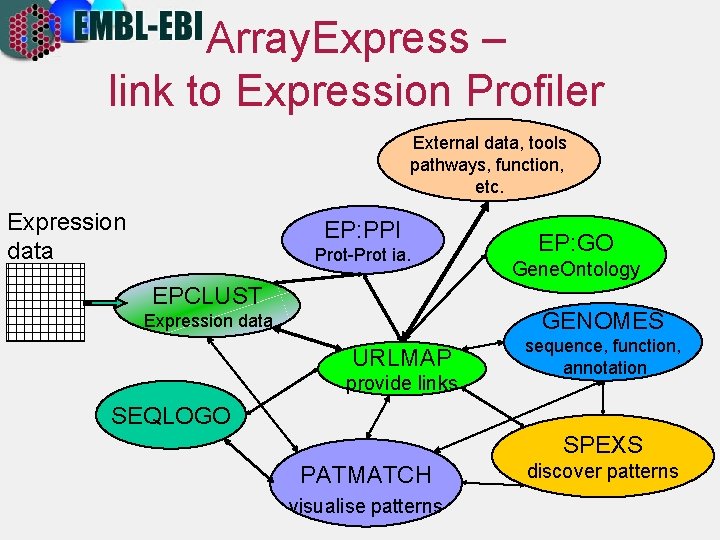
Array. Express – link to Expression Profiler External data, tools pathways, function, etc. Expression data EP: PPI Prot-Prot ia. EPCLUST EP: GO Gene. Ontology GENOMES Expression data URLMAP provide links sequence, function, annotation SEQLOGO SPEXS PATMATCH visualise patterns discover patterns

Array. Express – curation effort § User support and help documentation: • Ontologies and CV’s • Minimize free text, removal of synonyms • Help on MAGE-ML format and MAGE-OM § MIAME compliance-check § Curation at source (LIMS) § To provide high-quality, well-annotated data and allow automated data analysis

MIAMExpress - details § Submission and annotation tool: • Curators will monitor the submissions § Based on MIAME concepts: MIAMExpress • Experiment, Array and Protocol submissions • Generates MIAME-compliant information § Uses MGED Bio. Material Ontology terms: • Terms and required fields are explained § Allows user driven ontology development: • User can provide new terms and their sources § Allows browsing: • Array descriptions • Protocols

MIAMExpress - details § Version 1 launch in December 2002 § Expected users: MIAMExpress • Limited local bioinformatic support • No LIMS on site • Small scale users with custom made arrays § Can be installed as local version: • As a lab-book to annotate your experiment • As part of a LIMS § Interfaces: • Version 1 is general • Future versions, application specific interfaces - Species specific - Toxicogenomics specific (ILSI- HESI)


Array. Express - future § Load public data into Array. Express: • TIGR, EMBL, ILSI HESI, DESPRAD partners § Improve query interfaces § Launch MIAMExpress v. 1 (Dec. 2002) § MIAMExpress v. 2: • Extended according to the user needs • Integrated MGED ontology • Increased usability, flexibility and scalability § Develop curation tools

Acknowledgments § Microarray Informatics Team at EBI (19 members): • • • Alvis Brazma (Team Leader and MGED President) Helen Parkinson (Curation Coordinator) Mohammad Shojatalab (MIAMExpress Database Programmer) Ugis Sarkans (Array. Express Database development coordinator) Jaak Vilo (Expression Profiler) Curators and Programmers. § MGED members and working groups: • Alvis Brazma (MGED President, MIAME) • Chris Stoeckert, U. Penn. (MGED Ontology Working Group)

Resources and …. messages § Open sources resources: • • Array. Express and MIAMExpress schema-access to code MIAME document and glossary MAGE-ML dtd annotation examples MGED Ontology and other resources……… www. mged. org / www. ebi. ac. uk/microarray sansone@ebi. ac. uk § Be aware of MIAME ! • Nature, Lancet and have already expressed their interest • Founding agencies § Join MGED meetings, tutorials and mailing lists: • MGED-5 meeting in Japan (Sept. 2002) • Ontology for Bio. Sample description, EBI (Nov. 2002)