Unit 3 Enzymes Catalysis and enzyme kinetics OUTLINE

![3. 3. ENZYME KINETICS Steady-state Concentration Under experimental conditions [S]>>>[E]. The [ES] quickly reaches 3. 3. ENZYME KINETICS Steady-state Concentration Under experimental conditions [S]>>>[E]. The [ES] quickly reaches](https://slidetodoc.com/presentation_image_h2/07058868e10e95d868ac92ec781a0c43/image-26.jpg)
![3. 3. ENZYME KINETICS d[ ES ] = 0, dt so k 1 [ 3. 3. ENZYME KINETICS d[ ES ] = 0, dt so k 1 [](https://slidetodoc.com/presentation_image_h2/07058868e10e95d868ac92ec781a0c43/image-27.jpg)

- Slides: 56

Unit 3 Enzymes. Catalysis and enzyme kinetics.

OUTLINE 3. 1. Characteristics of biological catalysts. Coenzymes, cofactors, vitamins Enzyme nomenclature and classification 3. 2. Enzyme catalysis. Transition state Active site Enzyme-substrate complex Factors involved in enzyme catalysis 3. 3. Enzyme kinetics. Steady-state assumption and Michaelis-Menten equation Factors affecting the enzymatic activity Enzymatic inhibition • Reversible inhibition • Irreversible inhibition 3. 4. Enzyme regulation. Allosteric behaviour Covalent modification Proteolysis

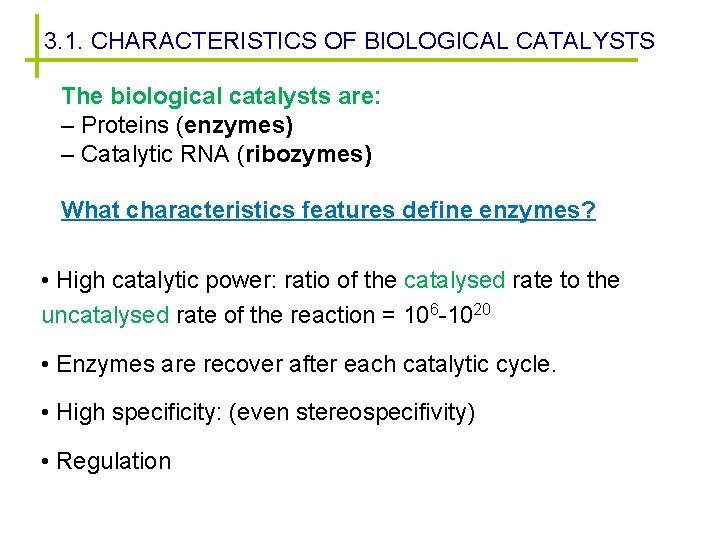
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS The biological catalysts are: – Proteins (enzymes) – Catalytic RNA (ribozymes) What characteristics features define enzymes? • High catalytic power: ratio of the catalysed rate to the uncatalysed rate of the reaction = 106 -1020 • Enzymes are recover after each catalytic cycle. • High specificity: (even stereospecifivity) • Regulation

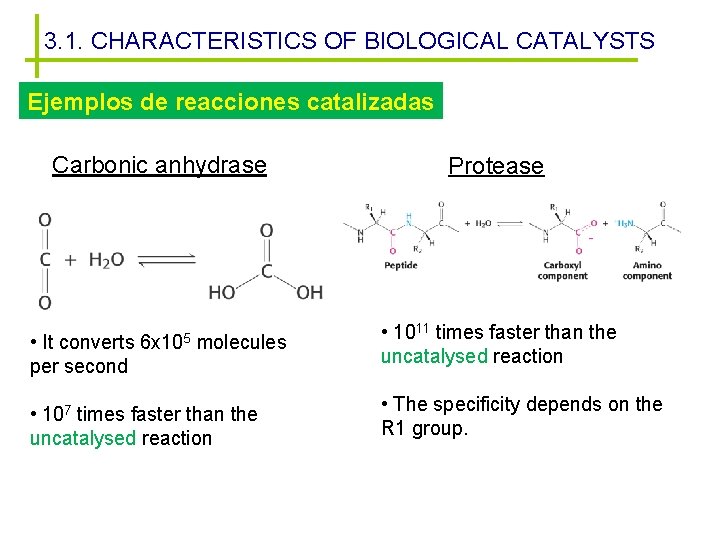
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS Ejemplos de reacciones catalizadas Carbonic anhydrase • It converts per second 6 x 105 molecules • 107 times faster than the uncatalysed reaction Protease • 1011 times faster than the uncatalysed reaction • The specificity depends on the R 1 group.

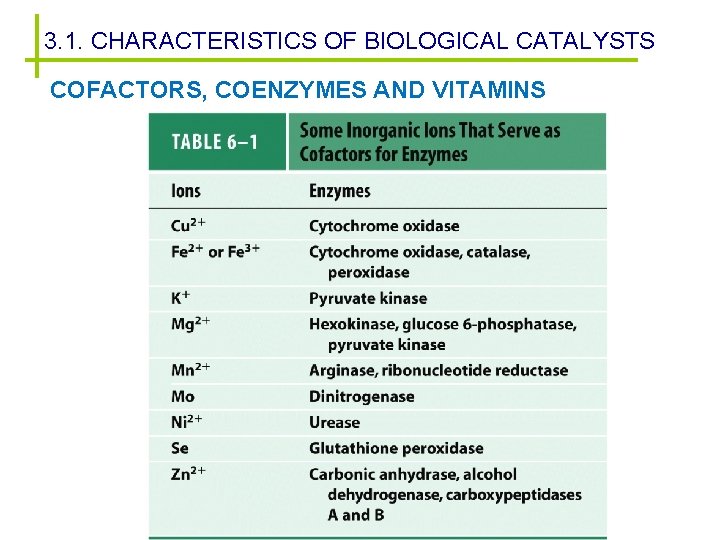
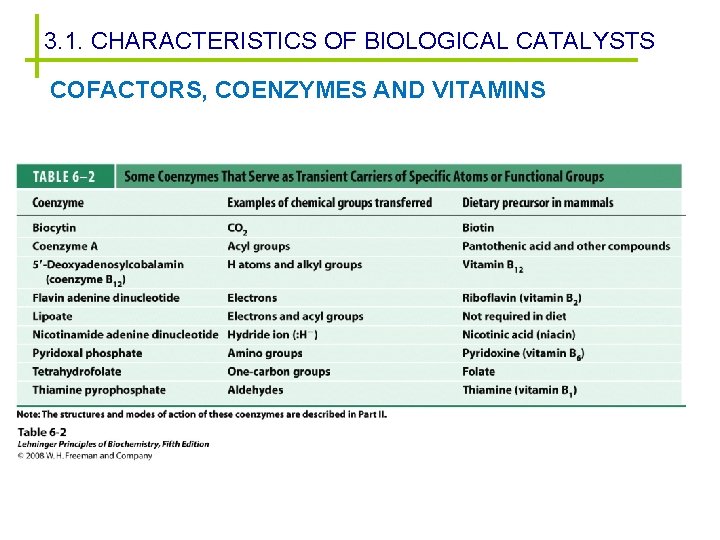
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS COFACTORS, COENZYMES AND VITAMINS Nonprotein components required for the enzymatic activity: cofactor – Apoenzyme + cofactor = holoenzyme – Two types of cofactors: • Metal ions: Mg 2+, Zn 2+, Cu 2+, Mn 2+, . . . • Coenzymes: small organic molecules synthesised from vitamins. Prosthetic groups: tightly bound coenzymes Cofactors deficiency promotes some health problems.

3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS COFACTORS, COENZYMES AND VITAMINS

3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS COFACTORS, COENZYMES AND VITAMINS

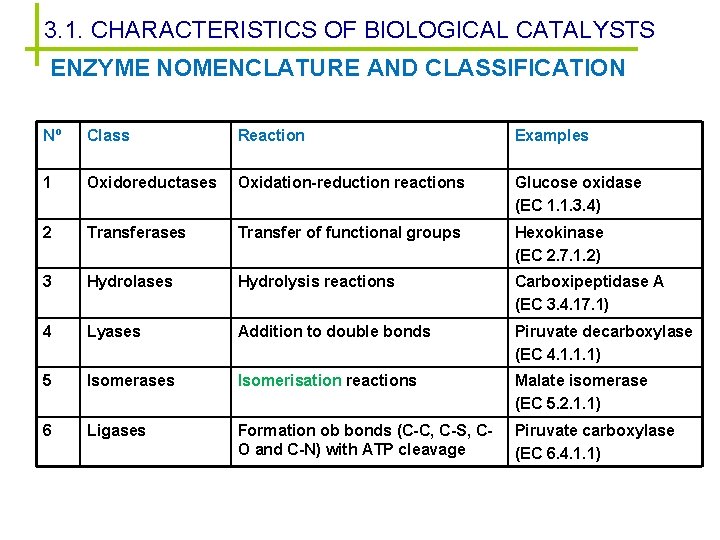
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS ENZYME NOMENCLATURE AND CLASSIFICATION Nº Class Reaction Examples 1 Oxidoreductases Oxidation-reduction reactions Glucose oxidase (EC 1. 1. 3. 4) 2 Transferases Transfer of functional groups Hexokinase (EC 2. 7. 1. 2) 3 Hydrolases Hydrolysis reactions Carboxipeptidase A (EC 3. 4. 17. 1) 4 Lyases Addition to double bonds Piruvate decarboxylase (EC 4. 1. 1. 1) 5 Isomerases Isomerisation reactions Malate isomerase (EC 5. 2. 1. 1) 6 Ligases Formation ob bonds (C-C, C-S, CO and C-N) with ATP cleavage Piruvate carboxylase (EC 6. 4. 1. 1)

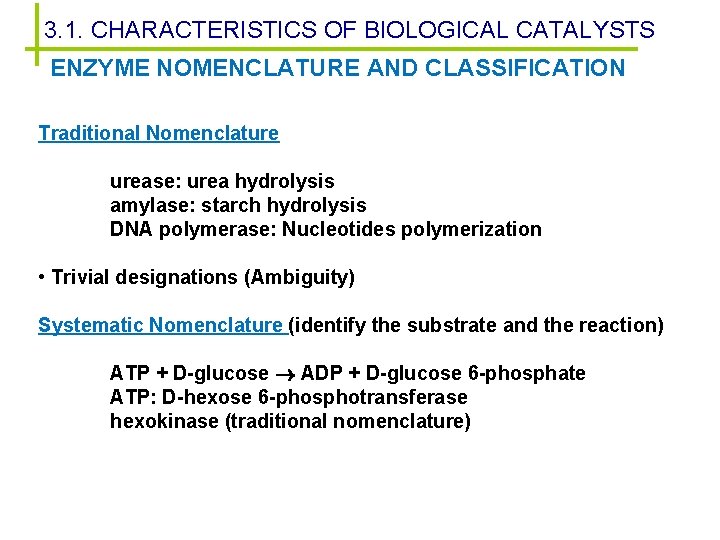
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS ENZYME NOMENCLATURE AND CLASSIFICATION Traditional Nomenclature urease: urea hydrolysis amylase: starch hydrolysis DNA polymerase: Nucleotides polymerization • Trivial designations (Ambiguity) Systematic Nomenclature (identify the substrate and the reaction) ATP + D-glucose ADP + D-glucose 6 -phosphate ATP: D-hexose 6 -phosphotransferase hexokinase (traditional nomenclature)

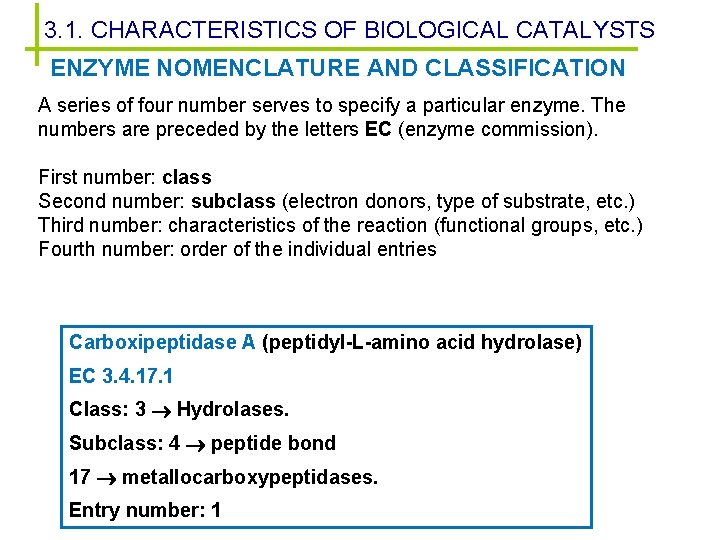
3. 1. CHARACTERISTICS OF BIOLOGICAL CATALYSTS ENZYME NOMENCLATURE AND CLASSIFICATION A series of four number serves to specify a particular enzyme. The numbers are preceded by the letters EC (enzyme commission). First number: class Second number: subclass (electron donors, type of substrate, etc. ) Third number: characteristics of the reaction (functional groups, etc. ) Fourth number: order of the individual entries Carboxipeptidase A (peptidyl-L-amino acid hydrolase) EC 3. 4. 17. 1 Class: 3 Hydrolases. Subclass: 4 peptide bond 17 metallocarboxypeptidases. Entry number: 1

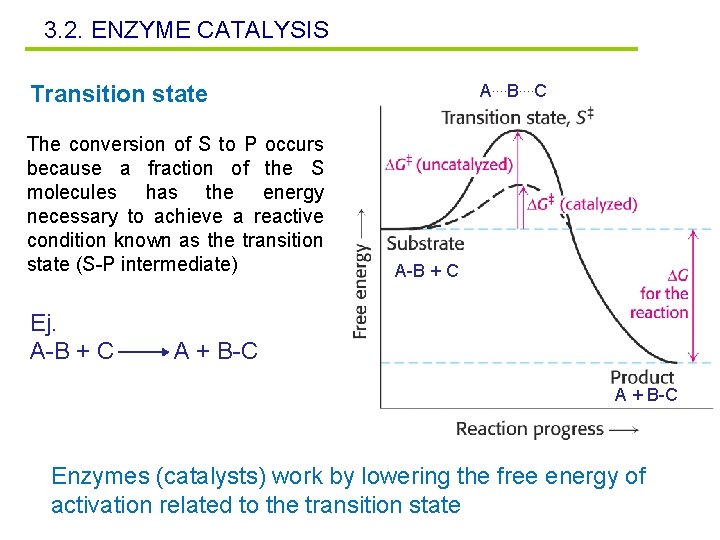
3. 2. ENZYME CATALYSIS Transition state The conversion of S to P occurs because a fraction of the S molecules has the energy necessary to achieve a reactive condition known as the transition state (S-P intermediate) Ej. A-B + C A…. B…. C A-B + C A + B-C Enzymes (catalysts) work by lowering the free energy of activation related to the transition state

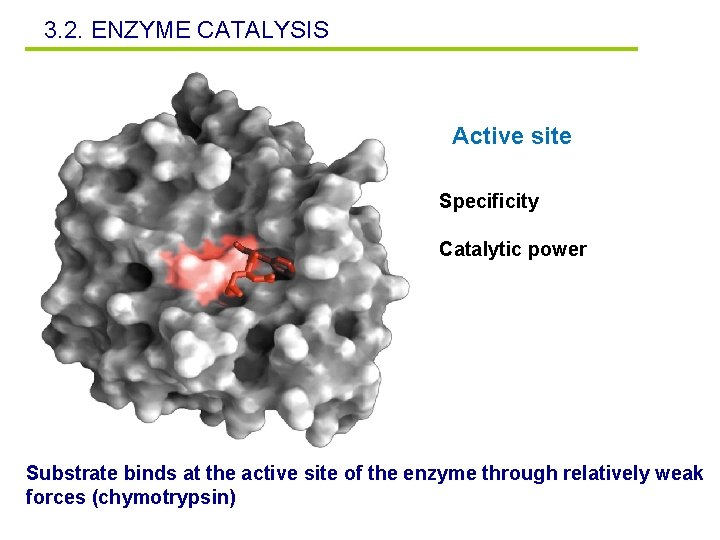
3. 2. ENZYME CATALYSIS Active site Specificity Catalytic power Substrate binds at the active site of the enzyme through relatively weak forces (chymotrypsin)

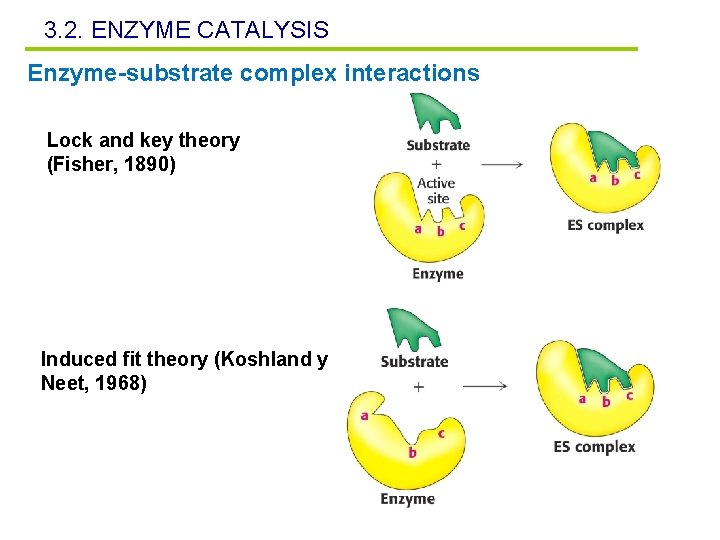
3. 2. ENZYME CATALYSIS Enzyme-substrate complex interactions Lock and key theory (Fisher, 1890) Induced fit theory (Koshland y Neet, 1968)

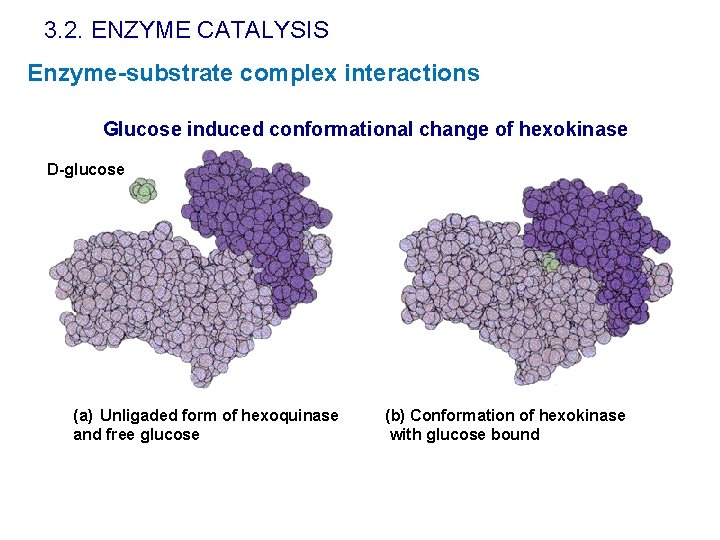
3. 2. ENZYME CATALYSIS Enzyme-substrate complex interactions Glucose induced conformational change of hexokinase D-glucose (a) Unligaded form of hexoquinase and free glucose (b) Conformation of hexokinase with glucose bound

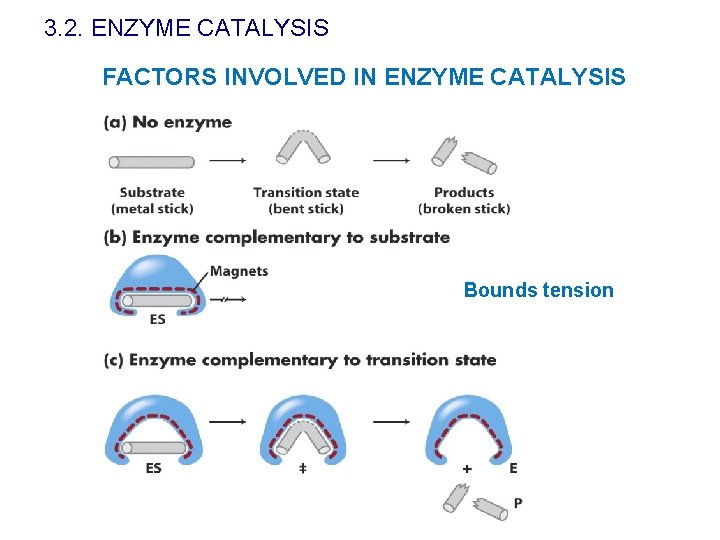
3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS • Proximity and orientation • Surface phenomena • Bounds tension • Presence of reactive groups

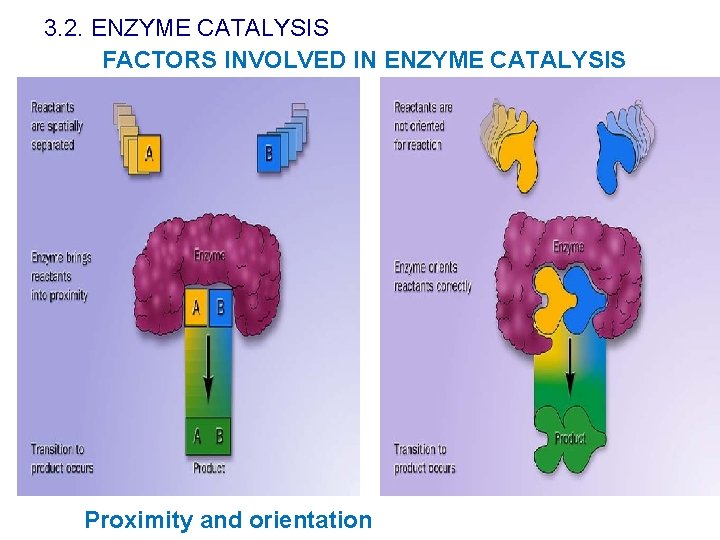
3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS Proximity and orientation

3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS Bounds tension

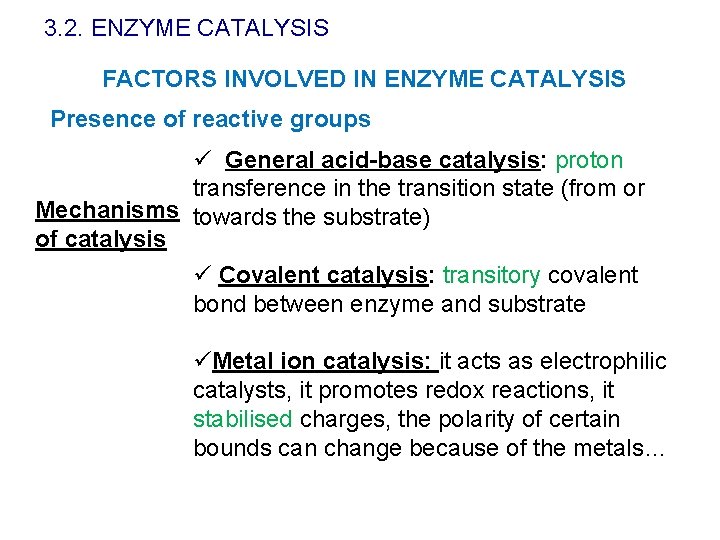
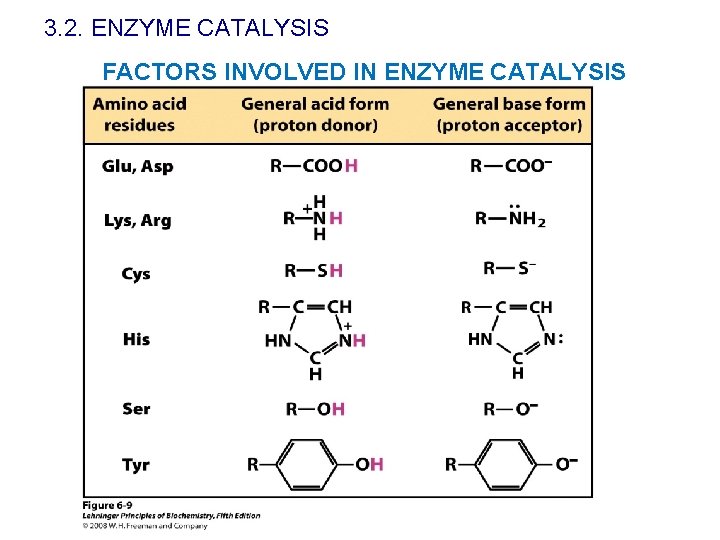
3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS Presence of reactive groups ü General acid-base catalysis: proton transference in the transition state (from or Mechanisms towards the substrate) of catalysis ü Covalent catalysis: transitory covalent bond between enzyme and substrate üMetal ion catalysis: it acts as electrophilic catalysts, it promotes redox reactions, it stabilised charges, the polarity of certain bounds can change because of the metals…

3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS

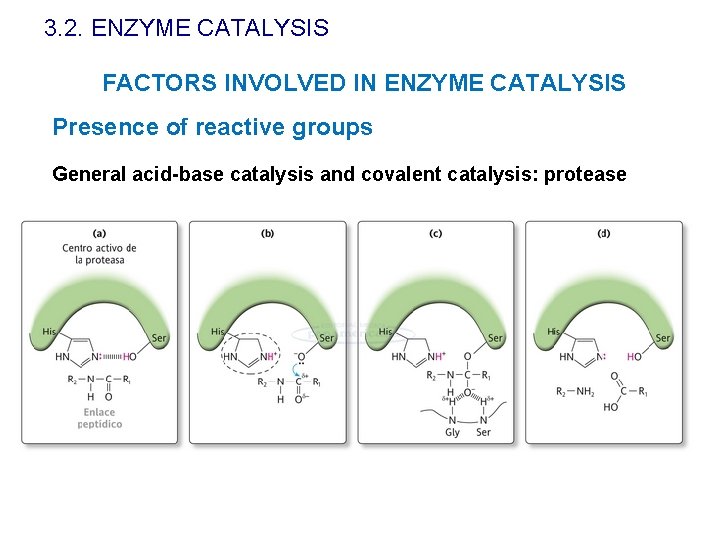
3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS Presence of reactive groups General acid-base catalysis and covalent catalysis: protease

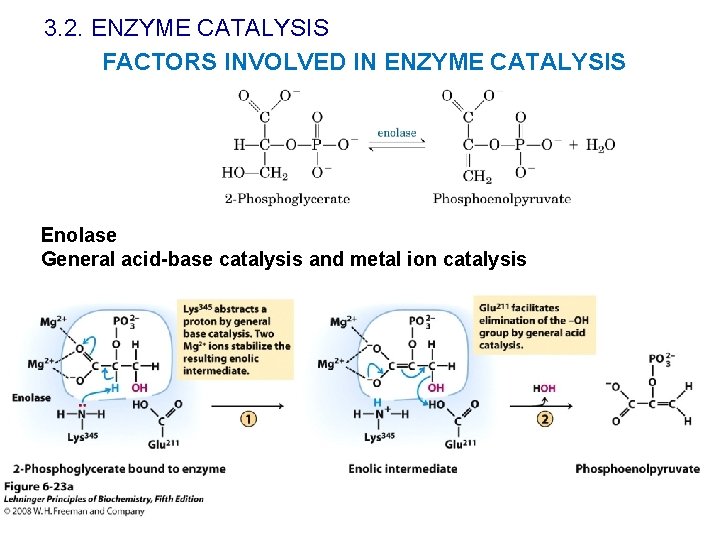
3. 2. ENZYME CATALYSIS FACTORS INVOLVED IN ENZYME CATALYSIS Enolase General acid-base catalysis and metal ion catalysis

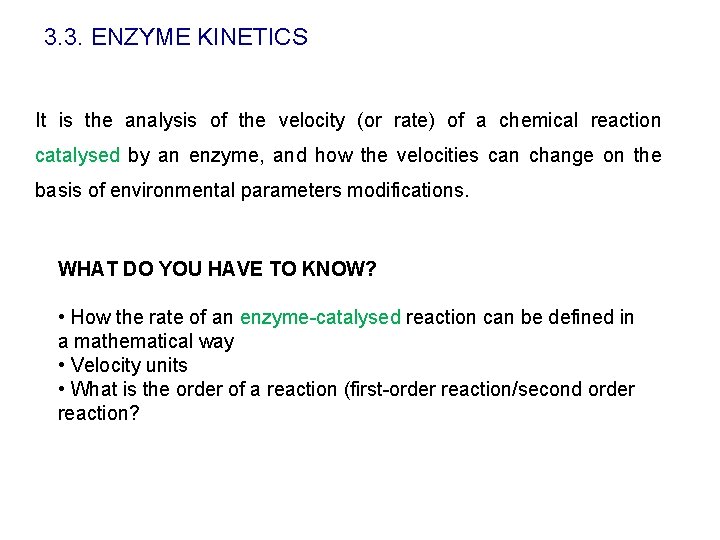
3. 3. ENZYME KINETICS It is the analysis of the velocity (or rate) of a chemical reaction catalysed by an enzyme, and how the velocities can change on the basis of environmental parameters modifications. WHAT DO YOU HAVE TO KNOW? • How the rate of an enzyme-catalysed reaction can be defined in a mathematical way • Velocity units • What is the order of a reaction (first-order reaction/second order reaction?

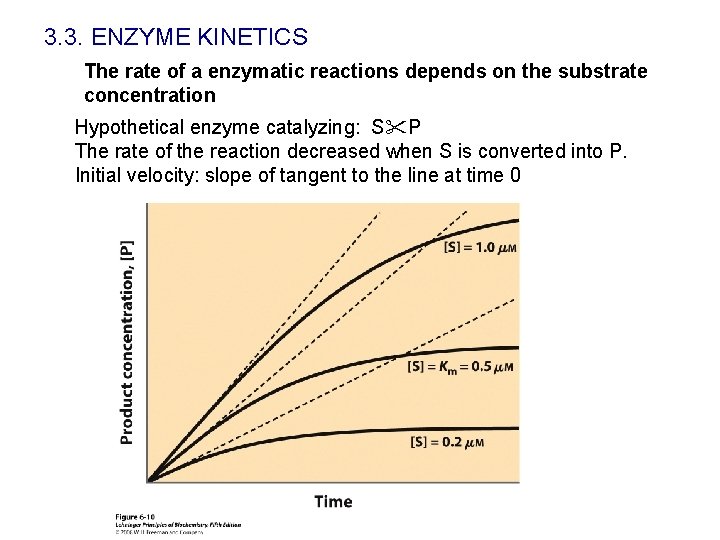
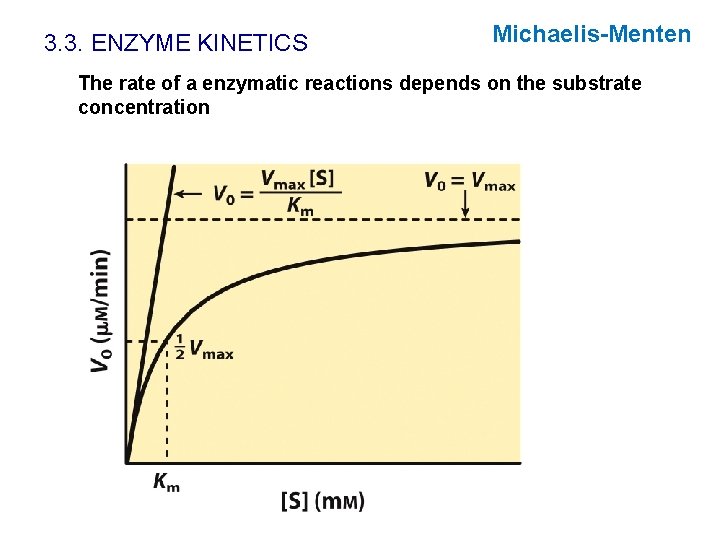
3. 3. ENZYME KINETICS The rate of a enzymatic reactions depends on the substrate concentration Hypothetical enzyme catalyzing: S P The rate of the reaction decreased when S is converted into P. Initial velocity: slope of tangent to the line at time 0

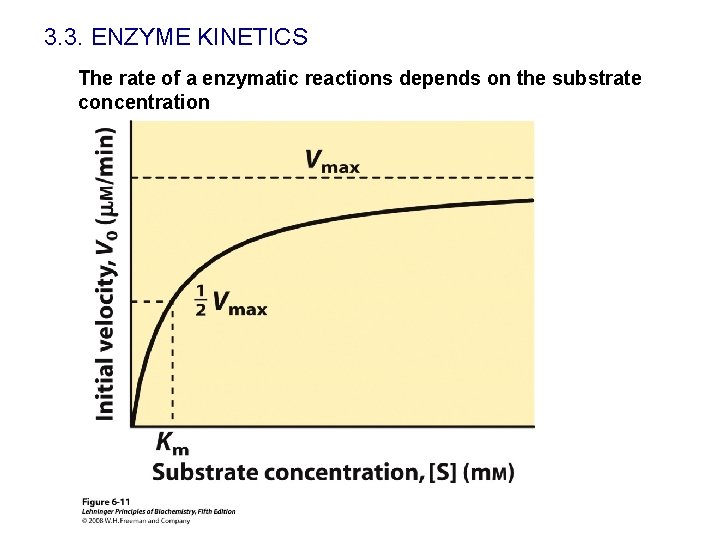
3. 3. ENZYME KINETICS The rate of a enzymatic reactions depends on the substrate concentration

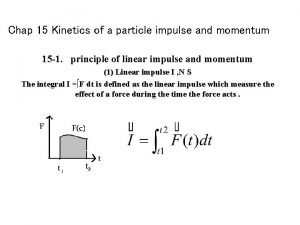
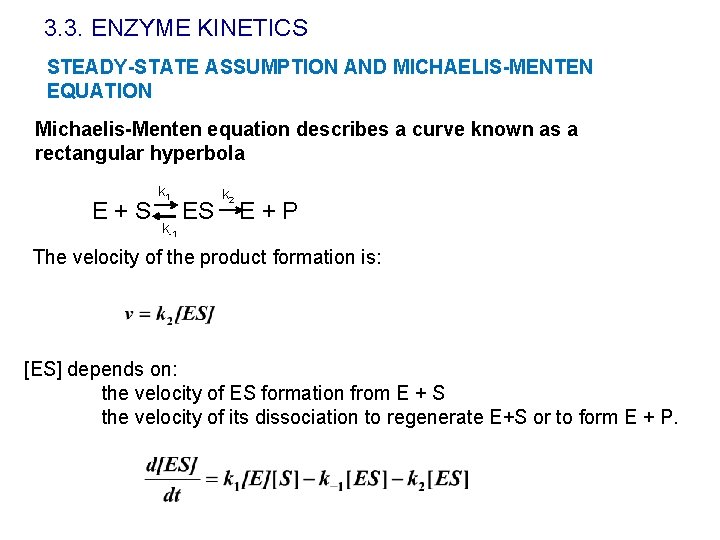
3. 3. ENZYME KINETICS STEADY-STATE ASSUMPTION AND MICHAELIS-MENTEN EQUATION Michaelis-Menten equation describes a curve known as a rectangular hyperbola E+S k 1 k-1 ES k 2 E+P The velocity of the product formation is: [ES] depends on: the velocity of ES formation from E + S the velocity of its dissociation to regenerate E+S or to form E + P.
![3 3 ENZYME KINETICS Steadystate Concentration Under experimental conditions SE The ES quickly reaches 3. 3. ENZYME KINETICS Steady-state Concentration Under experimental conditions [S]>>>[E]. The [ES] quickly reaches](https://slidetodoc.com/presentation_image_h2/07058868e10e95d868ac92ec781a0c43/image-26.jpg)
3. 3. ENZYME KINETICS Steady-state Concentration Under experimental conditions [S]>>>[E]. The [ES] quickly reaches a constant value in such dynamic system, and remains constant until complete P formation: Steady State assumption 0 Early stage ES formation Time Steady state [ES] is constant
![3 3 ENZYME KINETICS d ES 0 dt so k 1 3. 3. ENZYME KINETICS d[ ES ] = 0, dt so k 1 [](https://slidetodoc.com/presentation_image_h2/07058868e10e95d868ac92ec781a0c43/image-27.jpg)
3. 3. ENZYME KINETICS d[ ES ] = 0, dt so k 1 [ E ][ S ] = k-1 [ ES ] + k 2 [ ES ] Steady-state KM, Michaelis constant Maximal velocity is obtained when the enzyme is saturated: [E]T=[ES] Michaelis-Menten Equation

3. 3. ENZYME KINETICS

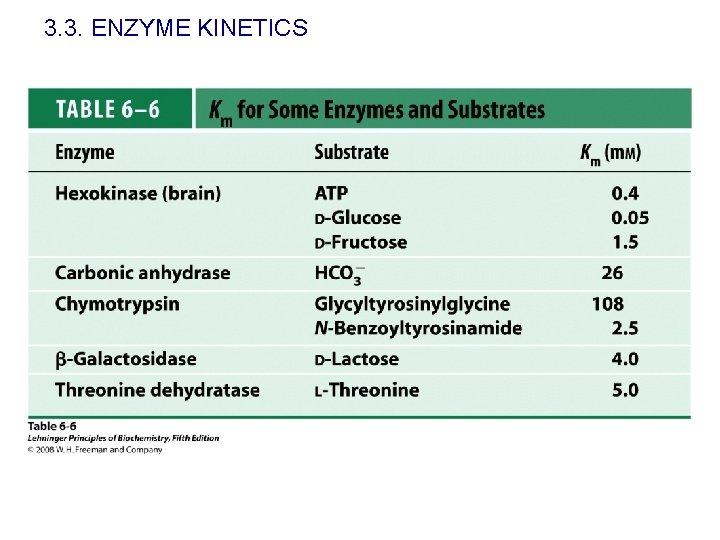
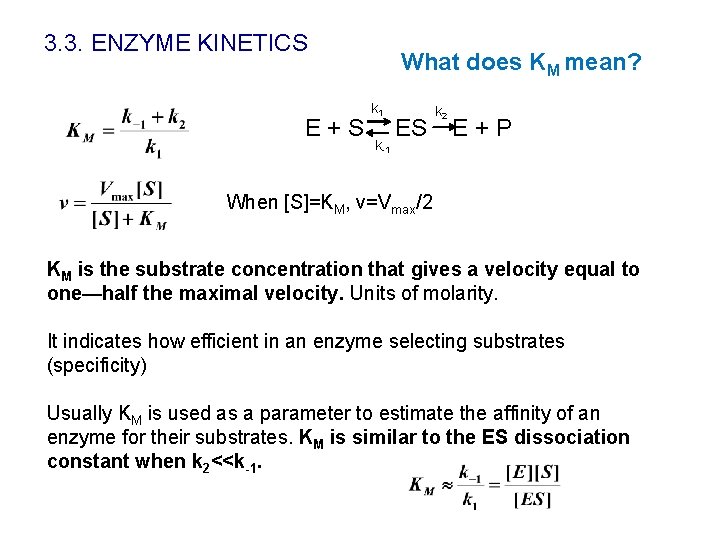
3. 3. ENZYME KINETICS E+S What does KM mean? k 1 k-1 ES k 2 E+P When [S]=KM, v=Vmax/2 KM is the substrate concentration that gives a velocity equal to one—half the maximal velocity. Units of molarity. It indicates how efficient in an enzyme selecting substrates (specificity) Usually KM is used as a parameter to estimate the affinity of an enzyme for their substrates. KM is similar to the ES dissociation constant when k 2<<k-1.

3. 3. ENZYME KINETICS Michaelis-Menten The rate of a enzymatic reactions depends on the substrate concentration

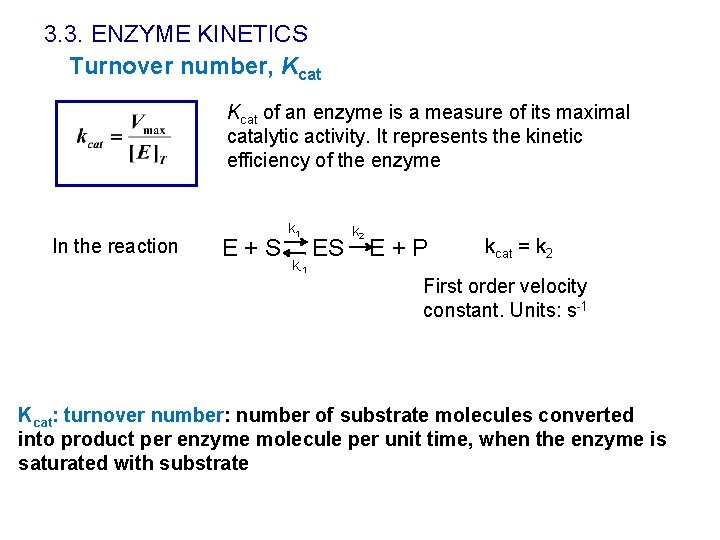
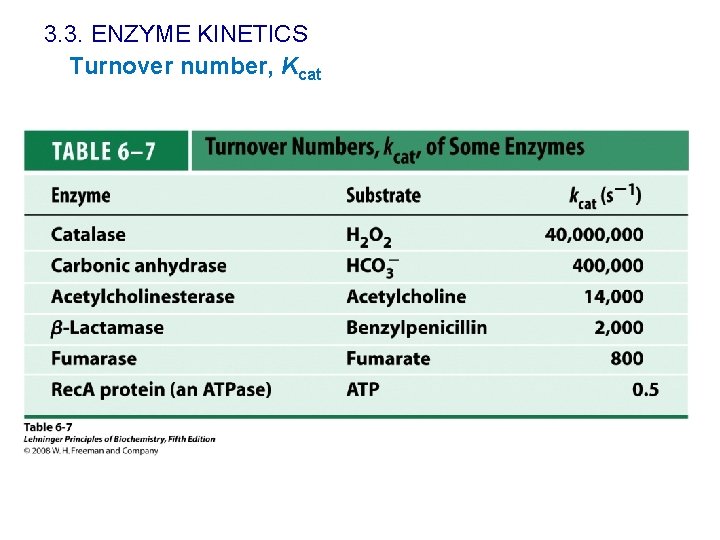
3. 3. ENZYME KINETICS Turnover number, Kcat of an enzyme is a measure of its maximal catalytic activity. It represents the kinetic efficiency of the enzyme In the reaction E+S k 1 k-1 ES k 2 E+P kcat = k 2 First order velocity constant. Units: s-1 Kcat: turnover number: number of substrate molecules converted into product per enzyme molecule per unit time, when the enzyme is saturated with substrate

3. 3. ENZYME KINETICS Turnover number, Kcat

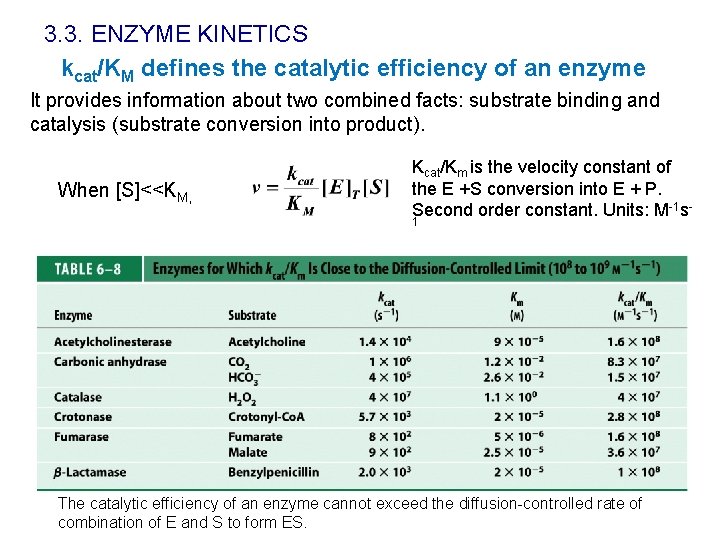
3. 3. ENZYME KINETICS kcat/KM defines the catalytic efficiency of an enzyme It provides information about two combined facts: substrate binding and catalysis (substrate conversion into product). When [S]<<KM, Kcat/Km is the velocity constant of the E +S conversion into E + P. Second order constant. Units: M-1 s 1 The catalytic efficiency of an enzyme cannot exceed the diffusion-controlled rate of combination of E and S to form ES.

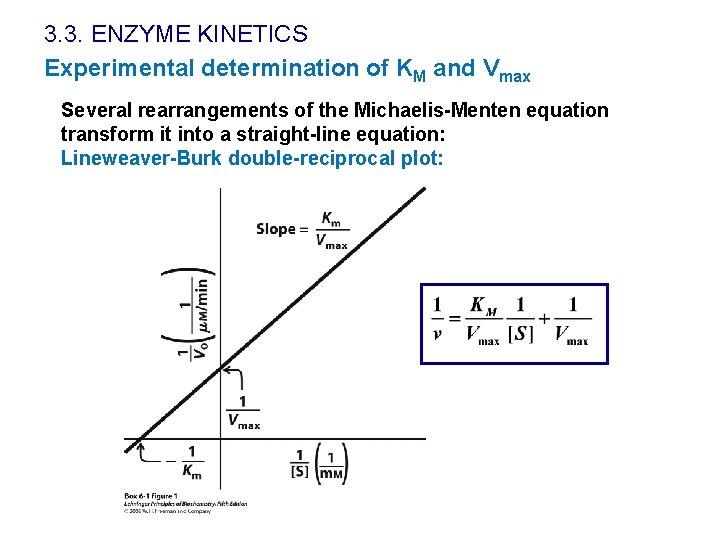
3. 3. ENZYME KINETICS Experimental determination of KM and Vmax Several rearrangements of the Michaelis-Menten equation transform it into a straight-line equation: Lineweaver-Burk double-reciprocal plot:

3. 3. ENZYME KINETICS Factors affecting the enzymatic activity Enzyme concentration -Enzymatic activity international unit (U): quantity of enzyme able to transform 1. 0 mol substrate per minute at 25ºC (under optimal conditions) - Specific enzymatic activity (U/mg): number of enzymatic unit per mg of purified protein. It indicates how pure the enzyme is. Balls: they represent proteins Red balls: enzyme molecules Both cylinders: same activity units Right cylinder shows higher specific activity than the left cylinder

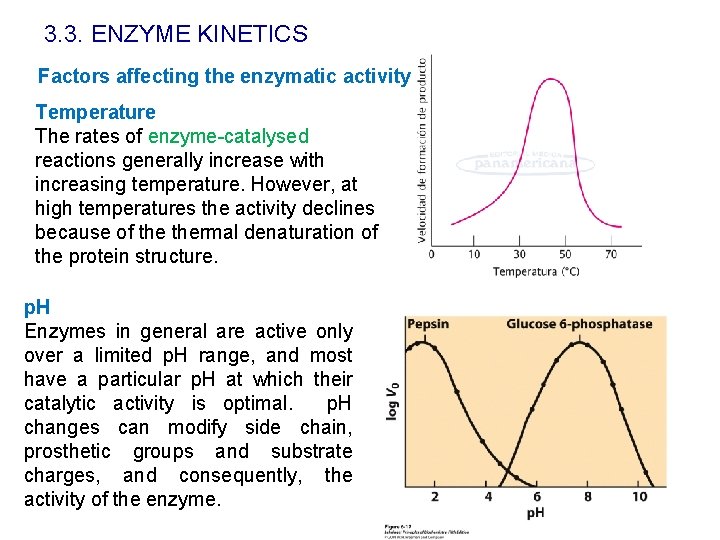
3. 3. ENZYME KINETICS Factors affecting the enzymatic activity Temperature The rates of enzyme-catalysed reactions generally increase with increasing temperature. However, at high temperatures the activity declines because of thermal denaturation of the protein structure. p. H Enzymes in general are active only over a limited p. H range, and most have a particular p. H at which their catalytic activity is optimal. p. H changes can modify side chain, prosthetic groups and substrate charges, and consequently, the activity of the enzyme.

3. 3. ENZYME KINETICS Enzymatic inhibition • Inhibition: velocity of an enzymatic reaction is decreased or inhibited by some agent (inhibitors) – Irreversible • Inhibitor causes stable, covalent alterations in the enzyme – Examples: » Ampicillin: causes covalent modification of a transpeptidase catalysing the synthesis of the bacterial cellular wall » Aspirin: causes covalent modification in a cyclooxygenase involved in inflammation – Reversible • Inhibitor interact with the enzyme through noncovalent association/dissociation reactions.

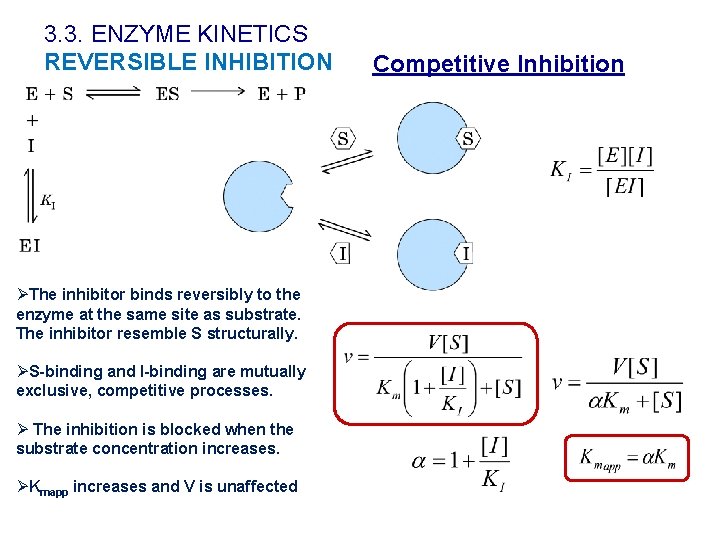
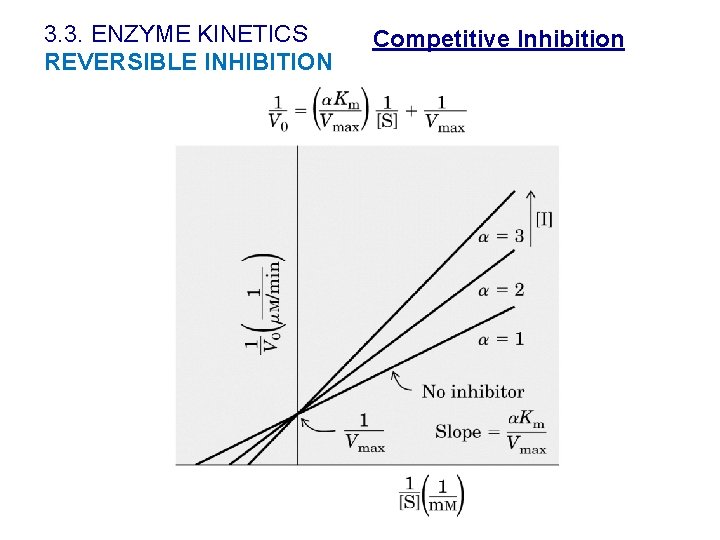
3. 3. ENZYME KINETICS REVERSIBLE INHIBITION ØThe inhibitor binds reversibly to the enzyme at the same site as substrate. The inhibitor resemble S structurally. ØS-binding and I-binding are mutually exclusive, competitive processes. Ø The inhibition is blocked when the substrate concentration increases. ØKmapp increases and V is unaffected Competitive Inhibition

3. 3. ENZYME KINETICS REVERSIBLE INHIBITION Competitive Inhibition

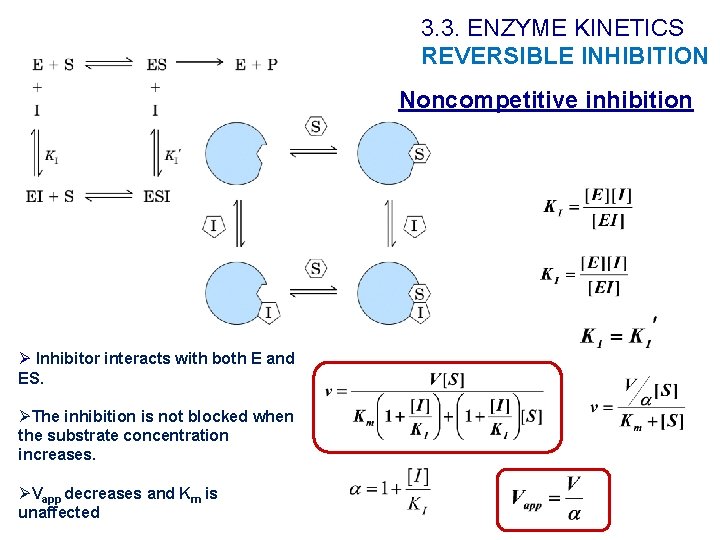
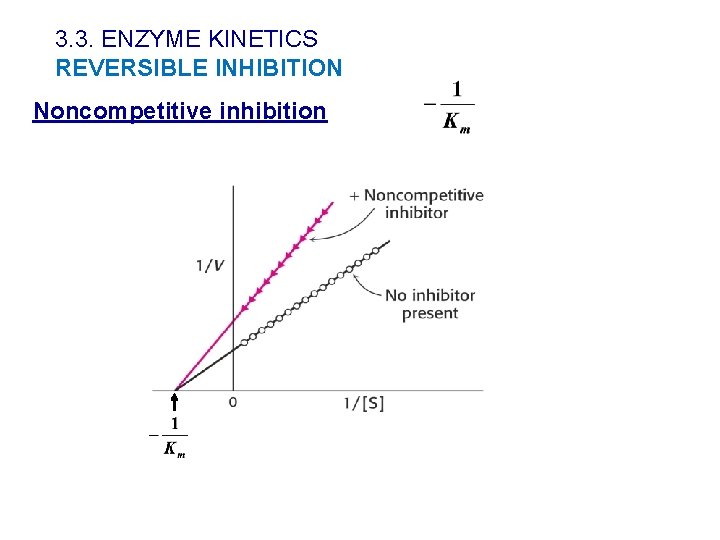
3. 3. ENZYME KINETICS REVERSIBLE INHIBITION Noncompetitive inhibition Ø Inhibitor interacts with both E and ES. ØThe inhibition is not blocked when the substrate concentration increases. ØVapp decreases and Km is unaffected

3. 3. ENZYME KINETICS REVERSIBLE INHIBITION Noncompetitive inhibition

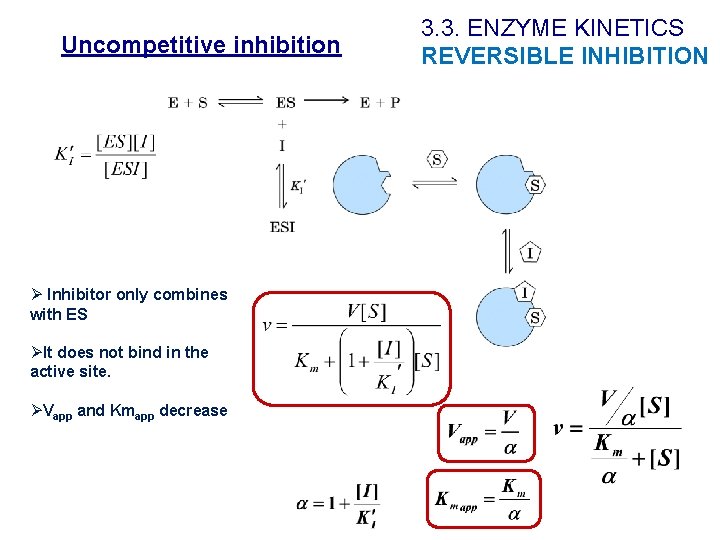
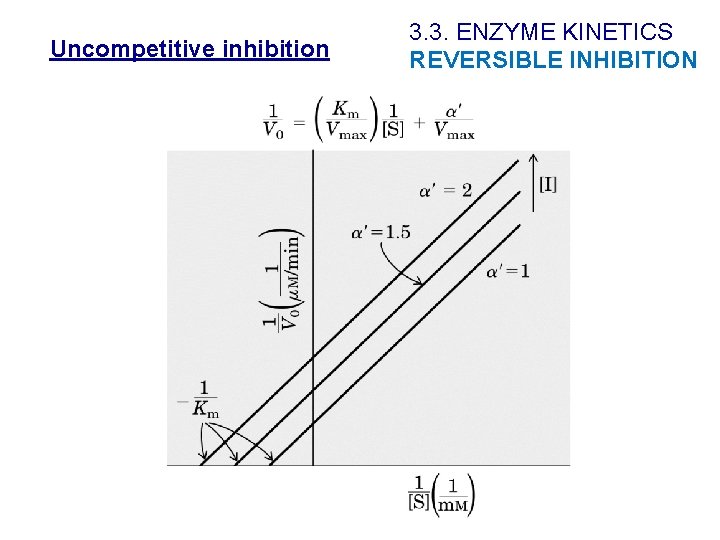
Uncompetitive inhibition Ø Inhibitor only combines with ES ØIt does not bind in the active site. ØVapp and Kmapp decrease 3. 3. ENZYME KINETICS REVERSIBLE INHIBITION

Uncompetitive inhibition 3. 3. ENZYME KINETICS REVERSIBLE INHIBITION


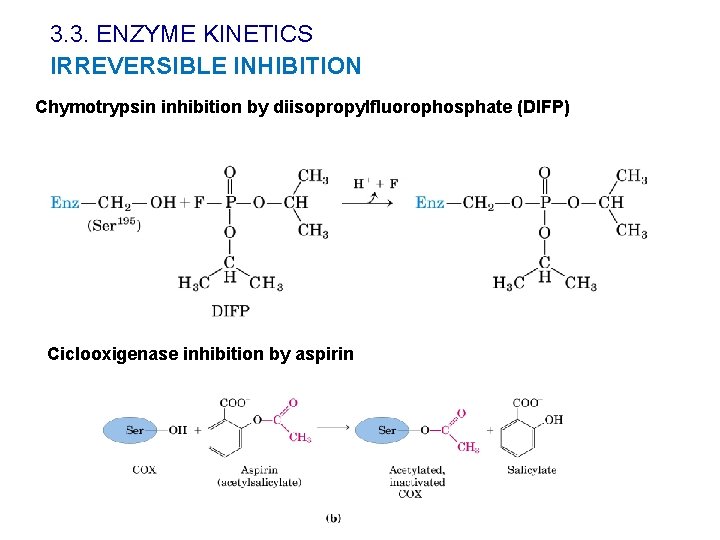
3. 3. ENZYME KINETICS IRREVERSIBLE INHIBITION Chymotrypsin inhibition by diisopropylfluorophosphate (DIFP) Ciclooxigenase inhibition by aspirin

3. 4. ENZYME REGULATION Living systems must regulate the enzymatic catalytic activity to: - Coordinate metabolic processes - Promote adaptations to environmental changes - Growth and complete the living cycle in the correct way Two mechanisms of regulation: 1. - Control of the enzyme availability 2. - Control of the enzymatic activity, by means of modifications of the conformation or structure

3. 4. ENZYME REGULATION ALLOSTERIC REGULATION Allosteric enzyme: Oligomeric organization (more than one active site and more than one effector-binding site) The regulatory effects exerted on the enzyme’s activity are achieved by conformational changes occurring in the protein when effector metabolites bind Conformational states for a protein (monomer): Taut state (T): Low substrate affinity Relaxed state (R) : High substrate affinity

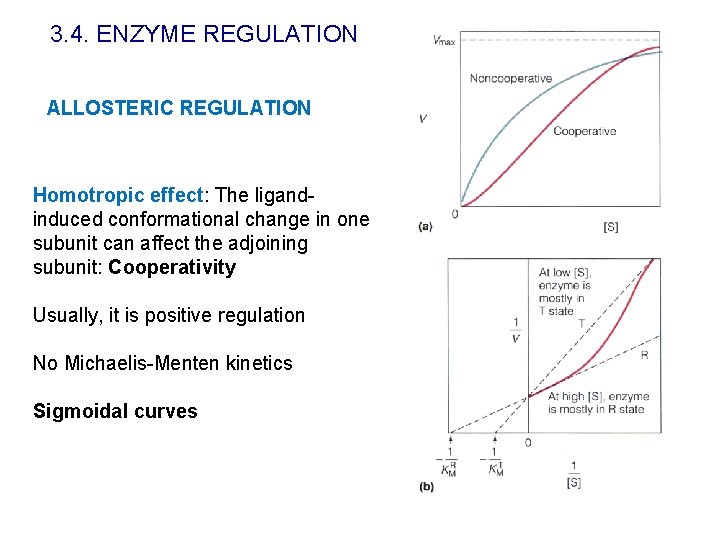
3. 4. ENZYME REGULATION ALLOSTERIC REGULATION Homotropic effect: The ligandinduced conformational change in one subunit can affect the adjoining subunit: Cooperativity Usually, it is positive regulation No Michaelis-Menten kinetics Sigmoidal curves

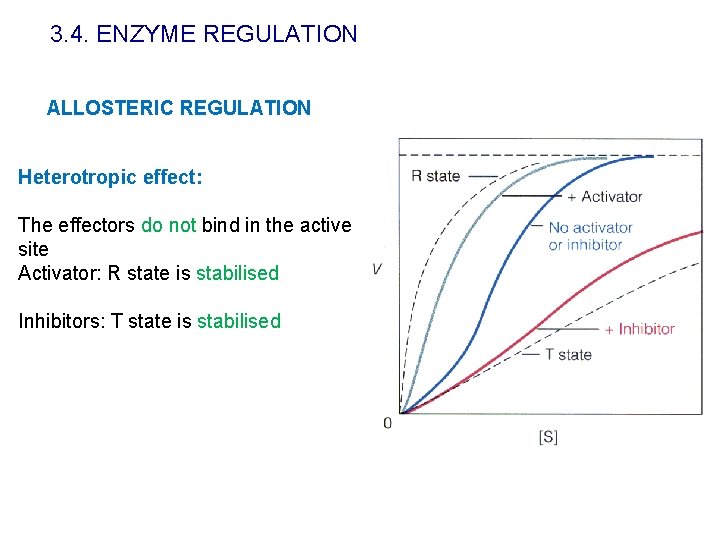
3. 4. ENZYME REGULATION ALLOSTERIC REGULATION Heterotropic effect: The effectors do not bind in the active site Activator: R state is stabilised Inhibitors: T state is stabilised

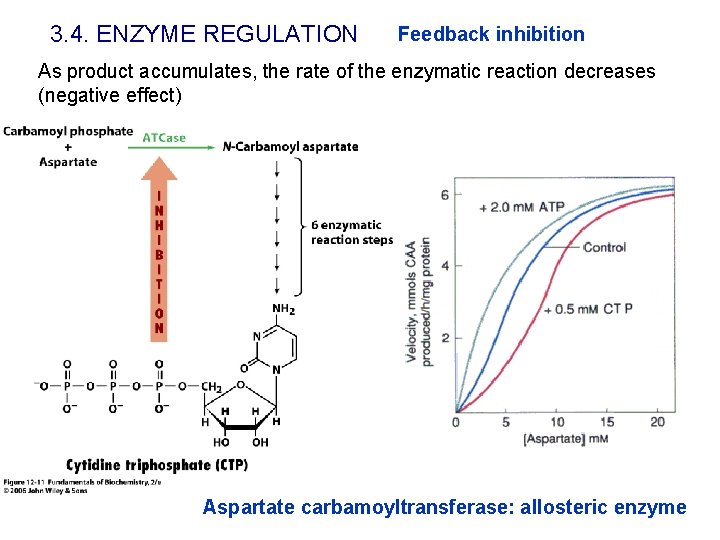
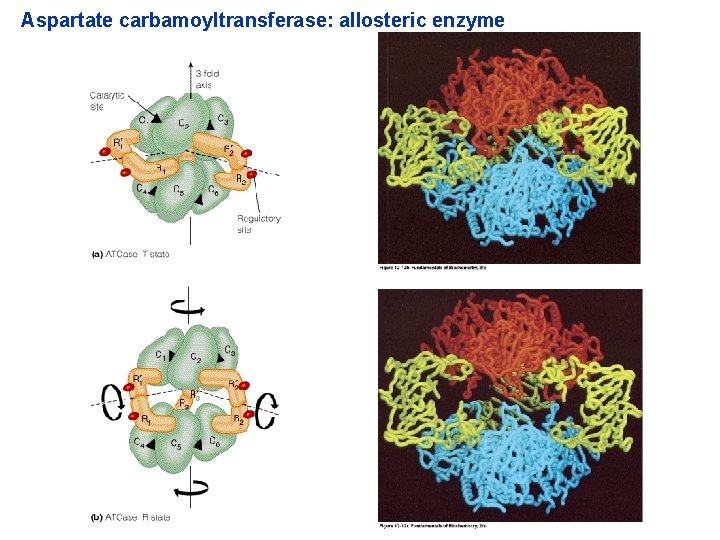
3. 4. ENZYME REGULATION Feedback inhibition As product accumulates, the rate of the enzymatic reaction decreases (negative effect) Aspartate carbamoyltransferase: allosteric enzyme

Aspartate carbamoyltransferase: allosteric enzyme

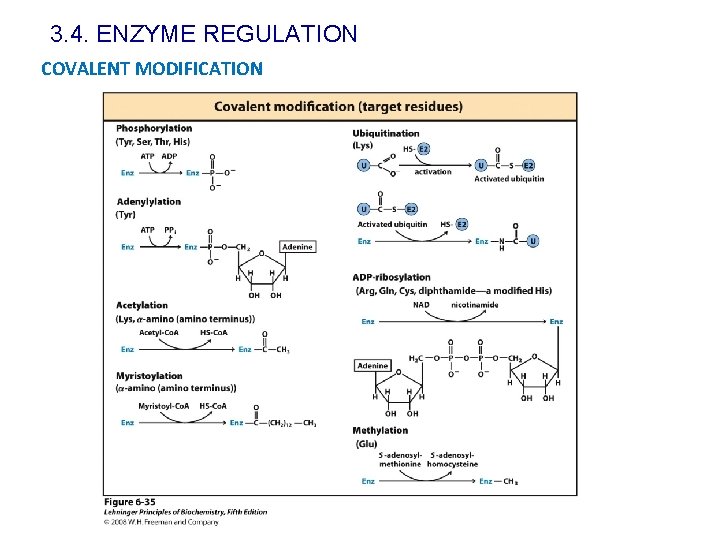
3. 4. ENZYME REGULATION COVALENT MODIFICATION

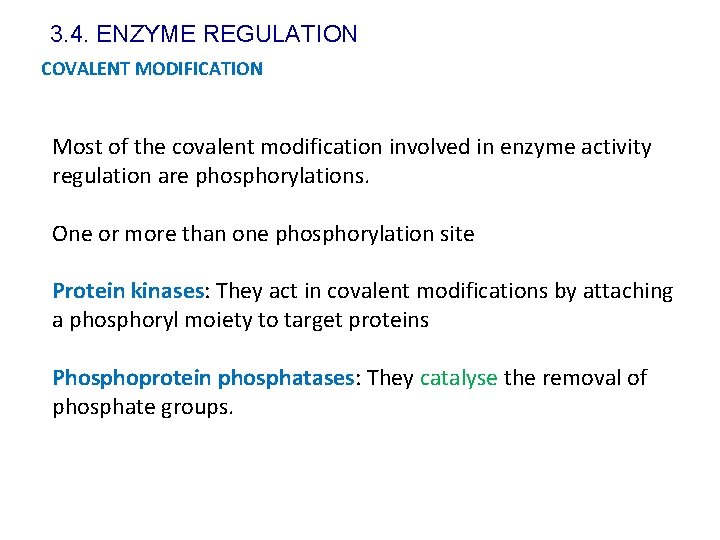
3. 4. ENZYME REGULATION COVALENT MODIFICATION Most of the covalent modification involved in enzyme activity regulation are phosphorylations. One or more than one phosphorylation site Protein kinases: They act in covalent modifications by attaching a phosphoryl moiety to target proteins Phosphoprotein phosphatases: They catalyse the removal of phosphate groups.

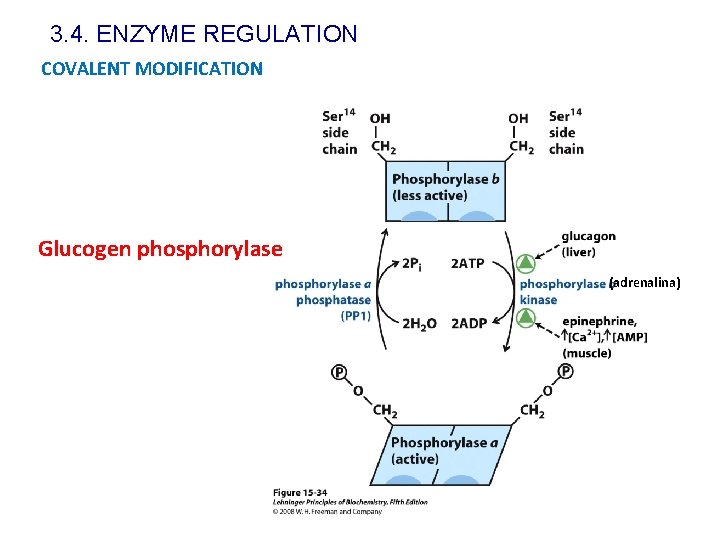
3. 4. ENZYME REGULATION COVALENT MODIFICATION Glucogen phosphorylase (adrenalina)

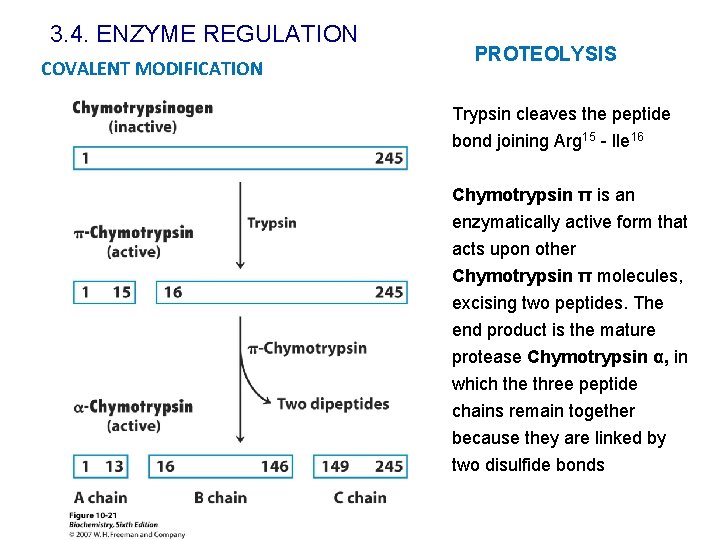
3. 4. ENZYME REGULATION PROTEOLYSIS Some proteins are synthesized as inactive precursors, called zymogens or proenzymes, that acquire full activity only upon specific proteolytic cleavage of one or several of their peptide bonds ü It is not energy dependent ü The peptide bond cleavage is irreversible Examples v Digestive enzymes v Blood clotting v Peptidic hormone (insulin) v Collagen v Caspases: apoptosis

3. 4. ENZYME REGULATION COVALENT MODIFICATION PROTEOLYSIS Trypsin cleaves the peptide bond joining Arg 15 - Ile 16 Chymotrypsin π is an enzymatically active form that acts upon other Chymotrypsin π molecules, excising two peptides. The end product is the mature protease Chymotrypsin α, in which the three peptide chains remain together because they are linked by two disulfide bonds
 Km in enzyme kinetics
Km in enzyme kinetics Competitive inhibition
Competitive inhibition Enzyme kinetics
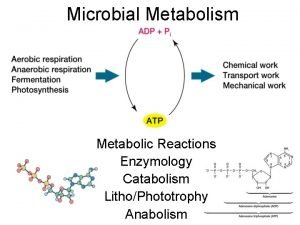
Enzyme kinetics Energy catalysis and biosynthesis
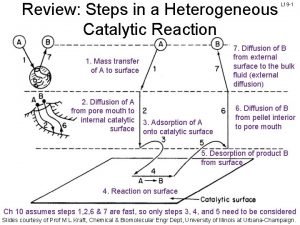
Energy catalysis and biosynthesis 7 steps of heterogeneous catalysis
7 steps of heterogeneous catalysis Apoenzyme
Apoenzyme Catalysis by approximation
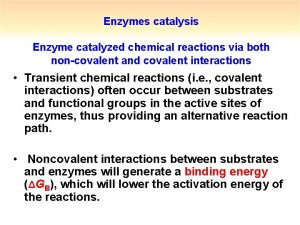
Catalysis by approximation What is covalent catalysis
What is covalent catalysis What is covalent catalysis
What is covalent catalysis Specific acid base catalysis
Specific acid base catalysis Specific acid base catalysis
Specific acid base catalysis Specific acid base catalysis
Specific acid base catalysis Langmuir-hinshelwood mechanism heterogeneous catalysis
Langmuir-hinshelwood mechanism heterogeneous catalysis Difference between atomic and molecular spectroscopy
Difference between atomic and molecular spectroscopy Site:slidetodoc.com
Site:slidetodoc.com Honh2 dissociation equation
Honh2 dissociation equation Chemistry grade 11 unit 4 chemical kinetics
Chemistry grade 11 unit 4 chemical kinetics It in a sentence
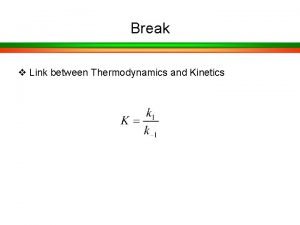
It in a sentence Kinetics and equilibrium
Kinetics and equilibrium Difference between 1st order and zero order kinetics
Difference between 1st order and zero order kinetics Difference between zero and first order kinetics
Difference between zero and first order kinetics Planar kinetics of a rigid body force and acceleration
Planar kinetics of a rigid body force and acceleration Kinetics of a particle: force and acceleration
Kinetics of a particle: force and acceleration Kinetics of a particle: impulse and momentum
Kinetics of a particle: impulse and momentum Hematology
Hematology Kinematics and kinetics of rigid bodies
Kinematics and kinetics of rigid bodies Planar kinetics of a rigid body work and energy
Planar kinetics of a rigid body work and energy Fermenter
Fermenter Unit 6 review questions
Unit 6 review questions Definition of chemical kinetics in chemistry
Definition of chemical kinetics in chemistry Factors affecting distribution of drugs
Factors affecting distribution of drugs Fbd and kd
Fbd and kd Collision theory of kinetics
Collision theory of kinetics Kinetics flotation chemicals
Kinetics flotation chemicals Chemical kinetics half life
Chemical kinetics half life Chemical kinetics definition
Chemical kinetics definition Kinetic of rigid body
Kinetic of rigid body Kinetics ap chemistry
Kinetics ap chemistry Kinetics of crystal violet fading
Kinetics of crystal violet fading Octet kinetics
Octet kinetics Kinetics of particles newton's second law
Kinetics of particles newton's second law Chemical kinetics experiment
Chemical kinetics experiment Dynafit kinetics
Dynafit kinetics Data kinetics ltd
Data kinetics ltd Vmax equation
Vmax equation Kinetics reaction
Kinetics reaction Applications of chemical kinetics
Applications of chemical kinetics Rate of growth
Rate of growth Steady state
Steady state Kinetics is the branch of
Kinetics is the branch of Vampnets for deep learning of molecular kinetics
Vampnets for deep learning of molecular kinetics Enzymes speed up chemical reactions by ____
Enzymes speed up chemical reactions by ____ Why are enzymes so important?
Why are enzymes so important? Enzyme activators examples
Enzyme activators examples Section 2-4 chemical reactions and enzymes
Section 2-4 chemical reactions and enzymes Bilateral cataract ketones and raised liver enzymes
Bilateral cataract ketones and raised liver enzymes Non functional plasma enzyme
Non functional plasma enzyme