Image Quantitation in Microarray Analysis More tomorrow Microarray
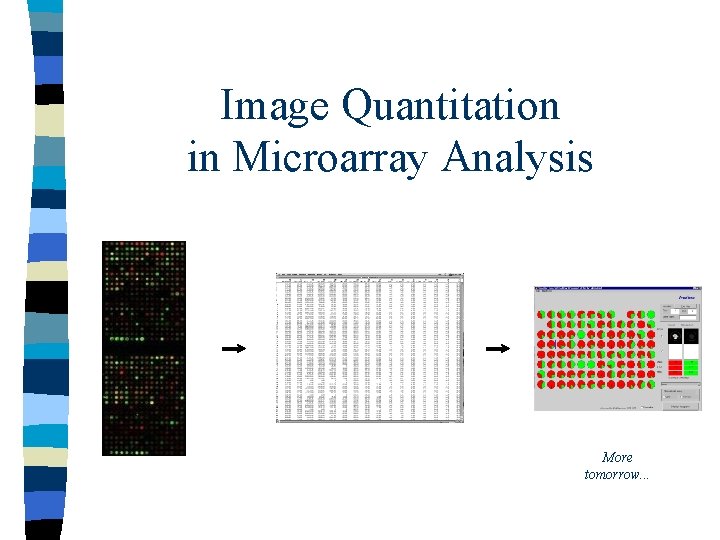
![Technical sample (labelled) probe (on chip) pseudo-colour image [image from Jeremy Buhler] Technical sample (labelled) probe (on chip) pseudo-colour image [image from Jeremy Buhler]](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-3.jpg)
![Images from scanner n Resolution – standard 10 m [currently, max 5 m] – Images from scanner n Resolution – standard 10 m [currently, max 5 m] –](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-5.jpg)
![Normalisation references [statistics] Normalization for c. DNA microarray data Yang et al. (2001) In Normalisation references [statistics] Normalization for c. DNA microarray data Yang et al. (2001) In](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-10.jpg)




- Slides: 19

Image Quantitation in Microarray Analysis More tomorrow. . .

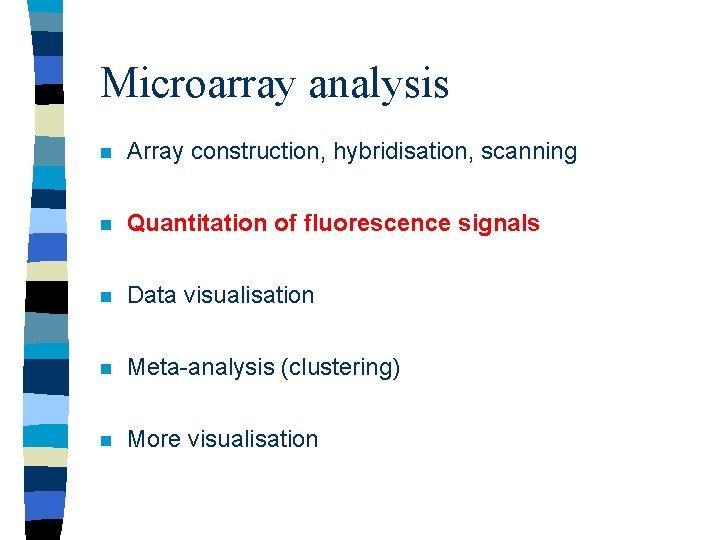
Microarray analysis n Array construction, hybridisation, scanning n Quantitation of fluorescence signals n Data visualisation n Meta-analysis (clustering) n More visualisation
![Technical sample labelled probe on chip pseudocolour image image from Jeremy Buhler Technical sample (labelled) probe (on chip) pseudo-colour image [image from Jeremy Buhler]](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-3.jpg)
Technical sample (labelled) probe (on chip) pseudo-colour image [image from Jeremy Buhler]

Experimental design n Track what’s on the chip • which spot corresponds to which gene n Duplicate experimental spots • reproducibility n Controls – DNAs spotted on glass • positive probe (induced or repressed) • negative probe (bacterial genes on human chip) – oligos on glass or synthesised on chip (Affymetrix) • point mutants (hybridisation plus/minus)
![Images from scanner n Resolution standard 10 m currently max 5 m Images from scanner n Resolution – standard 10 m [currently, max 5 m] –](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-5.jpg)
Images from scanner n Resolution – standard 10 m [currently, max 5 m] – 100 m spot on chip = 10 pixels in diameter n Image format – TIFF (tagged image file format) – can be compressed • (eg. Lempel-Ziv-Welch: ~ 5 x compression) – 1 cm x 1 cm image at 16 bit = 2 Mb (uncompressed) – other formats exist eg. SCN (used at Stanford University) n Separate image for each fluorescent sample – channel 1, channel 2, etc.

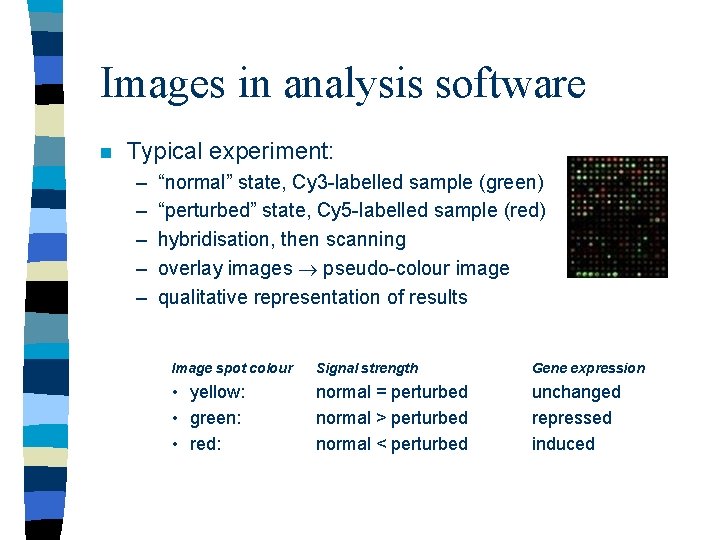
Images in analysis software n Typical experiment: – – – “normal” state, Cy 3 -labelled sample (green) “perturbed” state, Cy 5 -labelled sample (red) hybridisation, then scanning overlay images pseudo-colour image qualitative representation of results Image spot colour Signal strength Gene expression • yellow: • green: • red: normal = perturbed normal > perturbed normal < perturbed unchanged repressed induced

Quantitation process (1) Accurate representation of signal for each spot and determine ratio channel 1: channel 2 n Determine spot boundary – construct grid (dimensions of array / spot size) – iterative process to find spots n Measure signal – fluorescence • 8 bit = 256 shades • 16 bit = 65’ 536 shades – absolute output values vary from system to system

Quantitation process (2) n Measure background – local (usually best) – selected region – selected spots / probes from different species n Quality control – eg. fraction of pixels greater than background (Scan. Alyze) – flag aberrant spots n Determine ratio of signal strengths for each spot Ch 1/Ch 2 = (Ch 1 I-Ch 1 B)/Ch 2 I-Ch 2 B)

Normalisation n Eliminate systematic variation – correct for • dye incorporation • print-tip effects • hybridisation efficiencies • etc. n How? n Use STATISTICS!
![Normalisation references statistics Normalization for c DNA microarray data Yang et al 2001 In Normalisation references [statistics] Normalization for c. DNA microarray data Yang et al. (2001) In](https://slidetodoc.com/presentation_image_h2/a6e82539d6bb4e072a7df6c0ee22b258/image-10.jpg)
Normalisation references [statistics] Normalization for c. DNA microarray data Yang et al. (2001) In preparation Statistical methods for identifying differentially expressed genes in replicated c. DNA microarray experiments Dudoit et al. (2000) Technical report #578 Berkeley Statistics Dept. both available from ftp: //ftp. ch. embnet. org/pub/MAcourse/material/

Quantitation - problems n Reference signal is close zero – channels ratio (Ch 1/Ch 2) tends to infinity n Non-uniform background – “mean” background sometimes non-representative • bright particles • streaks on image – safer to use “median” (middle value) • less contribution by extreme values

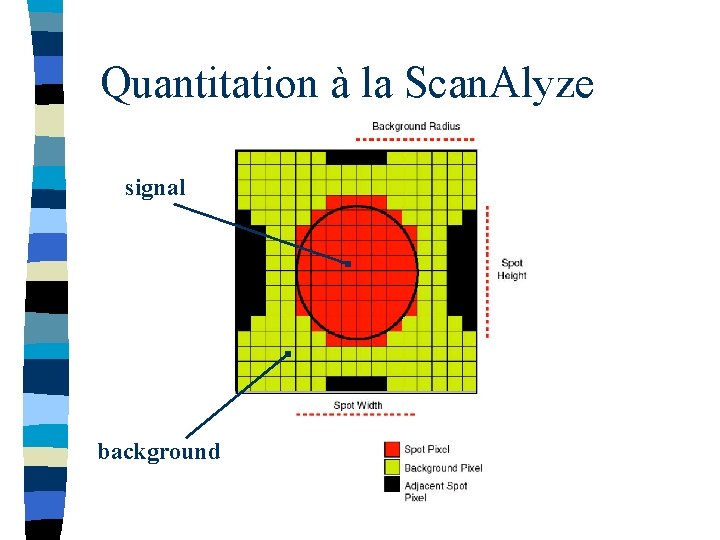
Quantitation à la Scan. Alyze signal background

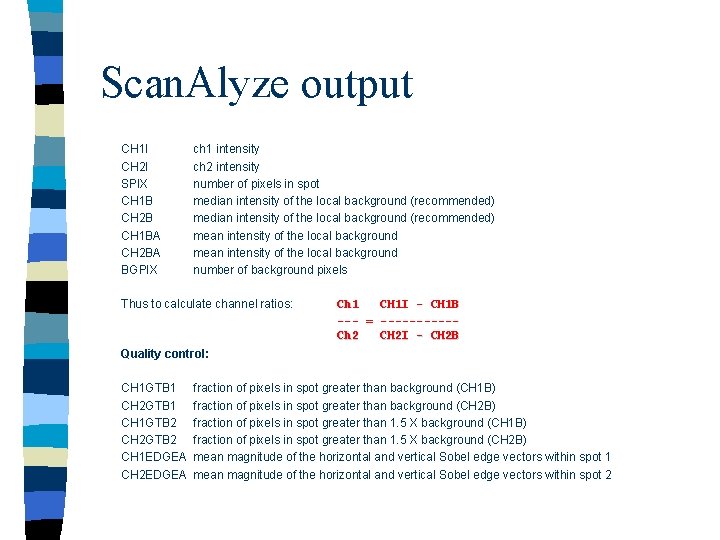
Scan. Alyze output CH 1 I CH 2 I SPIX CH 1 B CH 2 B CH 1 BA CH 2 BA BGPIX ch 1 intensity ch 2 intensity number of pixels in spot median intensity of the local background (recommended) mean intensity of the local background number of background pixels Thus to calculate channel ratios: Ch 1 CH 1 I - CH 1 B --- = -----Ch 2 CH 2 I - CH 2 B Quality control: CH 1 GTB 1 CH 2 GTB 1 CH 1 GTB 2 CH 2 GTB 2 CH 1 EDGEA CH 2 EDGEA fraction of pixels in spot greater than background (CH 1 B) fraction of pixels in spot greater than background (CH 2 B) fraction of pixels in spot greater than 1. 5 X background (CH 1 B) fraction of pixels in spot greater than 1. 5 X background (CH 2 B) mean magnitude of the horizontal and vertical Sobel edge vectors within spot 1 mean magnitude of the horizontal and vertical Sobel edge vectors within spot 2


Software packages - quantitation n Scan. Alyze – – n by Michael Eisen (Stanford University) quantitation of images no data visualisation free from http: //rana. lbl. gov/ Ima. Gene – Bio. Discovery Inc. – quantitation and some data visualisation – demo from http: //www. biodiscovery. com/ n plus many others - explore!

Making sense of raw data n Difficult to see results in tabulated data n Represent in graphical form Data visualisation examples from Ima. Gene. . .

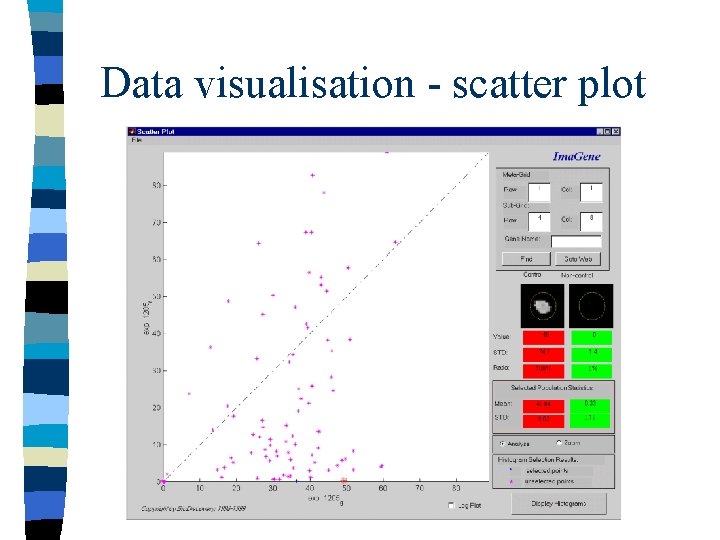
Data visualisation - scatter plot

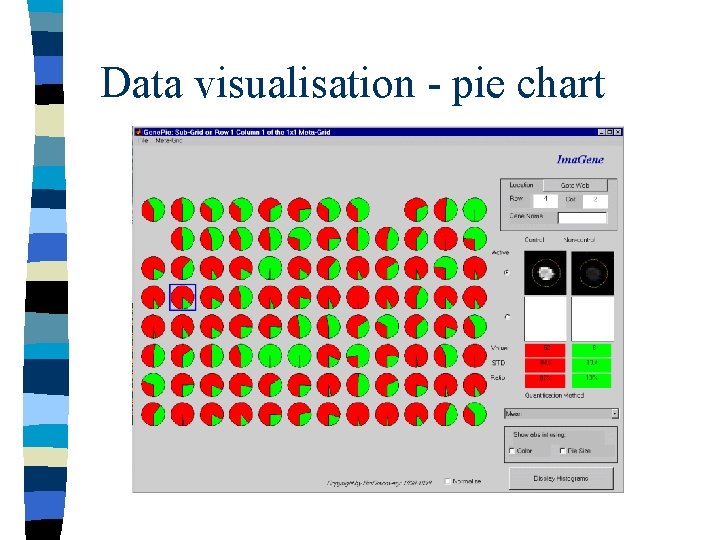
Data visualisation - pie chart

Scan. Alyze quick demo