Minimum Information About a Microarray Experiment MIAME Alvis

Minimum Information About a Microarray Experiment - MIAME Alvis Brazma European Bioinformatics Institute European Molecular Biology Laboratory

What is MIAME? F F A document, the goal of which is to specify the minimum information that must be reported about a microarray experiment in order to ensure its interpretability, as well as potential verification of the results Underlying motivation – – – to enable the establishment of public repositories for microarray data to serve as a basis for designing a microarray data exchange format

Acknowledgements F MIAME working group F MAML working group F MGED steering committee F John Aach, Wilhelm Ansorge, Pascal Hingamp, Frank Holstege, Alex Lash, John Quackenbush, Alan Robinson, Paul Spellman, Criss Stoeckert, Martin Vingron

MIAME history F F F A need to establish a public repository or repositories for microarray gene expression data became apparent in 1998 That requires data standards MGED 1 meeting in Cambridge in November, 1999 establishes five working groups, including the microarray data annotation group (MIAME) Several MIAME drafts produced by the group MGED steering committee meeting in November 2000 in Bethesda endorses a MIAME draft Last revision yesterday in MIAME working group meeting

Outline of this talk F Considerations behind the MIAME design – why F The MIAME details – what F Future developments and use of MIAME – how

How to think about MIAME What minimum information about a microarray gene expression measuring experiment should be recorded in a database for the database entries to be usable on stand-alone basis: – – – the users may not know any background information that is not recorded the database should be usable for automated data analysis and mining, i. e. not only on record-byrecord basis the data may be coming from different laboratories and different technology platforms
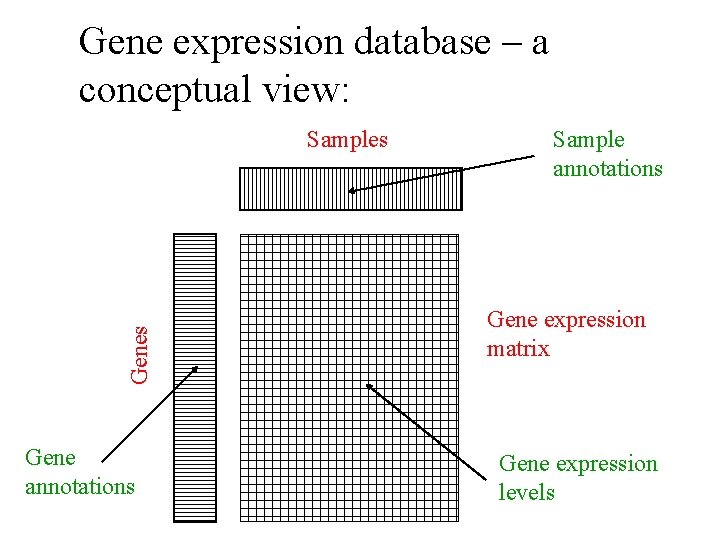
Gene expression database – a conceptual view: Genes Samples Gene annotations Sample annotations Gene expression matrix Gene expression levels
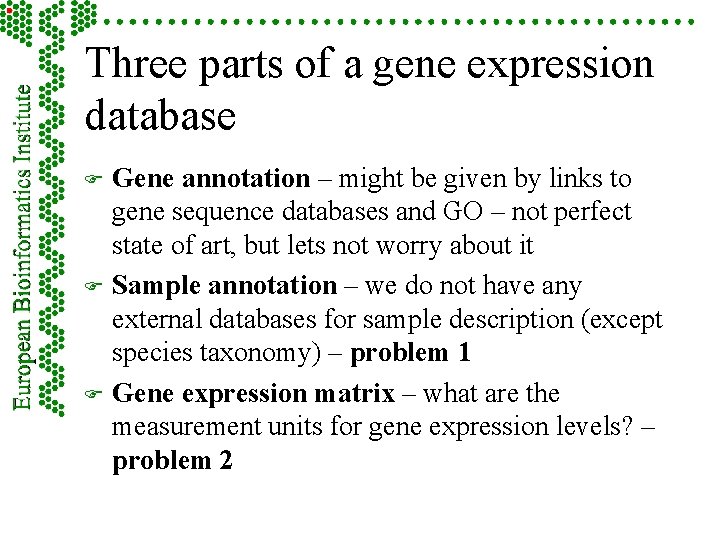
Three parts of a gene expression database F F F Gene annotation – might be given by links to gene sequence databases and GO – not perfect state of art, but lets not worry about it Sample annotation – we do not have any external databases for sample description (except species taxonomy) – problem 1 Gene expression matrix – what are the measurement units for gene expression levels? – problem 2
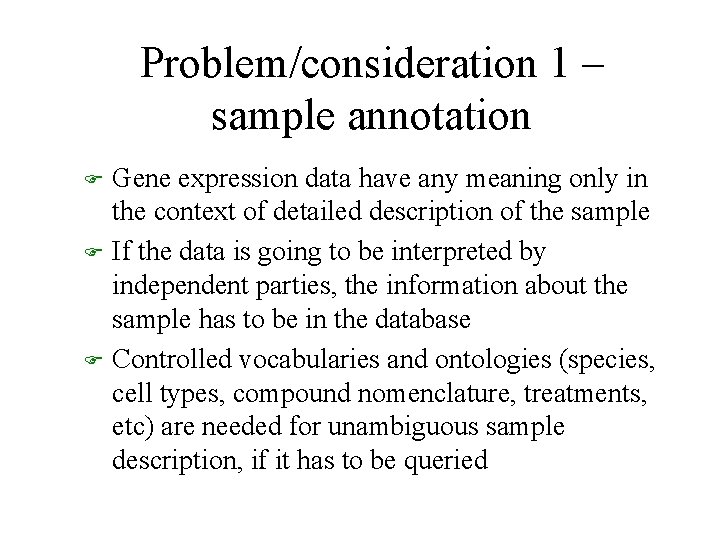
Problem/consideration 1 – sample annotation F F F Gene expression data have any meaning only in the context of detailed description of the sample If the data is going to be interpreted by independent parties, the information about the sample has to be in the database Controlled vocabularies and ontologies (species, cell types, compound nomenclature, treatments, etc) are needed for unambiguous sample description, if it has to be queried

Sample annotation – what can be done F Some use of free text descriptions are unavoidable F Controlled vocabularies and ontologies should be used wherever available F Externally defined controlled vocabularies and ontologies should be used whenever they exist
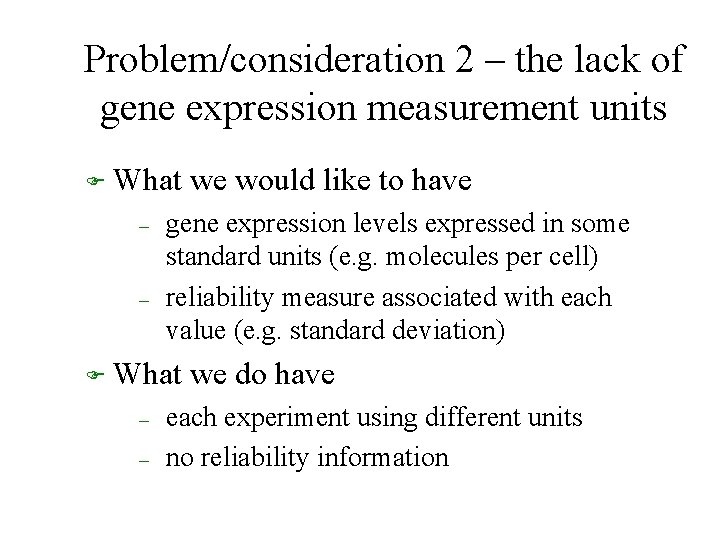
Problem/consideration 2 – the lack of gene expression measurement units F What – – gene expression levels expressed in some standard units (e. g. molecules per cell) reliability measure associated with each value (e. g. standard deviation) F What – – we would like to have we do have each experiment using different units no reliability information
Comparing expression data cm inc
Comparing expression data ? ?
Comparing expression data
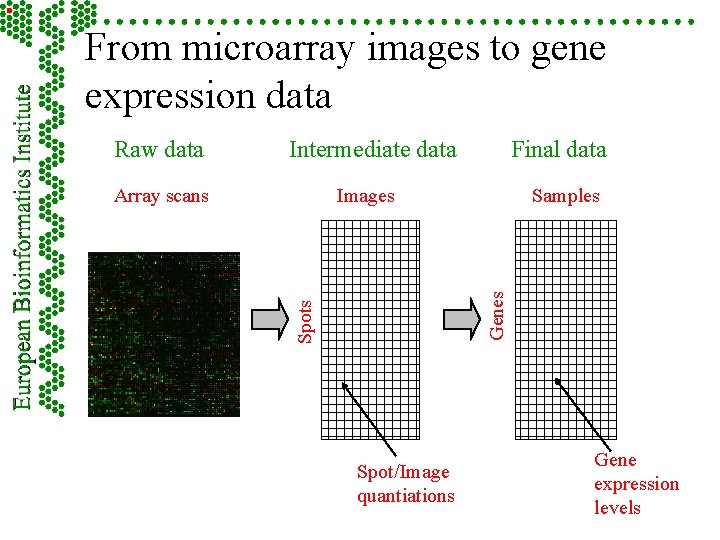
From microarray images to gene expression data Intermediate data Final data Images Spots Array scans Samples Genes Raw data Spot/Image quantiations Gene expression levels

What to do in the absence of standard measurement units? F Record raw, intermediate and final analysis data together with the detailed annotation how the analysis has been performed F This effectively passes on the responsibility about interpreting the final analysis data to the user
Measurement units F In perspective: – – standard controls for experiments (on chips and in the samples) should be introduced replicate measurements will become a norm F Temporary – solution: storing intermediate analysis results (including the images) and annotations of how they were obtained - i. e. , the evidence

Problem/consideration - 3 F We need to find a compromise found between the burden on the data producers to annotate and provide the data and the need of data to be sufficiently annotated for the database users – – F Too much detail may turn away the potential data providers and complicate the data submission and storage Too little detail may limit the usability of the data The current draft is a compromise between these two
Some more general principles F F F MIAME is aimed at a cooperative data provider, not as a legal document designed to close all loop -holes MIAME is an informal specification The concept of ‘qualifier, value, source’ triplets, e. g. , – – – F qualifier – cell type value – epithelial source – Human Anatomy (author, edition) The concept of ‘experimental protocol’

General principles - continued F F MIAME is not designed as a ‘questionnaire’ that can be filled in, but only as an informal specification based on which such a questionnaire, in fact, an annotation tool, can be based Although MIAME is conceptually independent on databases, the aim of establishing a microarray database should be kept in mind then reading MAIME

Outline of this talk F Considerations behind the MIAME design – why F The MIAME details – what
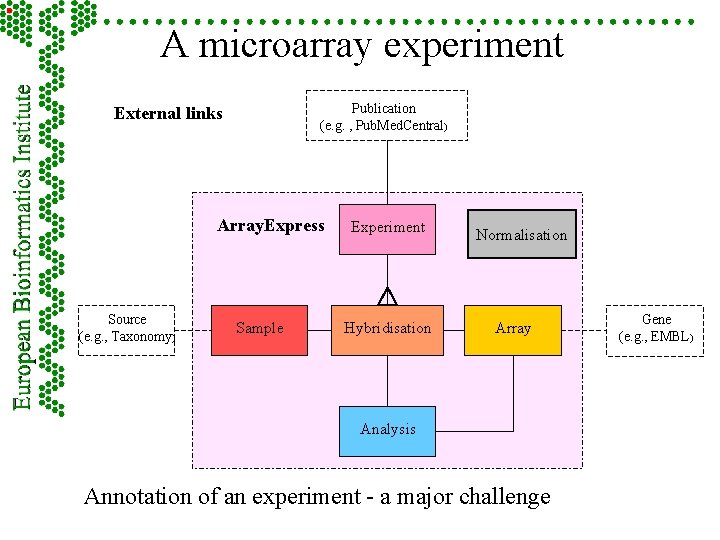
A microarray experiment Publication (e. g. , Pub. Med. Central) External links Array. Express Source (e. g. , Taxonomy) Sample Experiment Hybridisation Normalisation Array Analysis Annotation of an experiment - a major challenge Gene (e. g. , EMBL)
MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array 3. Samples: samples used, the extract preparation and labeling 4. Hybridizations: procedures and parameters 5. Measurements: images, quantitation, specifications 6. Controls: types, values, specifications www. mged. org

MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole
Part 1 - Experimental design: the set of the hybridisation experiments as a whole F Normally ‘an experiment’ should consist of one or more hybridisations that are in some way related and performed in a limited number of time, e. g. all related to the same publication – – – Author, contact information, citations Type of experiment (e. g. , time course, normal vs diseased comparison) Experimental factors – i. e. tested parameters in the experiment (e. g. time, dose, genetic variation, response to a compound) List of organisms used in the experiment List of platforms used
Experimental design - continued F List of samples, arrays and hybridisations and their relationships, e. g. : – – – Samples: S 1, S 2, S 3 Arrays: A 1, A 2, A 3 Hybridisations: • • • F H 1 is S 1 and S 2 on A 1 H 2 is S 2 and S 3 on A 2 H 3 is S 1 and S 2 on A 3 Which hybridisations are replicates, – e. g. H 1 and H 3 are replicates
Experimental design – continued 2 F Quality related indicators F Optional user defined ‘qualifier, value, source’ triplet – e. g. : – – – qualifier – survival data value – given source – user defined F Description publication of the experiment or link to a

MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array
Part 2 - Array design: each array used and each element (spot) on the array F This part is separate for each type of array used in the experiment F For the database, the array description should be normally submitted only once F For each physical array used in the experiment a unique ID and the array type are given
Array design – continued F F F Array design related information (e. g. platform type – insitu synthesized or spotted, array provider, surface type – glass, membrane, other, etc) Properties of each type of elements on the array, that are generated by similar protocols (e. g. synthesized oligos, PCR products, plasmids, colonies, others) – may be simple or composite (Affymetrix) Each element (spot) on the array
Array design – continued F Each element (spot) on the array – – – Elements may be simple or composite Each element must be identified by either the sequence, clone ID, PCR primer pair, or in any other unambiguous way Composite elements may be identified by a reference sequence May be linked to genes (preferably) Will normally be provided in a separate file (e. g. spreadsheet)

MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array 3. Samples: samples used, the extract preparation and labeling
Part 3 - Samples: samples used, the extract preparation and labeling F Sample source and treatment – – Organism (NCBI taxonomy) Additional ‘qualifier, value, source’ list • • cell source and type developmental sage organism part (tissue) animal/plant strain or line genetic variation disease state or normal … Typically only some of these qualifiers are relevant – an ontology tree is needed to implement the annotation tool for sample source and treatment
Sample - continued F Hybridisation – extract preparation Laboratory protocol, including extraction method, whether RNA, m. RNA, or genomic DNA is extracted, amplification method F Labelling – Laboratory protocol, including amount of nucleic acids labelled, label used (e. g. Cy 3, Cy 5, 33 P, etc)
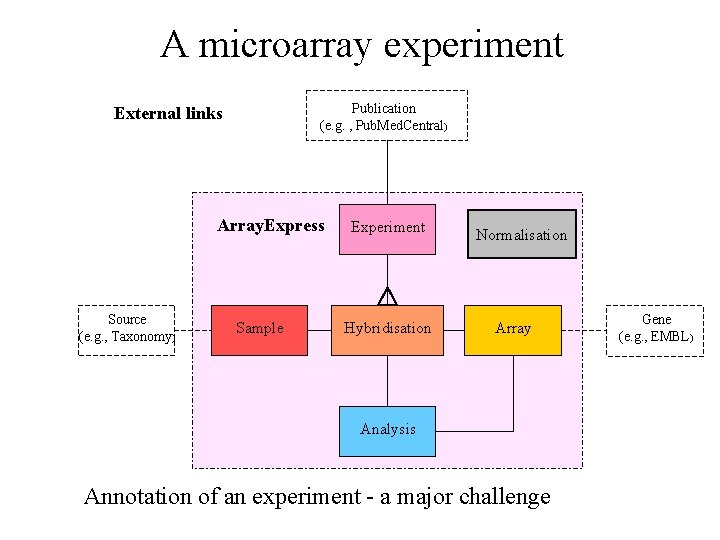
A microarray experiment Publication (e. g. , Pub. Med. Central) External links Array. Express Source (e. g. , Taxonomy) Sample Experiment Hybridisation Normalisation Array Analysis Annotation of an experiment - a major challenge Gene (e. g. , EMBL)
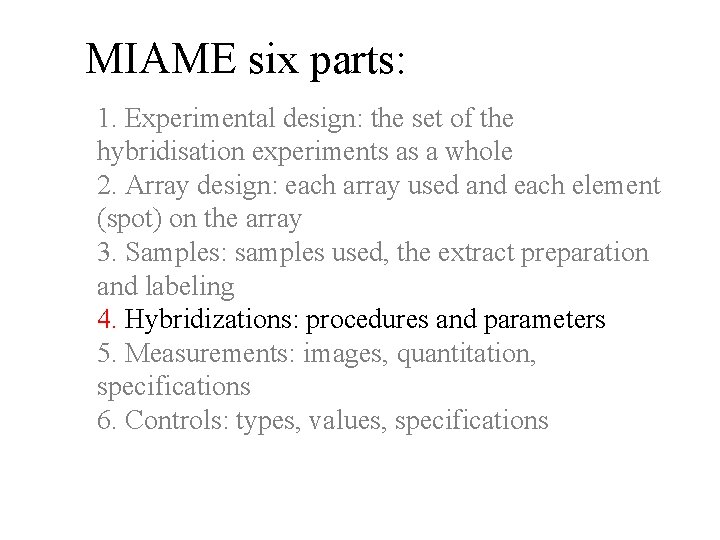
MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array 3. Samples: samples used, the extract preparation and labeling 4. Hybridizations: procedures and parameters 5. Measurements: images, quantitation, specifications 6. Controls: types, values, specifications
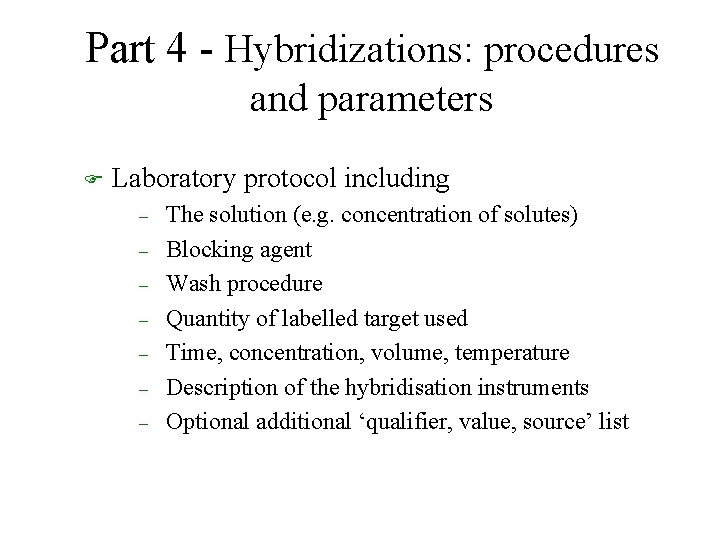
Part 4 - Hybridizations: procedures and parameters F Laboratory protocol including – – – – The solution (e. g. concentration of solutes) Blocking agent Wash procedure Quantity of labelled target used Time, concentration, volume, temperature Description of the hybridisation instruments Optional additional ‘qualifier, value, source’ list

MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array 3. Samples: samples used, the extract preparation and labeling 4. Hybridizations: procedures and parameters 5. Measurements: images, quantitation, specifications
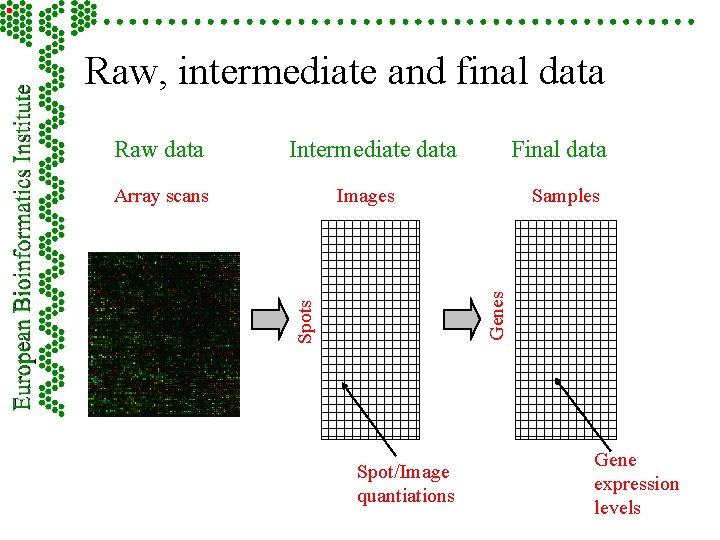
Raw, intermediate and final data Intermediate data Final data Images Spots Array scans Samples Genes Raw data Spot/Image quantiations Gene expression levels

Part 5 - Measurements: images, quantitation, specifications F Hybridisation scan raw data – image F Intermediate data – image analysis and quantiation F Final data – summarised information from possible replicates
From microarray images to gene expression data Raw data Array scans
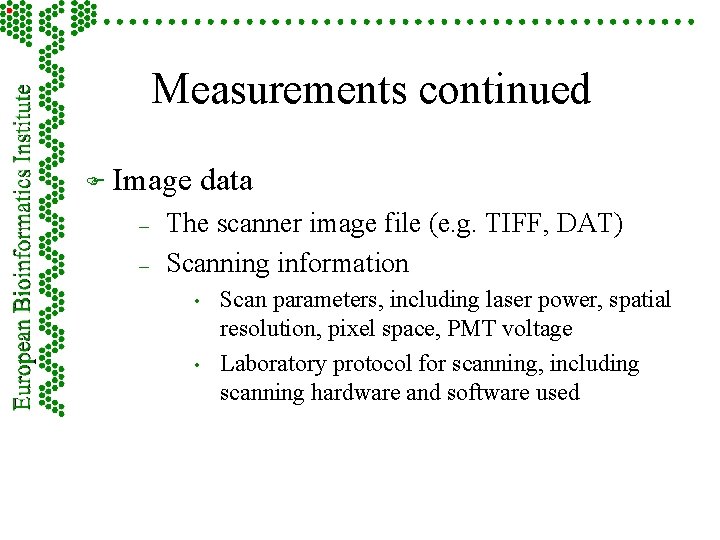
Measurements continued F Image – – data The scanner image file (e. g. TIFF, DAT) Scanning information • • Scan parameters, including laser power, spatial resolution, pixel space, PMT voltage Laboratory protocol for scanning, including scanning hardware and software used
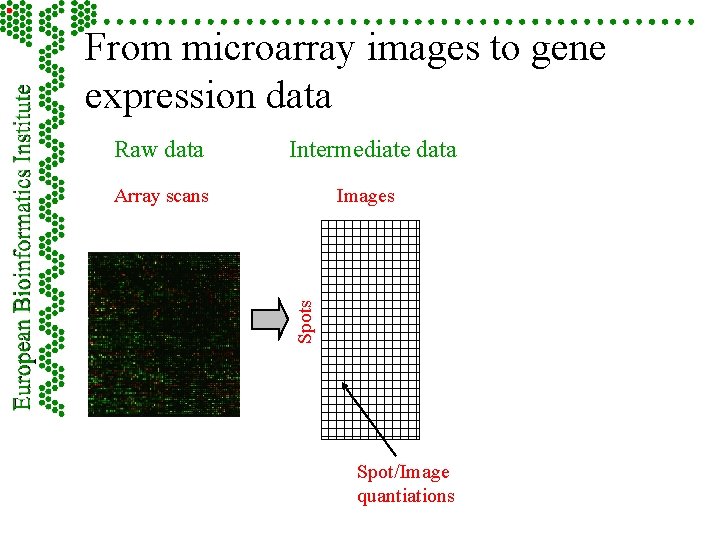
From microarray images to gene expression data Raw data Intermediate data Images Spots Array scans Spot/Image quantiations
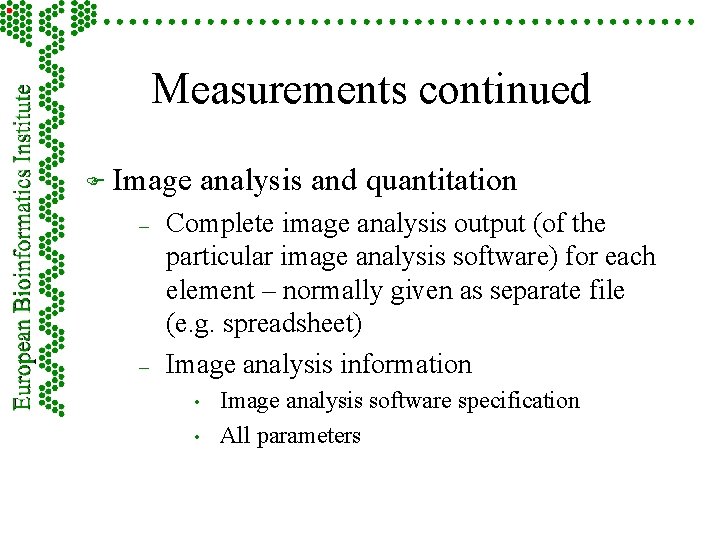
Measurements continued F Image – – analysis and quantitation Complete image analysis output (of the particular image analysis software) for each element – normally given as separate file (e. g. spreadsheet) Image analysis information • • Image analysis software specification All parameters
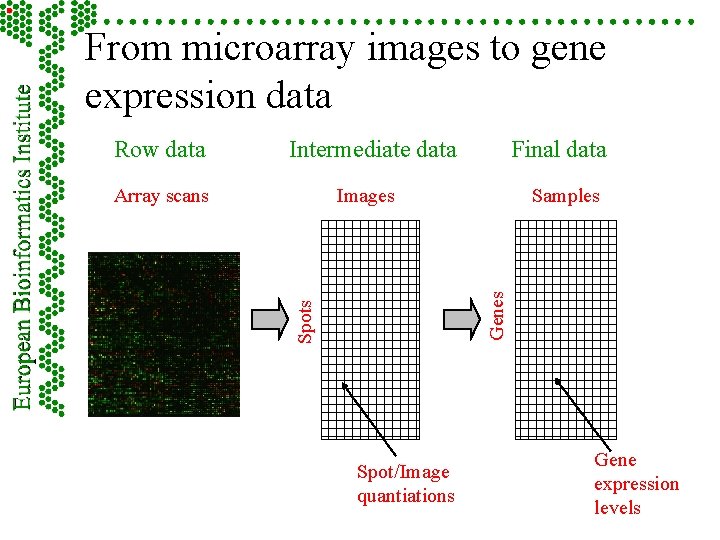
From microarray images to gene expression data Intermediate data Final data Images Spots Array scans Samples Genes Row data Spot/Image quantiations Gene expression levels
Measurements continued F Summarised replicates information from possible Derived measurement values summarising related elements as used by the author – Reliability information for these values, as used by the author (may be ‘unknown’) (these will be typically given in a spreadsheet) – Specifications of these two (e. g. , median value of the replicates, standard deviation) –
MIAME six parts: 1. Experimental design: the set of the hybridisation experiments as a whole 2. Array design: each array used and each element (spot) on the array 3. Samples: samples used, the extract preparation and labeling 4. Hybridizations: procedures and parameters 5. Measurements: images, quantitation, specifications 6. Controls: types, values, specifications

Part 6 - Controls: types, values, specifications F Normalisation strategy (spiking, housekeeping genes, total array, other) F Normalisation algorithm F Control array elements F Hybridisation extract preparation

Outline of this talk F Considerations behind the MIAME design – why F The MIAME details – what F Future developments and use of MIAME – why
How to use MIAME F Data exchange format (MAML) allowing to communicate MIAME information F Establishing MIAME compliant databases (e. g. Array. Express) F Developing annotation tools for generating MIAME compliant information F Journals and public funding agencies may establish MIAME related policies

Some questions F Is the current MIAME draft – – sufficiently detailed? (if not what are the queries it should support? ) or is it already excessive for data submitters? F Images – if they are required, what is the mechanism for their communication? – – – storage in the public repositories? responsibility of the data submitters? may be in the future they wont be necessary?

What is MIAME? F F A document, the goal of which is to specify the minimum information that must be reported about a microarray experiment in order to ensure its interpretability, as well as potential verification of the results Underlying motivation – – – to enable the establishment of public repositories for microarray data to serve as a basis for designing a microarray data exchange format

A proposal for MIAME future work F F F F To prepare the current MIAME draft for publishing To write up and post the MIAME glossary To collaborate with the data normalisation group for finalizing the MIAME part 6 and develop quality control section To collaborate with the Ontology group in developing the sample specification To collaborate with MAML group in the data exchange format To guide the development of MIAME compliant data annotation tools To update MIAME as the technology and our understanding of it develops To formulate a superset of recommended additional information

A proposal to MGED 3 F To endorse the current MIAME draft F To recommend journals and funding agencies to adopt policies promoting publishing MIAME compliant information F To facilitate the adoption of MIAME by the array technology providers and software companies

Outstanding questions F Does a natural, standard gene expression level measurement unit (units) exist? F If yes – – F If What is it (or they)? Is it (are they) achievable by microarrays? not – How much we will be abele to get out of gene expression databases?
- Slides: 55