CDC RealTime RTPCR Support Strategies and Updates Third




























- Slides: 59

CDC Real-Time RT-PCR Support Strategies and Updates Third Annual SARInet Meeting May 9 -11, 2016 San Juan, Puerto Rico Stephen Lindstrom, Ph. D. Diagnostics Development Team Virus Surveillance and Diagnosis Branch Influenza Division Centers for Disease Control and Prevention The findings and conclusions in this presentation are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention National Center for Immunization and Respiratory Diseases Influenza Division

Overview § Testing strategies & algorithms for surveillance and diagnostic testing § CDC r. RT-PCR assays for detection and characterization of influenza § Support strategies for r. RT-PCR testing § CLSIS – protocols/procedures § IRR - Reagent kits, control materials § A(H 1 N 1)pdm 09 reactivity and assay update

Influenza Viruses in Humans - 2016 Avian H 5 Avian H 9 Avian H 7 Avian H 10 Avian H 6 H 3 v Type A Potential Pandemic strains H 1 v H 1 H 2 H 1 1918 H 3 2009 1998/9 1940 1957 1968 1977 1997 2003 B/Yam Type B B/Vic 2016 Seasonal Strains

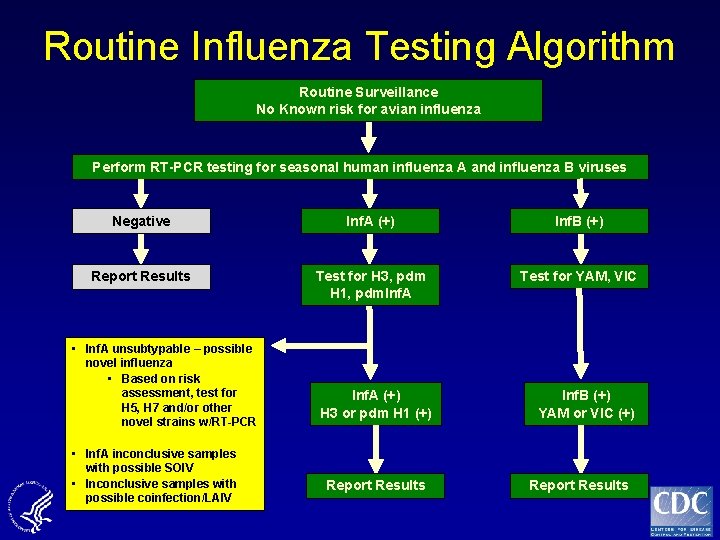
Routine Influenza Testing Algorithm Routine Surveillance No Known risk for avian influenza Perform RT-PCR testing for seasonal human influenza A and influenza B viruses Negative Inf. A (+) Inf. B (+) Report Results Test for H 3, pdm H 1, pdm. Inf. A Test for YAM, VIC Inf. A (+) H 3 or pdm H 1 (+) Inf. B (+) YAM or VIC (+) • Inf. A unsubtypable – possible novel influenza • Based on risk assessment, test for H 5, H 7 and/or other novel strains w/RT-PCR • Inf. A inconclusive samples with possible SOIV • Inconclusive samples with possible coinfection/LAIV Report Results

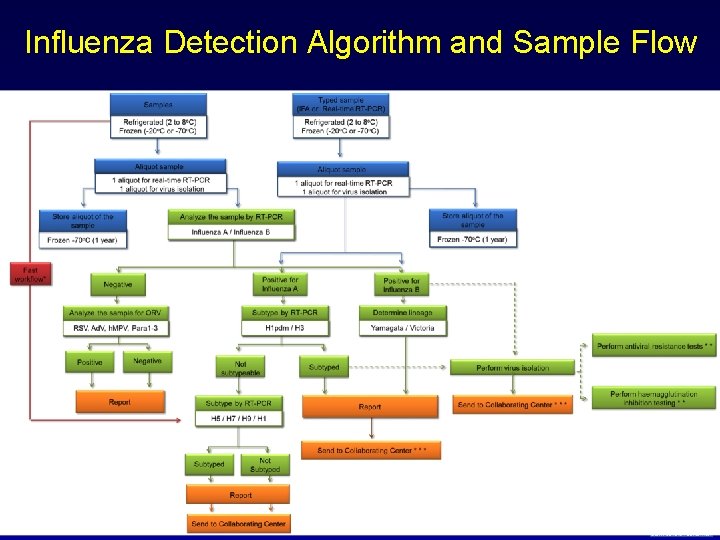
Influenza Detection Algorithm and Sample Flow

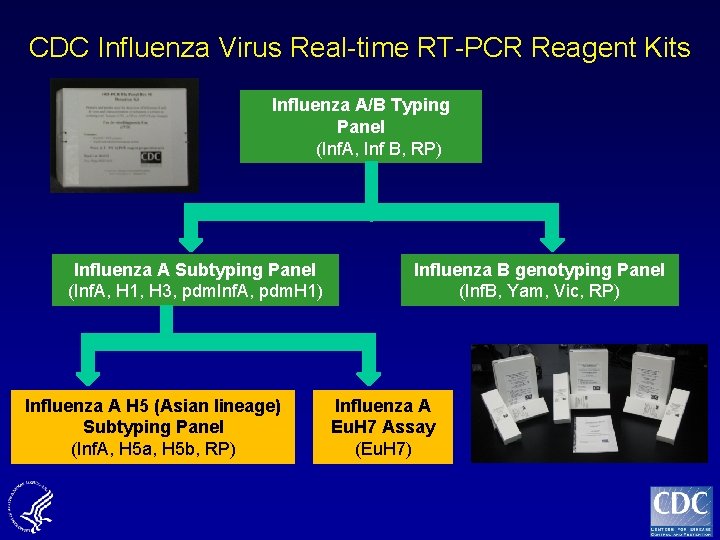
CDC Influenza Virus Real-time RT-PCR Reagent Kits Influenza A/B Typing Panel (Inf. A, Inf B, RP) Influenza A Subtyping Panel (Inf. A, H 1, H 3, pdm. Inf. A, pdm. H 1) Influenza A H 5 (Asian lineage) Subtyping Panel (Inf. A, H 5 a, H 5 b, RP) Influenza B genotyping Panel (Inf. B, Yam, Vic, RP) Influenza A Eu. H 7 Assay (Eu. H 7)

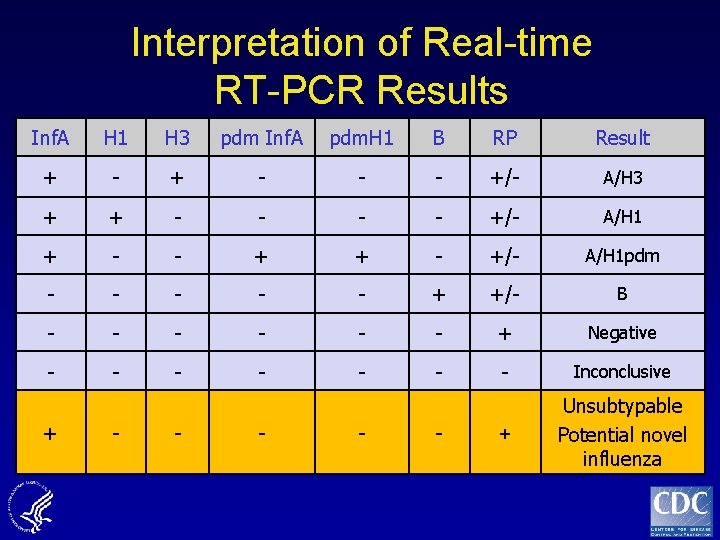
Interpretation of Real-time RT-PCR Results Inf. A H 1 H 3 pdm Inf. A pdm. H 1 B RP Result + - - - +/- A/H 3 + + - - +/- A/H 1 + - - + + - +/- A/H 1 pdm - - - + +/- B - - - + Negative - - - - Inconclusive + Unsubtypable Potential novel influenza + - - -

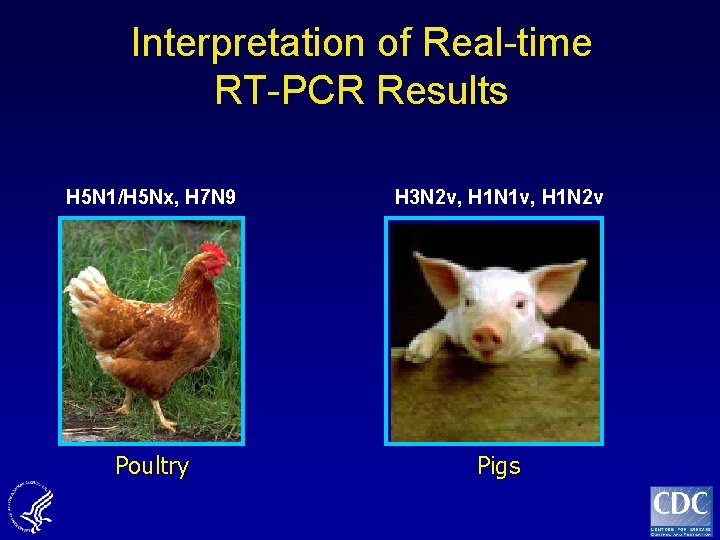
Interpretation of Real-time RT-PCR Results H 5 N 1/H 5 Nx, H 7 N 9 H 3 N 2 v, H 1 N 1 v, H 1 N 2 v Poultry Pigs




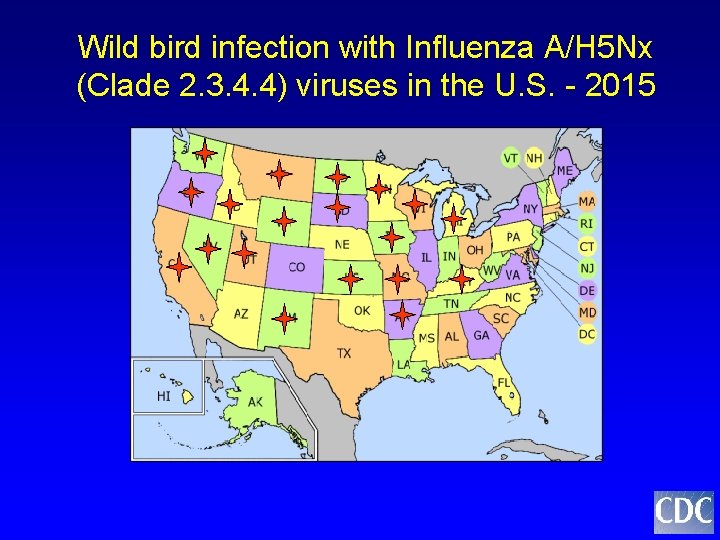
Wild bird infection with Influenza A/H 5 Nx (Clade 2. 3. 4. 4) viruses in the U. S. - 2015


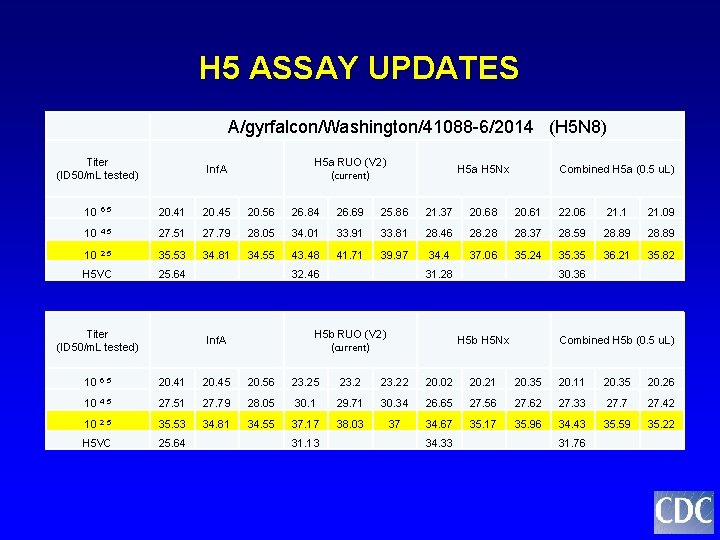
H 5 ASSAY UPDATES A/gyrfalcon/Washington/41088 -6/2014 (H 5 N 8) Titer (ID 50/m. L tested) H 5 a RUO (V 2) (current) Inf. A H 5 a H 5 Nx Combined H 5 a (0. 5 u. L) 10 6. 5 20. 41 20. 45 20. 56 26. 84 26. 69 25. 86 21. 37 20. 68 20. 61 22. 06 21. 1 21. 09 10 4. 5 27. 51 27. 79 28. 05 34. 01 33. 91 33. 81 28. 46 28. 28 28. 37 28. 59 28. 89 10 2. 5 35. 53 34. 81 34. 55 43. 48 41. 71 39. 97 34. 4 37. 06 35. 24 35. 35 36. 21 35. 82 H 5 VC 25. 64 32. 46 31. 28 30. 36 Titer (ID 50/m. L tested) H 5 b RUO (V 2) (current) Inf. A H 5 b H 5 Nx Combined H 5 b (0. 5 u. L) 10 6. 5 20. 41 20. 45 20. 56 23. 25 23. 22 20. 02 20. 21 20. 35 20. 11 20. 35 20. 26 10 4. 5 27. 51 27. 79 28. 05 30. 1 29. 71 30. 34 26. 65 27. 56 27. 62 27. 33 27. 7 27. 42 10 2. 5 35. 53 34. 81 34. 55 37. 17 38. 03 37 34. 67 35. 17 35. 96 34. 43 35. 59 35. 22 H 5 VC 25. 64 31. 13 34. 33 31. 76

Recommendations for H 5 Nx influenza testing The recommendations for testing for suspect novel influenza cases, including suspect human infection with clade 2. 3. 4. 4 HPAI viruses such as H 5 N 2 and H 5 N 8, remain the same as in the current guidance outlined in the CDC Flu Dx Panel package insert. Ø Testing should be considered in suspect human cases with a recent history of contact with poultry (e. g. poultry farm, household raising poultry, or bird market) either in the United States or during travel to a country with documented HPAI avian influenza in poultry and/or humans. Ø

Confirmed Human H 7 N 9 Cases as of April 20, 2016 (FAO)

Human cases of avian influenza A (H 7 N 9) Number of confirmed cases* 779 Number of fatal confirmed cases 300 * As of April, 2016

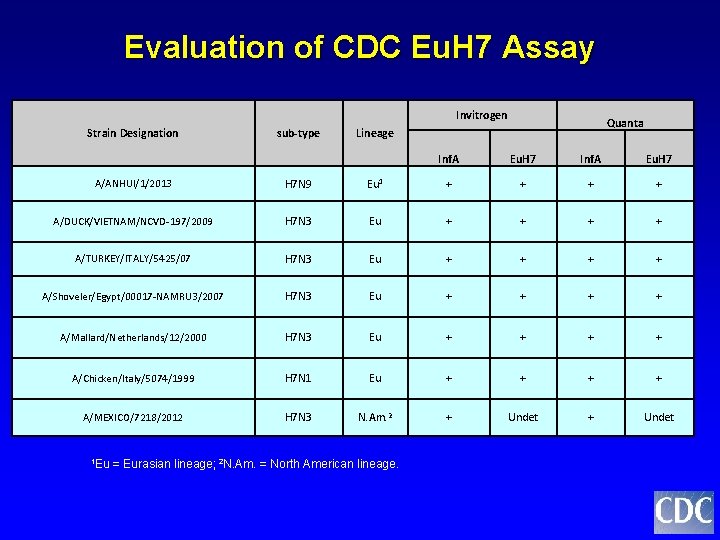
Evaluation of CDC Eu. H 7 Assay Invitrogen Strain Designation sub-type Quanta Lineage Inf. A Eu. H 7 A/ANHUI/1/2013 H 7 N 9 Eu 1 + + A/DUCK/VIETNAM/NCVD-197/2009 H 7 N 3 Eu + + A/TURKEY/ITALY/5425/07 H 7 N 3 Eu + + A/Shoveler/Egypt/00017 -NAMRU 3/2007 H 7 N 3 Eu + + A/Mallard/Netherlands/12/2000 H 7 N 3 Eu + + A/Chicken/Italy/5074/1999 H 7 N 1 Eu + + A/MEXICO/7218/2012 H 7 N 3 N. Am. 2 + Undet 1 Eu = Eurasian lineage; 2 N. Am. = North American lineage.

Human cases of “variant” Influenza infection in the United States

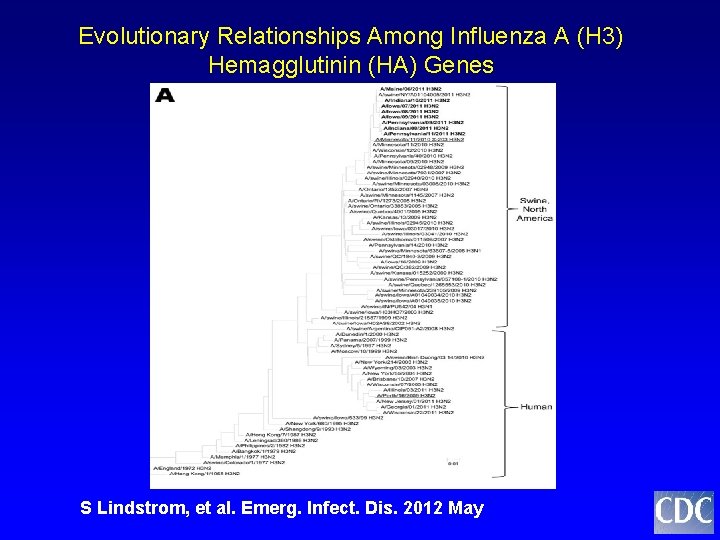
Evolutionary Relationships Among Influenza A (H 3) Hemagglutinin (HA) Genes S Lindstrom, et al. Emerg. Infect. Dis. 2012 May

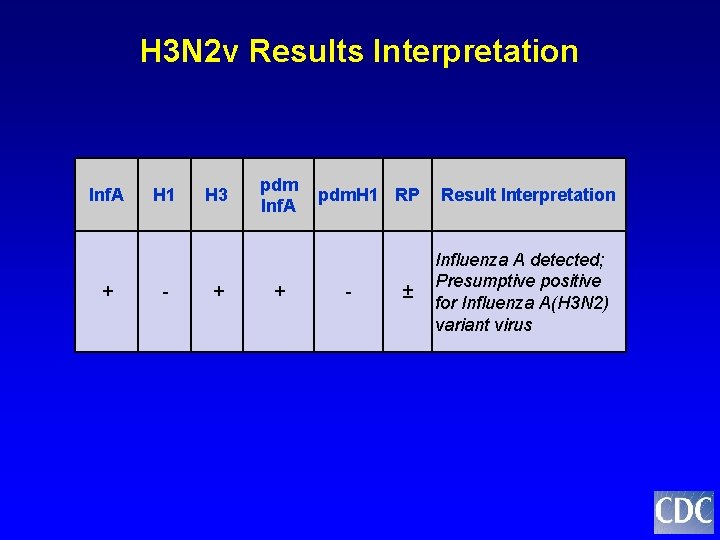
H 3 N 2 v Results Interpretation Inf. A + H 1 - H 3 + pdm. H 1 RP Inf. A + - ± Result Interpretation Influenza A detected; Presumptive positive for Influenza A(H 3 N 2) variant virus

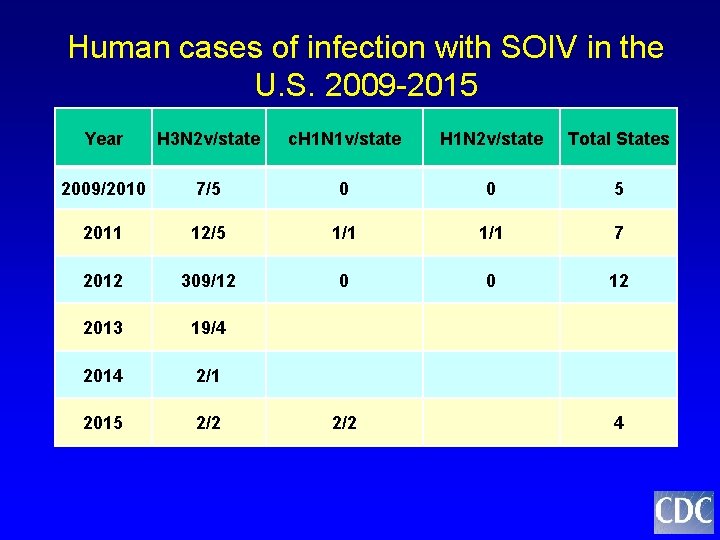
Human cases of infection with SOIV in the U. S. 2009 -2015 Year H 3 N 2 v/state c. H 1 N 1 v/state H 1 N 2 v/state Total States 2009/2010 7/5 0 0 5 2011 12/5 1/1 7 2012 309/12 0 0 12 2013 19/4 2014 2/1 2015 2/2 4

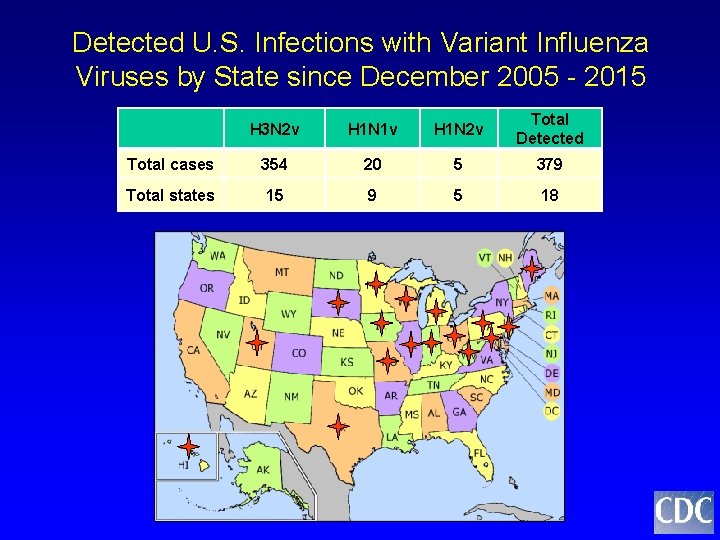
Detected U. S. Infections with Variant Influenza Viruses by State since December 2005 - 2015 H 3 N 2 v H 1 N 1 v H 1 N 2 v Total Detected Total cases 354 20 5 379 Total states 15 9 5 18

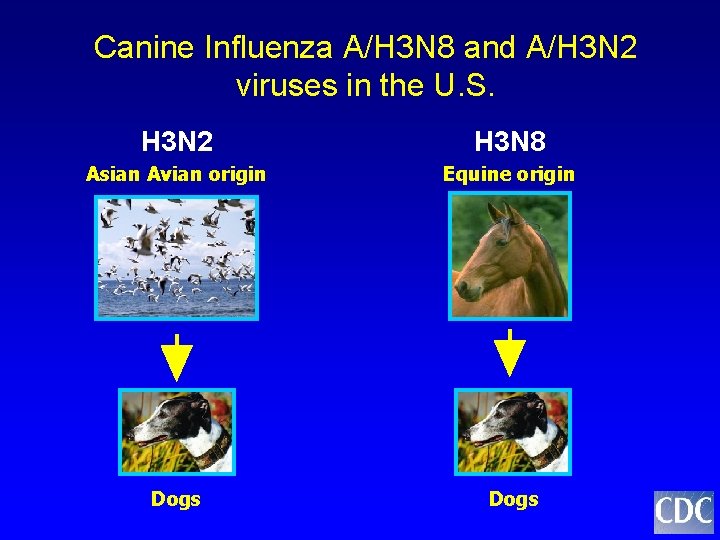
Canine Influenza A/H 3 N 8 and A/H 3 N 2 viruses in the U. S. H 3 N 2 H 3 N 8 Asian Avian origin Equine origin Dogs

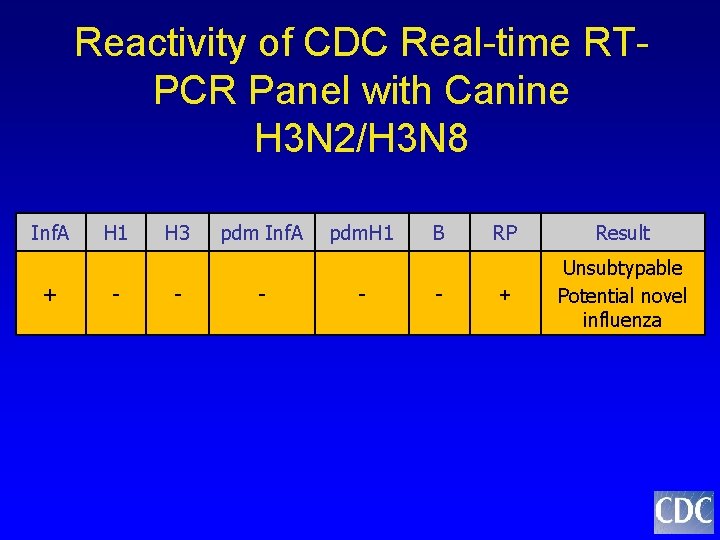
Reactivity of CDC Real-time RTPCR Panel with Canine H 3 N 2/H 3 N 8 Inf. A + H 1 - H 3 - pdm Inf. A - pdm. H 1 - B - RP Result + Unsubtypable Potential novel influenza

Support Strategies for Public Health and Research Laboratories

Reagent ordering - IRR www. influenzareagentresource. org

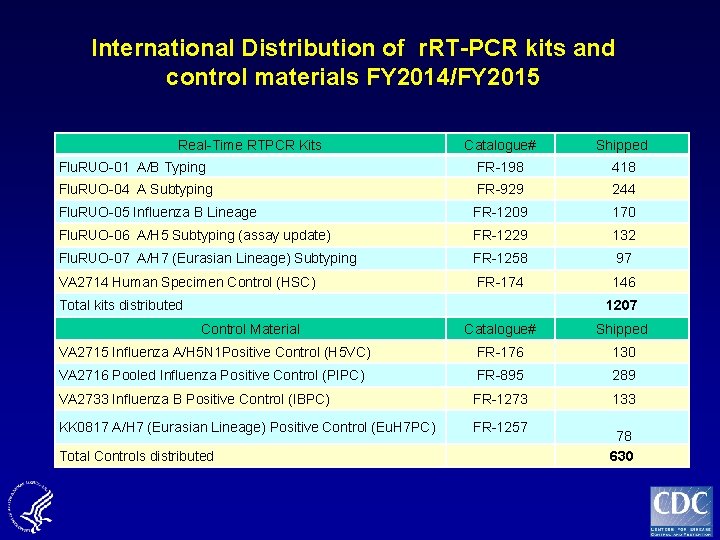
International Distribution of r. RT-PCR kits and control materials FY 2014/FY 2015 Real-Time RTPCR Kits Catalogue# Shipped Flu. RUO-01 A/B Typing FR-198 418 Flu. RUO-04 A Subtyping FR-929 244 Flu. RUO-05 Influenza B Lineage FR-1209 170 Flu. RUO-06 A/H 5 Subtyping (assay update) FR-1229 132 Flu. RUO-07 A/H 7 (Eurasian Lineage) Subtyping FR-1258 97 VA 2714 Human Specimen Control (HSC) FR-174 146 Total kits distributed 1207 Control Material Catalogue# Shipped VA 2715 Influenza A/H 5 N 1 Positive Control (H 5 VC) FR-176 130 VA 2716 Pooled Influenza Positive Control (PIPC) FR-895 289 VA 2733 Influenza B Positive Control (IBPC) FR-1273 KK 0817 A/H 7 (Eurasian Lineage) Positive Control (Eu. H 7 PC) FR-1257 133 78 630 Total Controls distributed

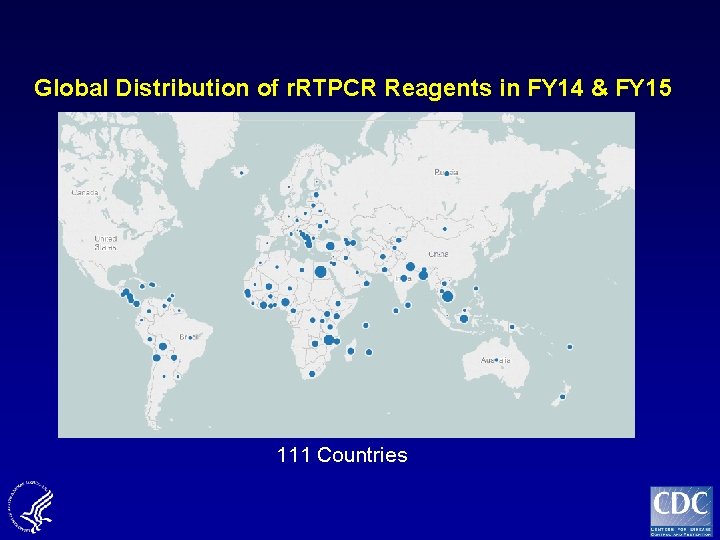
Global Distribution of r. RTPCR Reagents in FY 14 & FY 15 111 Countries

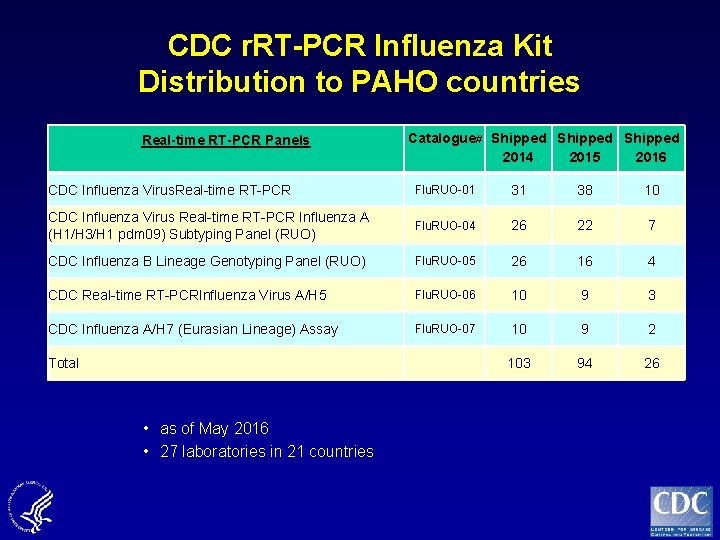
CDC r. RT-PCR Influenza Kit Distribution to PAHO countries Real-time RT-PCR Panels Catalogue# Shipped 2014 2015 2016 CDC Influenza Virus. Real-time RT-PCR Flu. RUO-01 31 38 10 CDC Influenza Virus Real-time RT-PCR Influenza A (H 1/H 3/H 1 pdm 09) Subtyping Panel (RUO) Flu. RUO-04 26 22 7 CDC Influenza B Lineage Genotyping Panel (RUO) Flu. RUO-05 26 16 4 CDC Real-time RT-PCRInfluenza Virus A/H 5 Flu. RUO-06 10 9 3 CDC Influenza A/H 7 (Eurasian Lineage) Assay Flu. RUO-07 10 9 2 103 94 26 Total • as of May 2016 • 27 laboratories in 21 countries

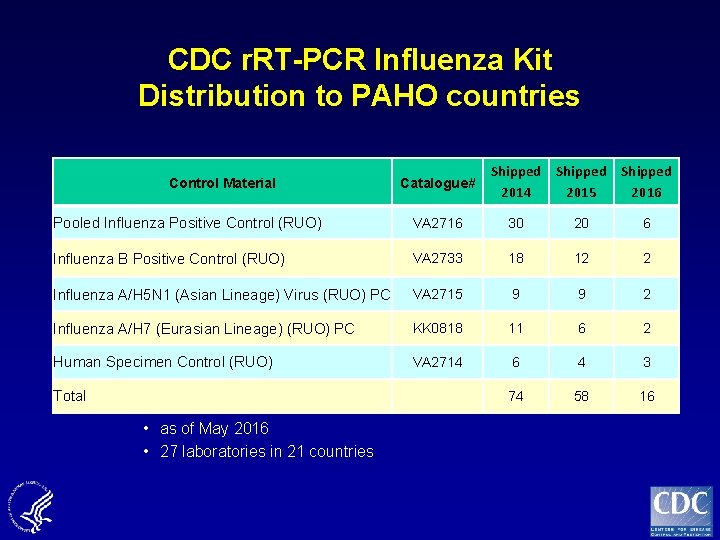
CDC r. RT-PCR Influenza Kit Distribution to PAHO countries Control Material Catalogue# Shipped 2014 2015 2016 Pooled Influenza Positive Control (RUO) VA 2716 30 20 6 Influenza B Positive Control (RUO) VA 2733 18 12 2 Influenza A/H 5 N 1 (Asian Lineage) Virus (RUO) PC VA 2715 9 9 2 Influenza A/H 7 (Eurasian Lineage) (RUO) PC KK 0818 11 6 2 Human Specimen Control (RUO) VA 2714 6 4 3 74 58 16 Total • as of May 2016 • 27 laboratories in 21 countries

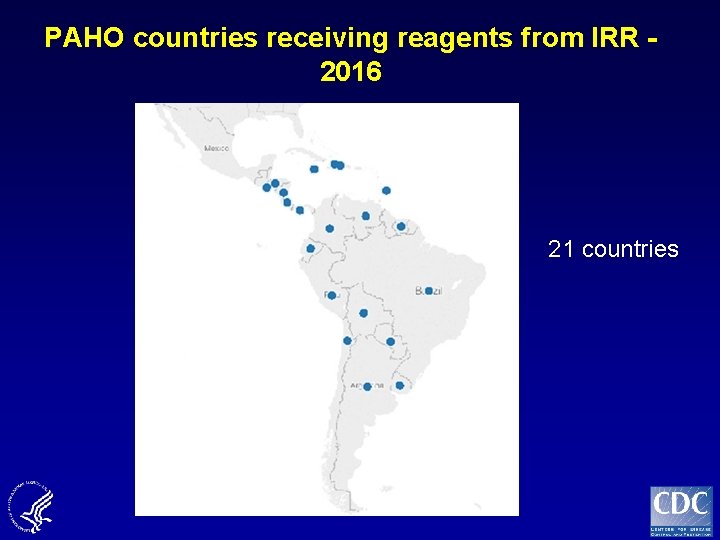
PAHO countries receiving reagents from IRR - 2016 21 countries

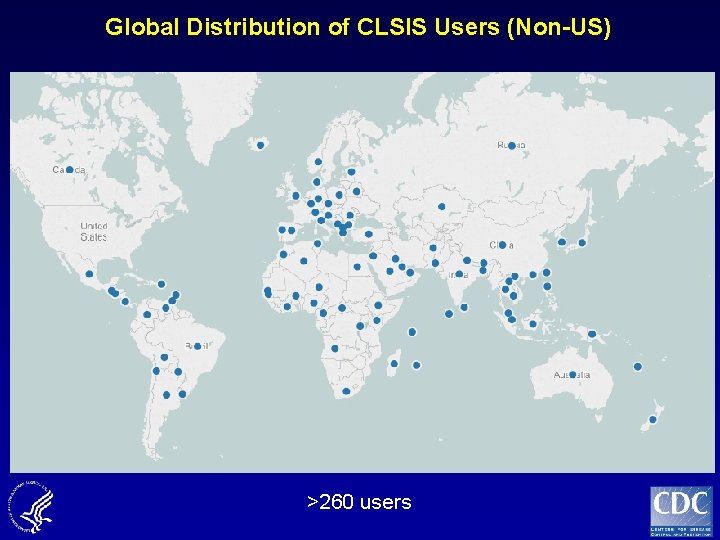
CDC Sharepoint Site for Laboratory Support for Influenza Surveillance >260 users § § § Provide public health and research laboratories access to multiple assays, procedures and methods depending on need (equipment, chemistry, etc. ) Allow for coordinated communication with registered laboratories Provide timely notification of assay updates www. cdc. gov/flu/clsis

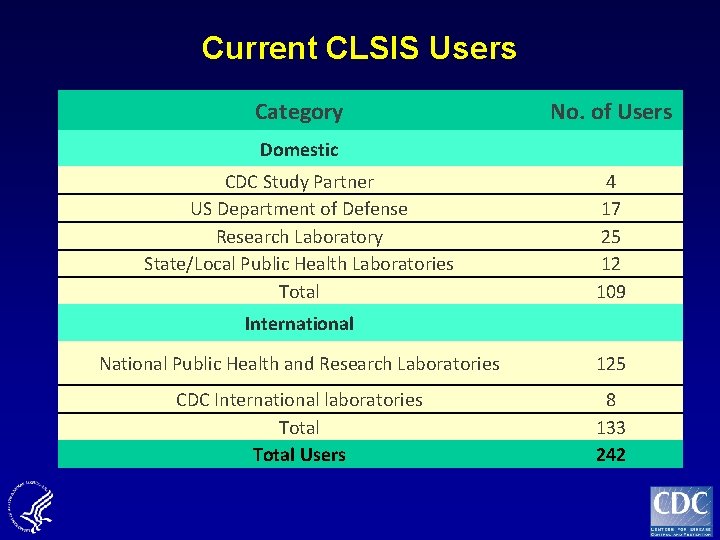
Current CLSIS Users Category No. of Users Domestic CDC Study Partner US Department of Defense Research Laboratory State/Local Public Health Laboratories Total 4 17 25 12 109 International National Public Health and Research Laboratories 125 CDC International laboratories Total Users 8 133 242

Global Distribution of CLSIS Users (Non-US) >260 users

CDC Sharepoint Site for Laboratory Support for Influenza Surveillance Real-time r. RT-PCR laboratory protocols for influenza RNA extraction procedures (manual & automated) Package inserts for kits and controls Enzyme options Equipment options Primer/probe sequence information Chemistry options for probes manufacturing (ZEN, BHQ) www. cdc. gov/flu/clsis

CDC Sharepoint Site for Laboratory Support for Influenza Surveillance UPCOMING UPDATES: § Updates to r. RTPCR procedures - Assay specific protocols for Flu. A subtyping and Flu. B genotyping. § Package insert for updated Flu. A subtyping kit (Ver 2) § Assay sequence updates to pdm. H 1 primers/probe § Flu. B YAM/VIC genoyping primer/probe sequences www. cdc. gov/flu/clsis

PAHO countries registered on CLSIS - 2016 44 registrants 19 countries

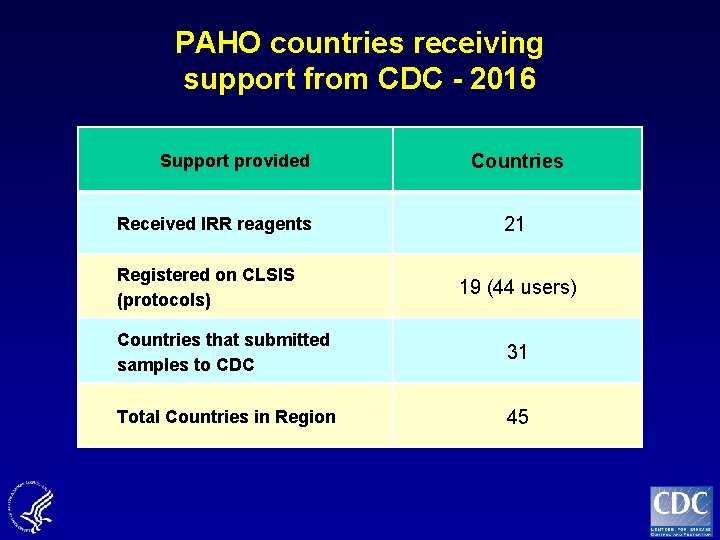
PAHO countries receiving support from CDC - 2016 Support provided Received IRR reagents Registered on CLSIS (protocols) Countries 21 19 (44 users) Countries that submitted samples to CDC 31 Total Countries in Region 45

ASSAY UPDATE Influenza A (H 3/H 1 pdm 09) Subtyping Panel (RUO) Ver 2 – pdm. H 1 assay

Reports of “unsubtypables” • March 1 – Report from Utah, Arizona & West Virginia of “Unsubtypable” specimens Inf. A H 1 H 3 pdm Inf. A pdm. H 1 B RP Result + - - + Inconclusive

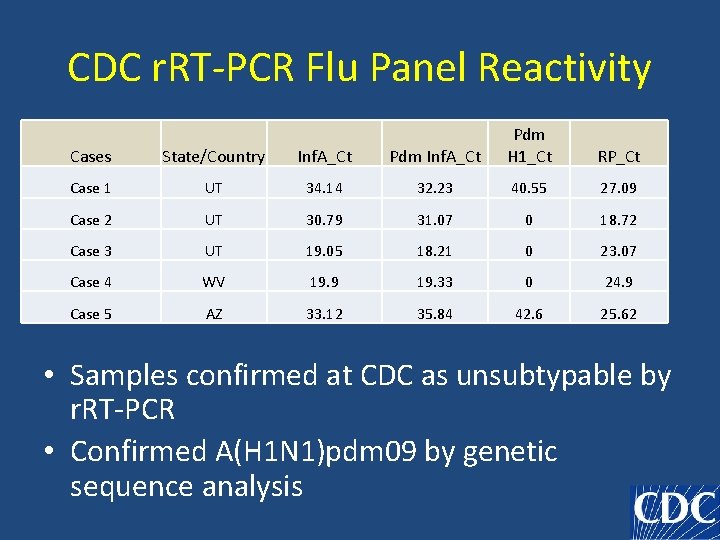
CDC r. RT-PCR Flu Panel Reactivity Cases State/Country Inf. A_Ct Pdm H 1_Ct Case 1 UT 34. 14 32. 23 40. 55 27. 09 Case 2 UT 30. 79 31. 07 0 18. 72 Case 3 UT 19. 05 18. 21 0 23. 07 Case 4 WV 19. 9 19. 33 0 24. 9 Case 5 AZ 33. 12 35. 84 42. 6 25. 62 RP_Ct • Samples confirmed at CDC as unsubtypable by r. RT-PCR • Confirmed A(H 1 N 1)pdm 09 by genetic sequence analysis

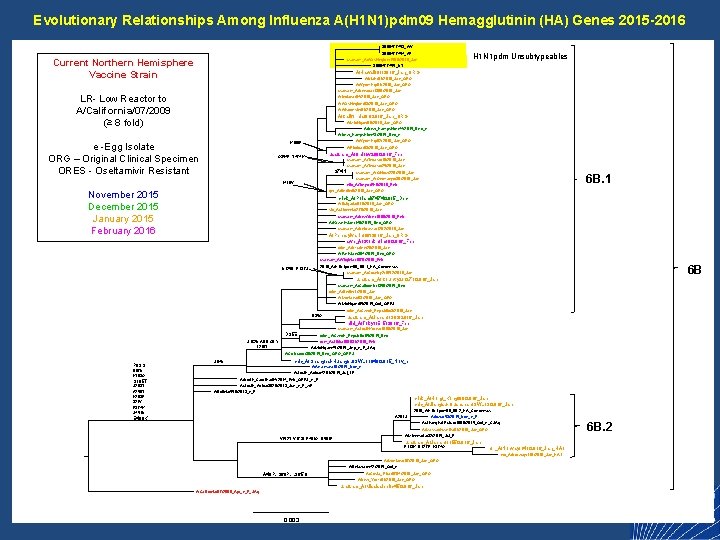
Evolutionary Relationships Among Influenza A(H 1 N 1)pdm 09 Hemagglutinin (HA) Genes 2015 -2016 3000477453_WV 3000477454_AR H 1 N 1 pdm Unsubtypeables usafsam_A/Washington/1589/2016_Jan Current Northern Hemisphere Vaccine Strain 3000477455_UT A/Hawaii/01/2016_Jan_ORG A/Utah/01/2016_Jan_ORG A/Wyoming/01/2016_Jan_ORG usafsam_A/Nevada/1369/2016_Jan LR- Low Reactor to A/California/07/2009 (≥ 8 fold) A/Indiana/04/2016_Jan_ORG A/Washington/03/2016_Jan_ORG A/Wisconsin/01/2016_Jan_ORG A/California/01/2016_Jan_ORG A/Michigan/06/2016_Jan_ORG A/New_Hampshire/44/2015_Dec_e A/New_Hampshire/43/2015_Dec_e A/Wyoming/02/2016_Jan_ORG K 160 R e -Egg Isolate ORG – Original Clinical Specimen ORES - Oseltamivir Resistant A/Florida/03/2016_Jan_ORG usafsam_A/Ohio/w 288/2016_Feb Q 354 R, T 474 K usafsam_A/Texas/x 18/2016_Jan usafsam_A/Texas/x 25/2016_Jan S 74 N usafsam_A/Ohio/w 228/2016_Jan 6 B. 1 usafsam_A/Germany/x 36/2016_Jan I 418 V niid_A/Tokyo/Eh 6/2016_Feb ger_A/Berlin/6/2016_Jan_ORG November 2015 December 2015 January 2015 February 2016 crick_A/Poland/7474/2015_Dec A/Bulgaria/010/2016_Jan_ORG slo_A/Slovenia/278/2016_Jan usafsam_A/New. York/1680/2016_Feb A/Kazakhstan/146/2015_Dec_ORG usafsam_A/Nebraska/1292/2016_Jan A/Pennsylvania/06/2016_Jan_ORG swe_A/Stockholm/8/2016_Feb crick_A/Ukraine/20/2016_Jan A/Pakistan/394/2015_Dec_ORG usafsam_A/Virginia/1679/2016_Feb 6 B 2016_A/H 1 N 1 pdm 09_6 B. 1_HA_Concensus N 125 D, P 137 S usafsam_A/Country 2/1652/2016_Jan usafsam_A/Country 2/1271/2016_Jan usafsam_A/California/1350/2015_Dec crick_A/Berlin/1/2016_Jan A/Montana/03/2016_Jan_ORG A/Michigan/65/2015_Oct_ORES crick_A/Czech_Republic/3/2016_Jan D 35 G usafsam_A/Japan/1393/2016_Jan niid_A/Tokyo/Eh 5/2016_Feb usafsam_A/South. Korea/1688/2016_Jan D 35 E crick_A/Czech_Republic/95/2015_Dec rom_A/Sibiu/190931/2016_Feb S 162 N ADD GLY I 216 T A/Michigan/45/2015_Sep_e_F_SAg A/Colorado/30/2015_Dec_ORG_ORES P 83 S D 97 N K 163 Q S 185 T S 203 T A 256 T K 283 E 321 V E 374 K S 451 N cnic_A/Shanghai-Huangpu/SWL 11940/2015_Nov_e S 84 N A/Arkansas/10/2015_Nov_e A/South_Africa/4291/2015_Jul_LR A/North_Carolina/04/2014_Feb_ORES_e_F A/South_Africa/3626/2013_Jun_e_F_w. F A/Bolivia/559/2013_e_F crick_A/Hong_Kong/89/2016_Jan cnic_A/Jiangsu-Quanshan/SWL 13/2016_Jan 2016_A/H 1 N 1 pdm 09_6 B. 2_HA_Concensus A 261 S E 499 K A/Iowa/53/2015_Nov_e_F 6 B. 2 A/Shanghai-Putuo/1860/2015_Oct_e_CSAg A/Massachusetts/01/2016_Jan_ORG A/Minnesota/32/2015_Jul_F V 152 T, V 173 I, E 491 G, D 501 E usafsam_A/Japan/1155/2016_Jan R 113 K, D 127 E, K 374 Q nor_A/Norway/141/2016_Jan_HA 1 nor_A/Norway/116/2016_Jan_HA 1 A/Montana/07/2016_Jan_ORG A/Delaware/42/2015_Oct_e A 48 P, S 69 P, L 365 Q A/Costa_Rica/0784/2016_Jan_ORG A/New_York/01/2016_Jan_ORG usafsam_A/Mississippi/x 45/2016_Jan A/California/07/2009_Apr_e_F_SAg 0. 003

Primer/probe sequence identities to unsub A(H 1 N 1)pdm 09 pdm. H 1 Primer. Probes Updated pdm. H 1 Prime A/California/07/200 UT 3000477455_4(Neg) AR 3000416491_4(Neg) AR 3000477454_4(Neg) WV 3000416492_4(Neg) WV 3000416493_4(Neg) WV 3000416494_4(Neg) WV 3000416495_4(Neg) 900 910 920 930 940 950 960 970 980 990 1000 1010 1020. . |. . . . |. . For-GTGCTATAAACACCAGCCTCCCATT---Pro-ATACATCCGATCACMATTGGAAAATGTCC------------------CAGGATTGAGGAATRTCCCGTCT-Rev ----------------. . . . R. . . A. . . R. . . . AAA----------------. . . G. . R AA. G. . . TCAG. AT. . . . AAAATATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. AT. . . . A. . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GA. G. . . . . . . TCAG. AT. . . . A. . . . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. AT. . . . A. . . . G. . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. . -. . .

Announcement Broadcast to US Public Health Labs (March 30) • A(H 1 N 1)pdm 09 viruses with specific substitutions are identified as influenza A unsubtypable with the CDC Flu r. RT-PCR Dx Panel with Inf. A assay Ct values less than 35. • Public health laboratories are reminded that specimens with influenza A unsubtypable results with Inf. A Ct values less than 35 should be immediately sent to CDC for further characterization. • Specimens that are identified as influenza A(H 1 N 1)pdm 09, but show a discrepancy of greater than 5 Ct between the Inf. A and pdm. H 1 subtyping assay may be observed, and may also be submitted for further characterization. • Please do NOT send diagnostic specimens to your respective designated influenza surveillance reference center.

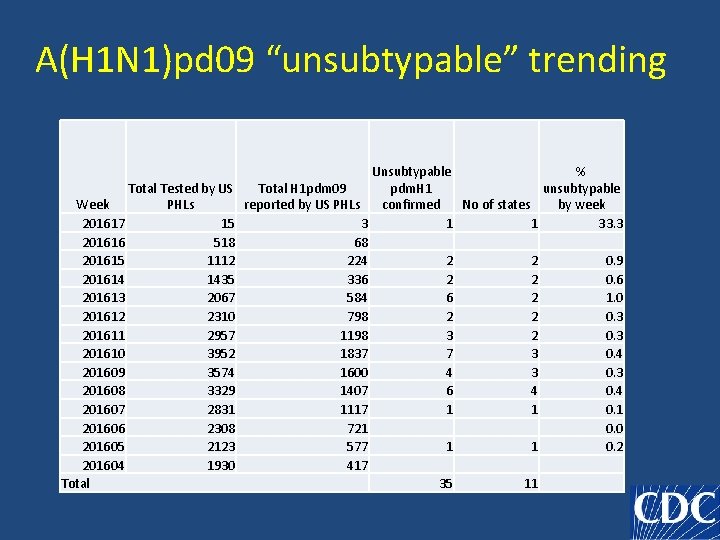
A(H 1 N 1)pdm 09 “unsubtypable” cases in US • 35 cases • 11 states • As of May 2, 216

A(H 1 N 1)pd 09 “unsubtypable” trending Total Tested by US Week PHLs 201617 15 201616 518 201615 1112 201614 1435 201613 2067 201612 2310 201611 2957 201610 3952 201609 3574 201608 3329 201607 2831 201606 2308 201605 2123 201604 1930 Total Unsubtypable % Total H 1 pdm 09 pdm. H 1 unsubtypable reported by US PHLs confirmed No of states by week 3 1 1 33. 3 68 224 2 2 0. 9 336 2 2 0. 6 584 6 2 1. 0 798 2 2 0. 3 1198 3 2 0. 3 1837 7 3 0. 4 1600 4 3 0. 3 1407 6 4 0. 4 1117 1 1 0. 1 721 0. 0 577 1 1 0. 2 417 35 11

Primer/probe sequence identities to unsubtypable A(H 1 N 1)pdm 09 pdm. H 1 Primer. Probes Updated pdm. H 1 Prime A/California/07/200 UT 3000477456_4(Neg) MN 3000477928 -4(Neg) MN 3000477929_4(Neg) MN 3000477739_4(Neg) MN 3000477955_4(Neg) MN 3000477956_4(Neg) UT 3000477455_4(Neg) AR 3000416491_4(Neg) AR 3000477454_4(Neg) TN 3000477508_4(Neg) TN 3000477509_4(Neg) TN 3000477511_4(Neg) TN 3000477512_4(Neg) WV 3000416493_4(Neg) WV 3000416494_4(Neg) WV 3000416495_4(Neg) WV 3000477453_4(Neg) TN 3000477748_4(Neg) DE 3000478049_4(Neg) MN 3000477919_4(Late MN 3000477920_4(Late MN 3000477921_4(Late UT 3000477742_4(Late BR 3000418142_4(Late BR 3000478143_4(Late BR 3000478144_4(Late Br 3000478145_4(Late BR 3000478146_4(Late 900 910 920 930 940 950 960 970 980 990 1000 1010 1020. . |. . . . |. . For-GTGCTATAAACACCAGCCTCCCATT---Pro-ATACATCCGATCACMATTGGAAAATGTCC------------------CAGGATTGAGGAATRTCCCGTCT-Rev ----------------. . . . R. . . A. . . R. . . . AAA----------------. . . G. . R AA. G. . . TCAG. AT. . . . AAAATATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. AT. . . . A. . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GA. G. . . . . . . TCAG. AT. . . . A. . . G. . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. AT. . . . A. . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . . . . . . . ATT. GA. G. . . . . . . . . . . . . TCAG. AT. . . . A. . A. . . . G. . . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GA. G. . . . . . . TCAG. AT. . . . A. . . . . G. . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. AT. . . . A. . . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GA. G. . . . . . . TCAG. AT. . . . A. . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GA. G. . . . . . . TCAG. ATG. . . . A. . . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GA. G. . . TCAG. ATG. . . . A. . . G. . . . AAAGTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. . -. . .

H 1 pdm Ver 2 primer/probe sequence identities to unsub A(H 1 N 1)pdm 09 900 910 pdm. H 1 Primer. Probes Updated pdm. H 1 Prime A/California/07/200 UT 3000477456_4(Neg) MN 3000477928 -4(Neg) MN 3000477929_4(Neg) MN 3000477739_4(Neg) MN 3000477955_4(Neg) MN 3000477956_4(Neg) UT 3000477455_4(Neg) AR 3000416491_4(Neg) AR 3000477454_4(Neg) TN 3000477508_4(Neg) TN 3000477509_4(Neg) TN 3000477511_4(Neg) TN 3000477512_4(Neg) WV 3000416493_4(Neg) WV 3000416494_4(Neg) WV 3000416495_4(Neg) WV 3000477453_4(Neg) TN 3000477748_4(Neg) DE 3000478049_4(Neg) MN 3000477919_4(Late MN 3000477920_4(Late MN 3000477921_4(Late UT 3000477742_4(Late BR 3000418142_4(Late BR 3000478143_4(Late BR 3000478144_4(Late Br 3000478145_4(Late BR 3000478146_4(Late 920 930 940 950 960 970 980 990 1000 1010 1020. . |. . . . |. . For-GTGCTATAAACACCAGCCTCCCATT---. . . G. . . M. . . A. . . . ------------------. . . R. . G. . . ----------------- Pro-ATACATCCRATCACAATTGGRAAATGTCCAAA---------------- CAGGATTGAGGAATGTCCCRTCT-Rev AAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . ATATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . . . G. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . G. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . G. . . . . . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . . . . . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . . . A. . . . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . A. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . A. . . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. AT. . . . . . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . . . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. ATG. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT. GAGGGTGCTATAAACACCAGCCTCCCATTTCAG. ATG. . . . GTATGTAAAAAGCACAAAATTGAGACTGGCCA. . . ATT

Update pdm. H 1 assay probe & reverse primer Confirm design and performance Assay qualification testing Consult with FDA re qualification requirements • Manufacturing qualification and QC • Performance validation testing • •

Update pdm. H 1 assay - Validation • Limit of detection – “range finding” 5 replicate Lo. D – 20 replicate Lo. D • Analytical reactivity – Inclusivity • Clinical performance – >50 negative specimens – >30 positive specimens • Reproducibility/precision

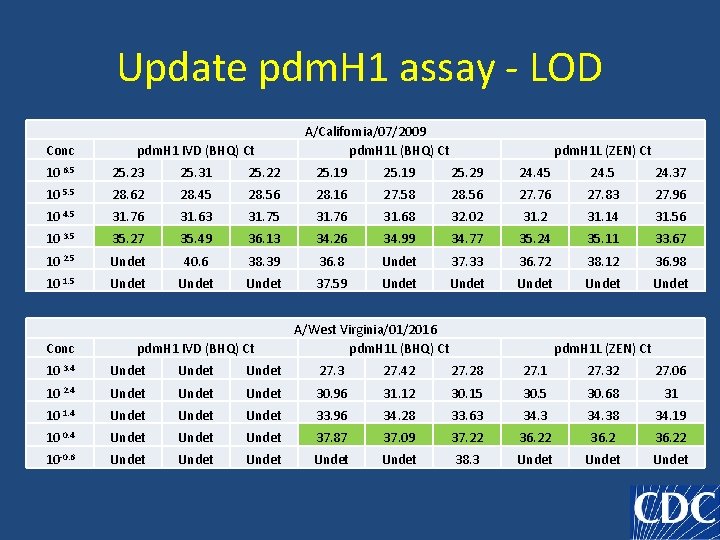
Update pdm. H 1 assay - LOD Conc pdm. H 1 IVD (BHQ) Ct A/California/07/2009 pdm. H 1 L (BHQ) Ct pdm. H 1 L (ZEN) Ct 10 6. 5 25. 23 25. 31 25. 22 25. 19 25. 29 24. 45 24. 37 10 5. 5 28. 62 28. 45 28. 56 28. 16 27. 58 28. 56 27. 76 27. 83 27. 96 10 4. 5 31. 76 31. 63 31. 75 31. 76 31. 68 32. 02 31. 14 31. 56 10 3. 5 35. 27 35. 49 36. 13 34. 26 34. 99 34. 77 35. 24 35. 11 33. 67 10 2. 5 Undet 40. 6 38. 39 36. 8 Undet 37. 33 36. 72 38. 12 36. 98 10 1. 5 Undet 37. 59 Undet Undet Conc pdm. H 1 IVD (BHQ) Ct A/West Virginia/01/2016 pdm. H 1 L (BHQ) Ct pdm. H 1 L (ZEN) Ct 10 3. 4 Undet 27. 3 27. 42 27. 28 27. 1 27. 32 27. 06 10 2. 4 Undet 30. 96 31. 12 30. 15 30. 68 31 10 1. 4 Undet 33. 96 34. 28 33. 63 34. 38 34. 19 10 0. 4 Undet 37. 87 37. 09 37. 22 36. 22 10 -0. 6 Undet Undet 38. 3 Undet

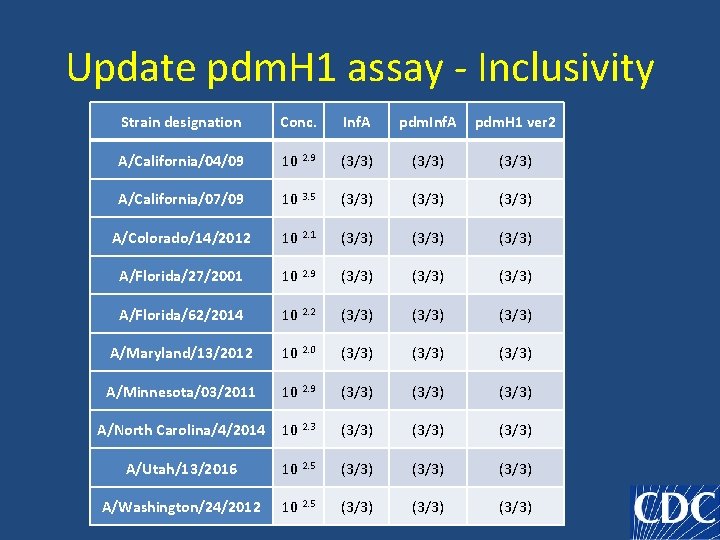
Update pdm. H 1 assay - Inclusivity Strain designation Conc. Inf. A pdm. H 1 ver 2 A/California/04/09 10 2. 9 (3/3) A/California/07/09 10 3. 5 (3/3) A/Colorado/14/2012 10 2. 1 (3/3) A/Florida/27/2001 10 2. 9 (3/3) A/Florida/62/2014 10 2. 2 (3/3) A/Maryland/13/2012 10 2. 0 (3/3) A/Minnesota/03/2011 10 2. 9 (3/3) A/North Carolina/4/2014 10 2. 3 (3/3) A/Utah/13/2016 10 2. 5 (3/3) A/Washington/24/2012 10 2. 5 (3/3)

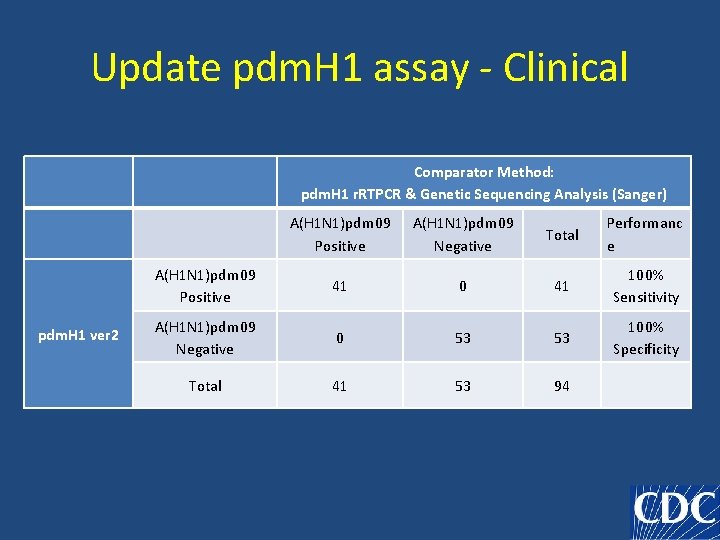
Update pdm. H 1 assay - Clinical Comparator Method: pdm. H 1 r. RTPCR & Genetic Sequencing Analysis (Sanger) pdm. H 1 ver 2 A(H 1 N 1)pdm 09 Positive A(H 1 N 1)pdm 09 Negative Total Performanc e A(H 1 N 1)pdm 09 Positive 41 0 41 100% Sensitivity A(H 1 N 1)pdm 09 Negative 0 53 53 100% Specificity Total 41 53 94

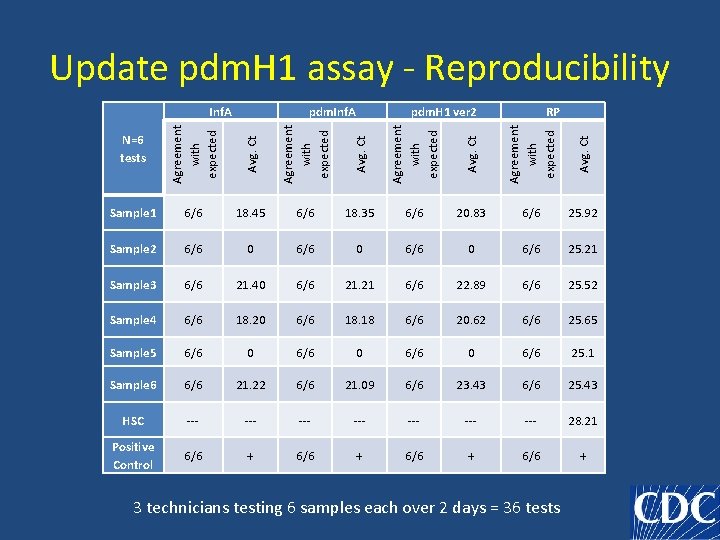
Update pdm. H 1 assay - Reproducibility N=6 tests Avg. Ct Agreement with expected Avg. Ct RP Agreement with expected pdm. H 1 ver 2 Avg. Ct pdm. Inf. A Agreement with expected Inf. A Sample 1 6/6 18. 45 6/6 18. 35 6/6 20. 83 6/6 25. 92 Sample 2 6/6 0 6/6 25. 21 Sample 3 6/6 21. 40 6/6 21. 21 6/6 22. 89 6/6 25. 52 Sample 4 6/6 18. 20 6/6 18. 18 6/6 20. 62 6/6 25. 65 Sample 5 6/6 0 6/6 25. 1 Sample 6 6/6 21. 22 6/6 21. 09 6/6 23. 43 6/6 25. 43 HSC --- --- 28. 21 Positive Control 6/6 + 3 technicians testing 6 samples each over 2 days = 36 tests

CDC Influenza Virus Real-time RT-PCR Influenza A (H 3/H 1 pdm 09) Subtyping Panel (RUO) Ver 2 • Rebuild current inventory of RUO subtyping kits (>300) in distribution - available mid-June • Remove H 1 (pre-2009) assay from panel • Announcement to be sent to users through CLISIS, IRR contact information and WHO

Diagnostics Development Team § § § Christine Warnes Kai-Hui Wu Nathelia (Tiki) Barnes Shannon Emery Ji Liu § § § Bo Shu Lashondra Berman Brian Lynch Lakshmi Malapati Marisela Rodriguez

Acknowledgements EPB: Lenee Blanton Desiree Mustaquim OD: Becky Garten Battelle Team: Stephen Burke (Project Manager) Jennifer Wess (Manufacturing) Janna Murray (IVD Product support) Chantelle Runnels (Quality Control) Yan Zheng (Quality Control) April Goodrich (Quality Assurance) DSR: Jan Pahl Nicky Sulaiman Kanwar Bedi Dennis Bagarozzi No Man island

Thank You