Other uses of DNA microarrays DNA microarray image
- Slides: 11

• Other uses of DNA microarrays • DNA microarray - image and data analysis • Proteomics - Chapter 4 p. 184 -212.

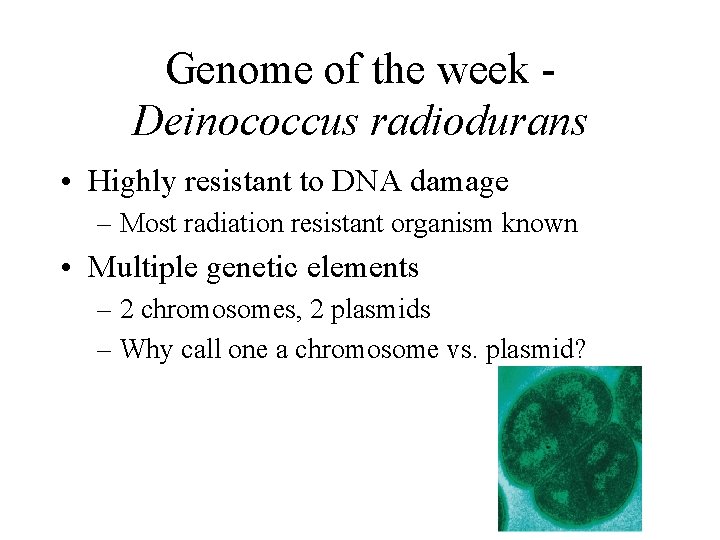
Genome of the week Deinococcus radiodurans • Highly resistant to DNA damage – Most radiation resistant organism known • Multiple genetic elements – 2 chromosomes, 2 plasmids – Why call one a chromosome vs. plasmid?

Why sequence D. radiodurans? • Learn how this bacterium is so resistant to DNA damage – This bacterium has nearly all known mechanisms for repairing DNA damage. – Redundancy of some DNA damage repair mechanisms. • Use this organism in bioremediation. – Sites contaminated with high levels of radioactivity – DOE (Department of Energy) sequences many microbial genomes - JGI

Applications of DNA microarrays • Monitor gene expression – – Study regulatory networks Drug discovery - mechanism of action Diagnostics - tumor diagnosis etc. • Genomic DNA hybridizations – – • ? Explore microbial diversity Whole genome comparisons - genome evolution Identify DNA binding sites Diagnostics - tumor diagnosis

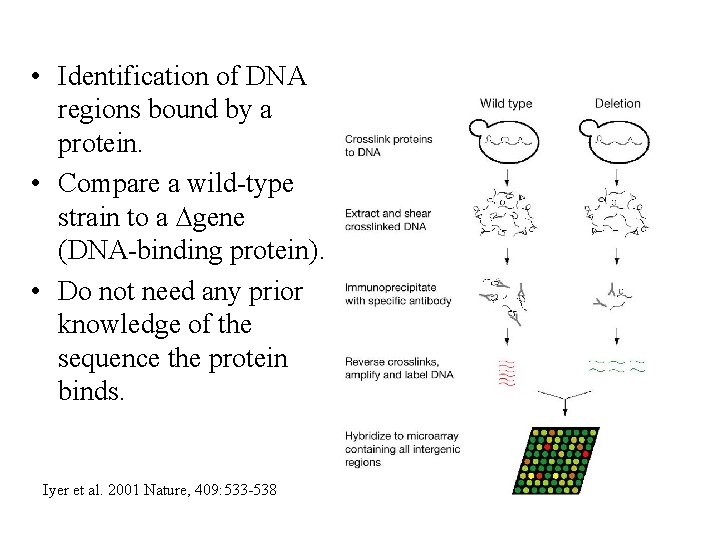
• Identification of DNA regions bound by a protein. • Compare a wild-type strain to a ∆gene (DNA-binding protein). • Do not need any prior knowledge of the sequence the protein binds. Iyer et al. 2001 Nature, 409: 533 -538

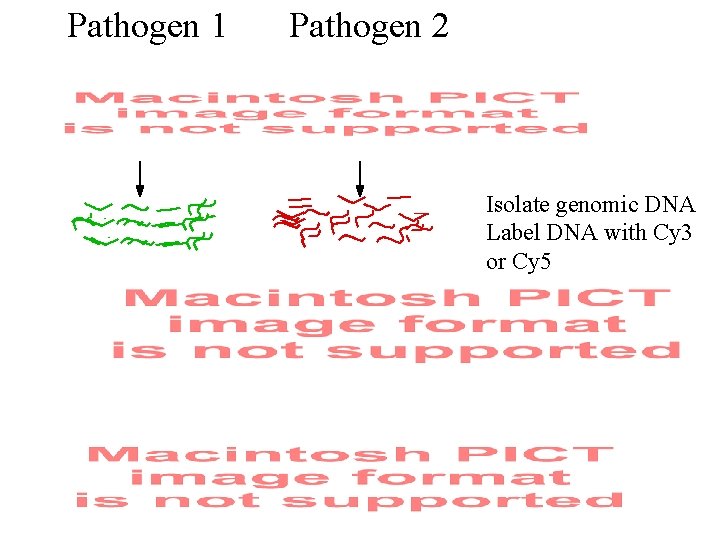
Pathogen 1 Pathogen 2 Isolate genomic DNA Label DNA with Cy 3 or Cy 5

Genomic DNA hybridizations • Determine the similarity between two different strains. – Example - E. coli outbreak • Can only detect presence or loss of genes from the sequenced strain. Will NOT detect acquisition of new genes into strain being tested.

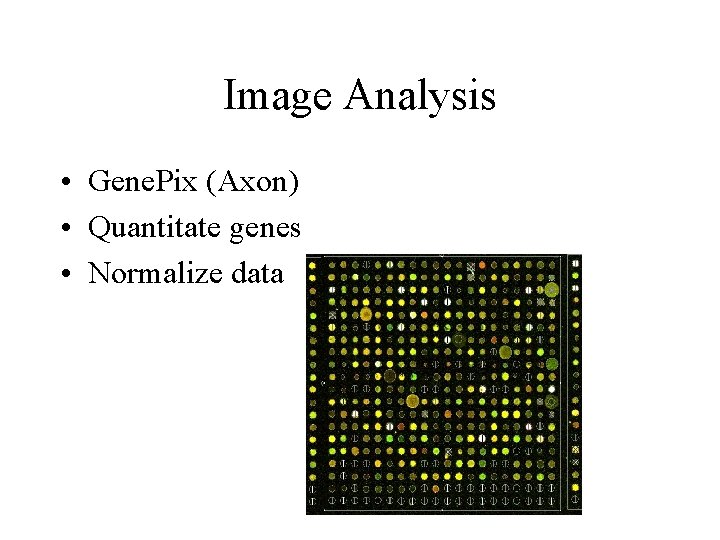
Image Analysis • Gene. Pix (Axon) • Quantitate genes • Normalize data

Scanning • How do we detect the Cy 3 and Cy 5 labeled c. DNA hybridized to the slide? • Cy 3 and Cy 5 are fluorophores – Absorb light at one wavelength (excitation) and emit light at another wavelength (emission). – The excitation and emission wavelengths of two fluorophores should not overlap • Laser based scanners - 532 nm laser (Cy 3) and 635 nm laser (Cy 5) for excitation. – Also can use microscopes for detection using excitation and emission filters.

Data normalization • Why do we need to normalize microarray data? – Correct for experimental errors • Northern blot example • Microbial microarrays – Assume the expression of most genes don’t change – We know every gene - sum the intensity in both channels and make the equal. – Many other ways of normalizing data - not one standard way. Area of active research.

Data analysis • How do we determine what is a differentially expressed gene? – Fold change? • What is a significant fold change? – Statistical analysis of the data • Multiple repeats of an experiment • Iterative outlier analysis • SAM - Significance Analysis of Microarrays - applies t-test statistics and false discovery rates to analyze microarray data. – Log 2 transformation of data -often used in analysis of array data. • Cy 5/Cy 3 = 1 = no difference in expression • Cy 5/Cy 3 = 4 or 0. 25 = 4 -fold change (4 -1=3 and 1 -0. 25 = 0. 75) • Log 2 transformed data - no difference = 0 • Log 2 of 4 = 2 Log 2 of 0. 25 = -2. • Data not bounded by zero