Deep learning enhanced Markov State Models MSMs Wei





- Slides: 35

Deep learning enhanced Markov State Models (MSMs) Wei Wang Feb 20, 2019

Outline • General protocol of building MSM • Challenges with MSM • VAMPnets • Time-lagged auto-encoder 2

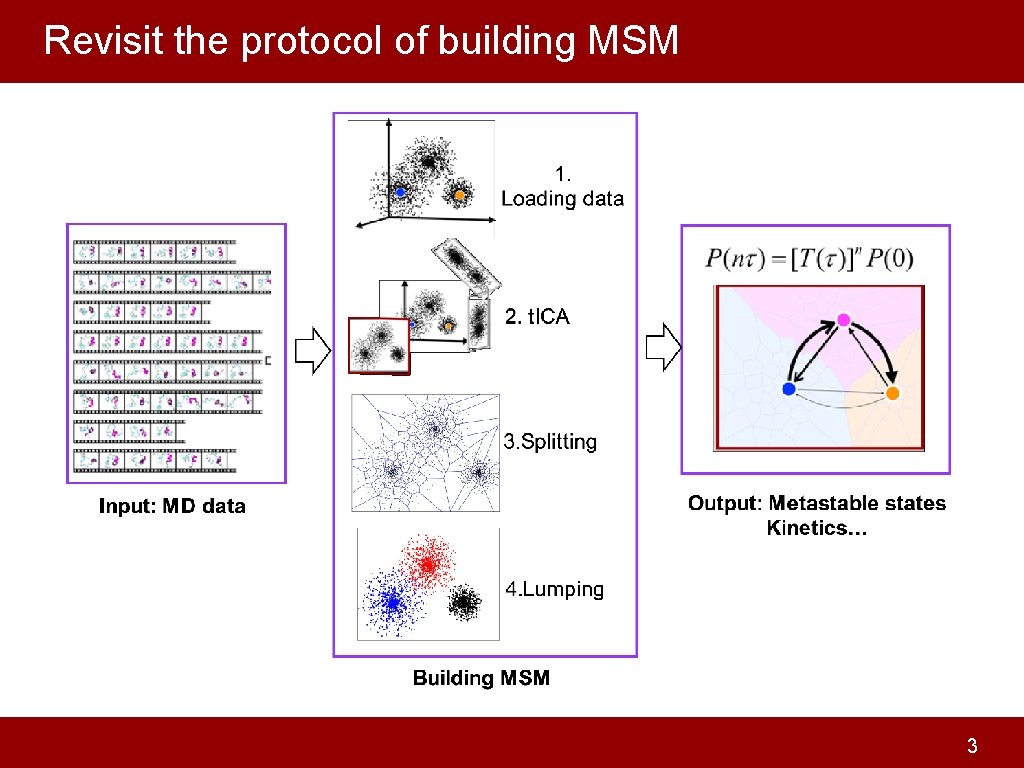
Revisit the protocol of building MSM 3

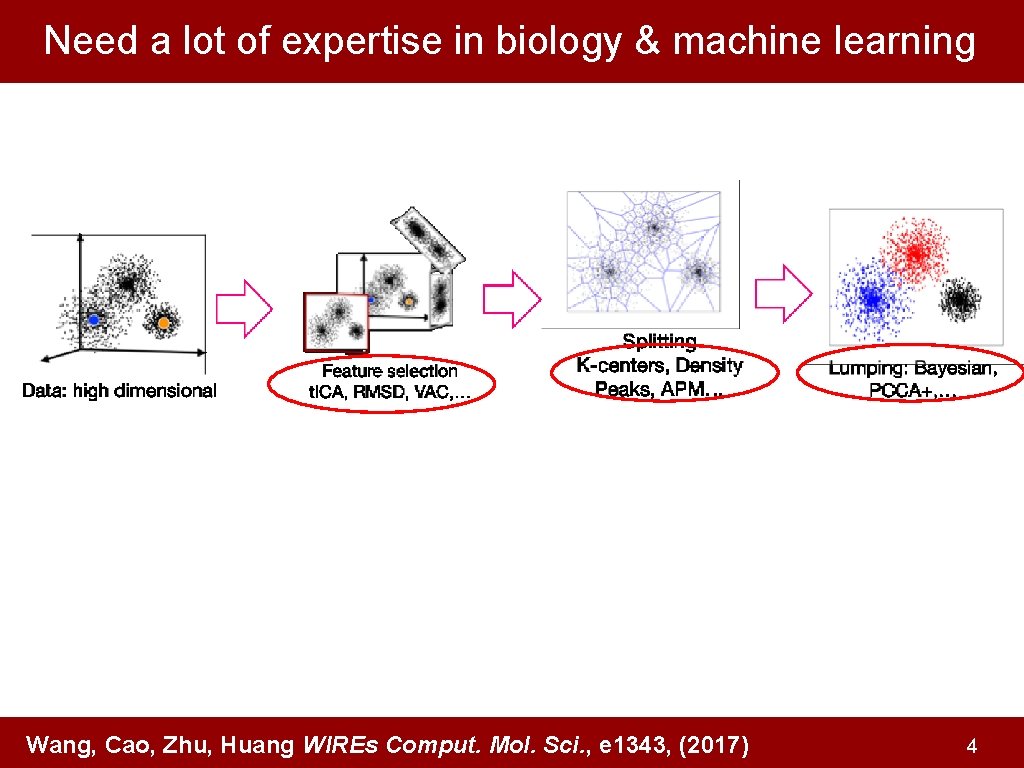
Need a lot of expertise in biology & machine learning Wang, Cao, Zhu, Huang WIREs Comput. Mol. Sci. , e 1343, (2017) 4

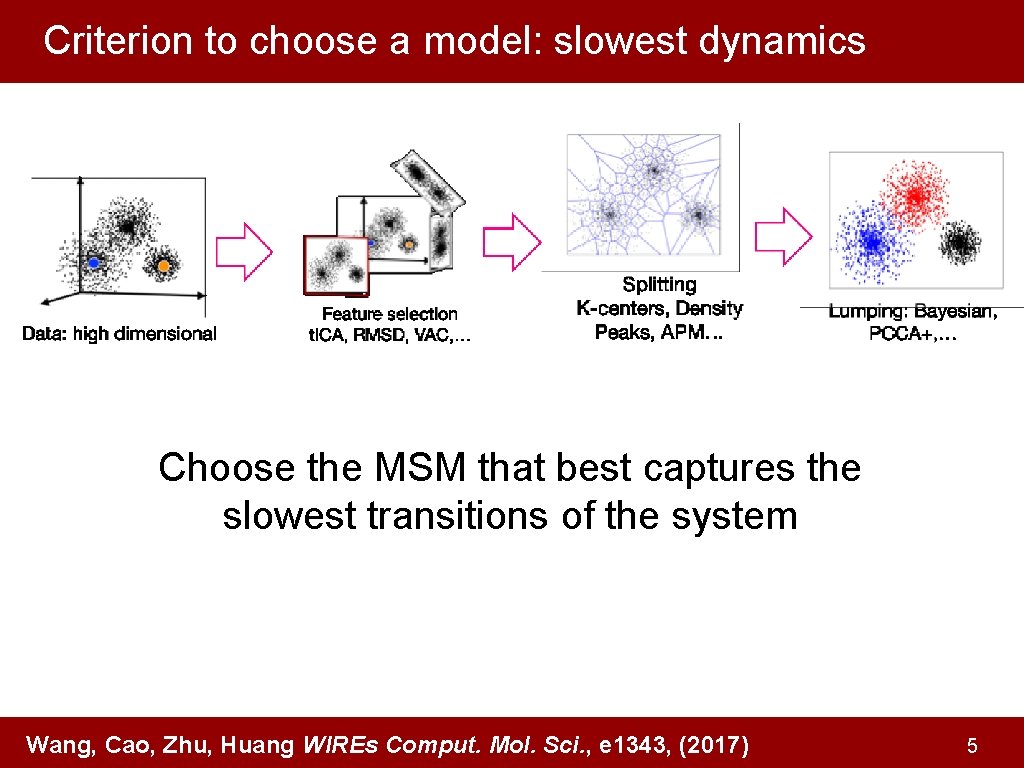
Criterion to choose a model: slowest dynamics Choose the MSM that best captures the slowest transitions of the system Wang, Cao, Zhu, Huang WIREs Comput. Mol. Sci. , e 1343, (2017) 5

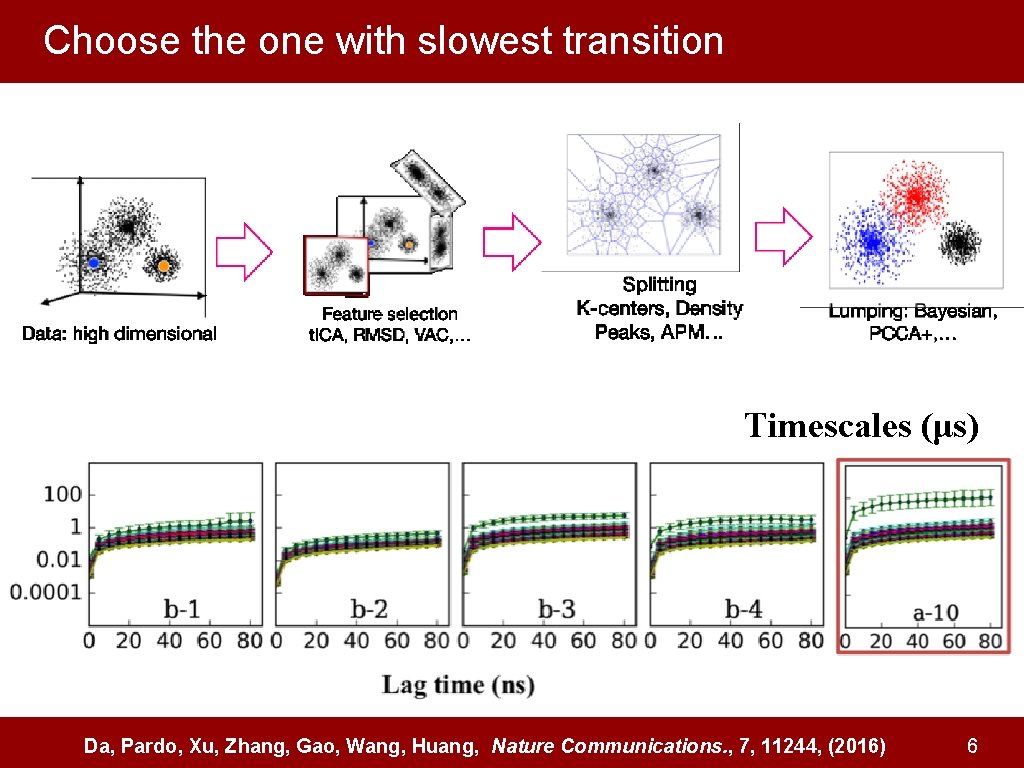
Choose the one with slowest transition Timescales (μs) Da, Pardo, Xu, Zhang, Gao, Wang, Huang, Nature Communications. , 7, 11244, (2016) 6

Perform this cumbersome work: search • • Propose good clustering algorithms & features Parametric search using good strategies http: //msmbuilder. org/osprey/1. 1. 0 7

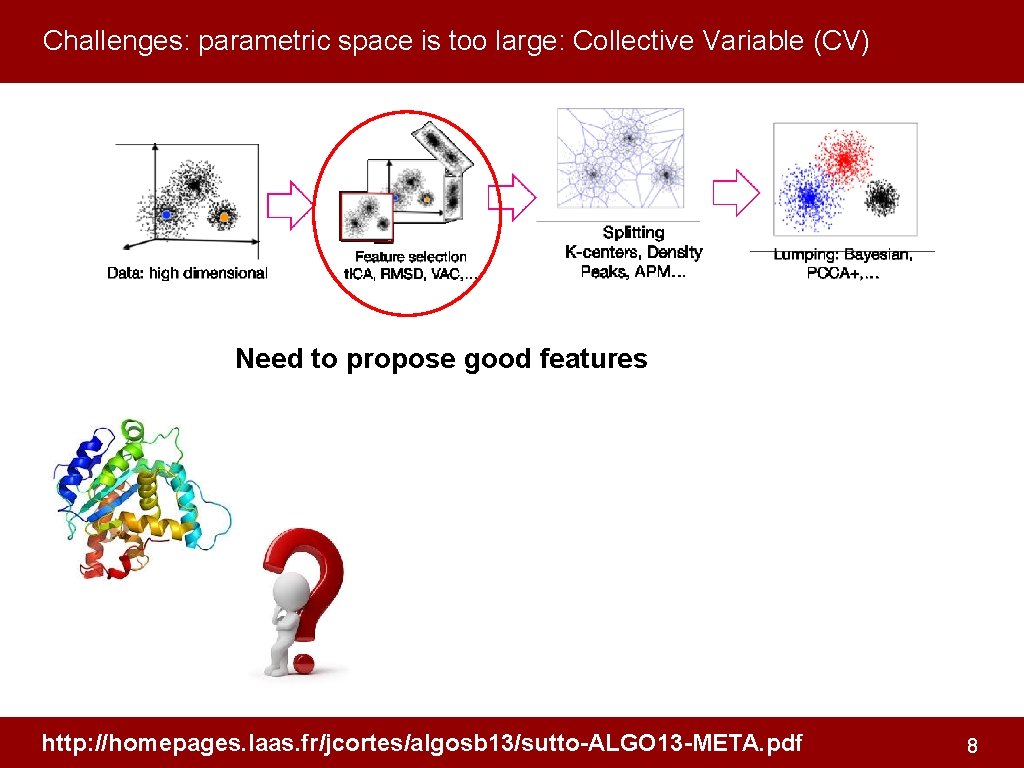
Challenges: parametric space is too large: Collective Variable (CV) Need to propose good features http: //homepages. laas. fr/jcortes/algosb 13/sutto-ALGO 13 -META. pdf 8

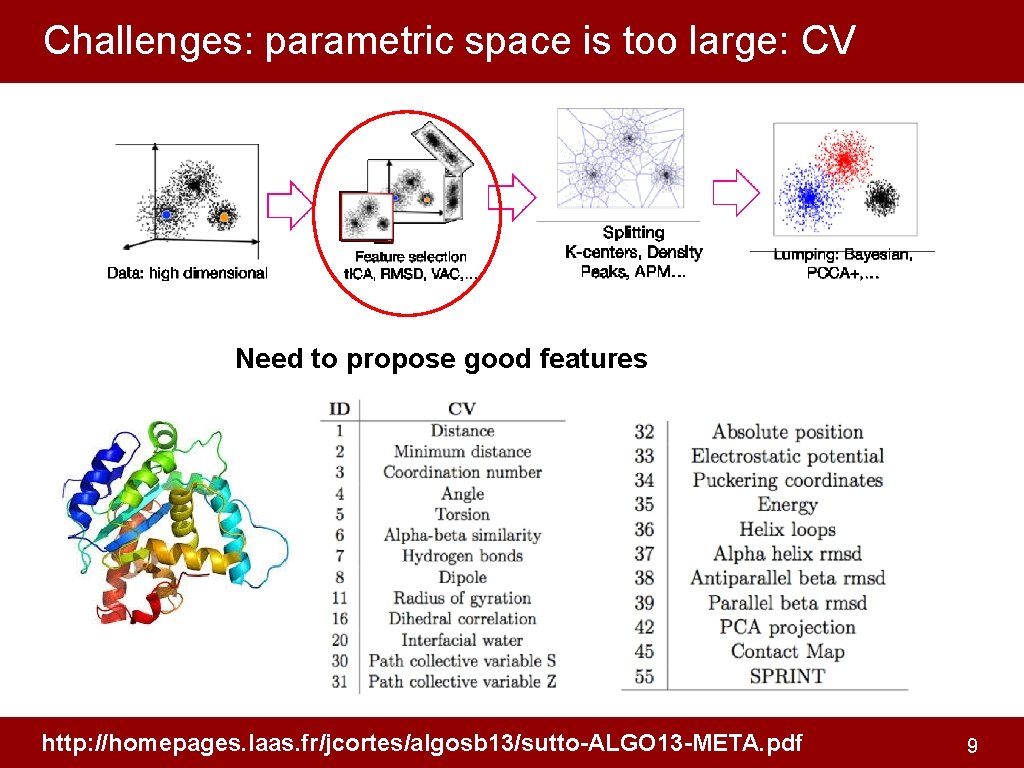
Challenges: parametric space is too large: CV Need to propose good features http: //homepages. laas. fr/jcortes/algosb 13/sutto-ALGO 13 -META. pdf 9

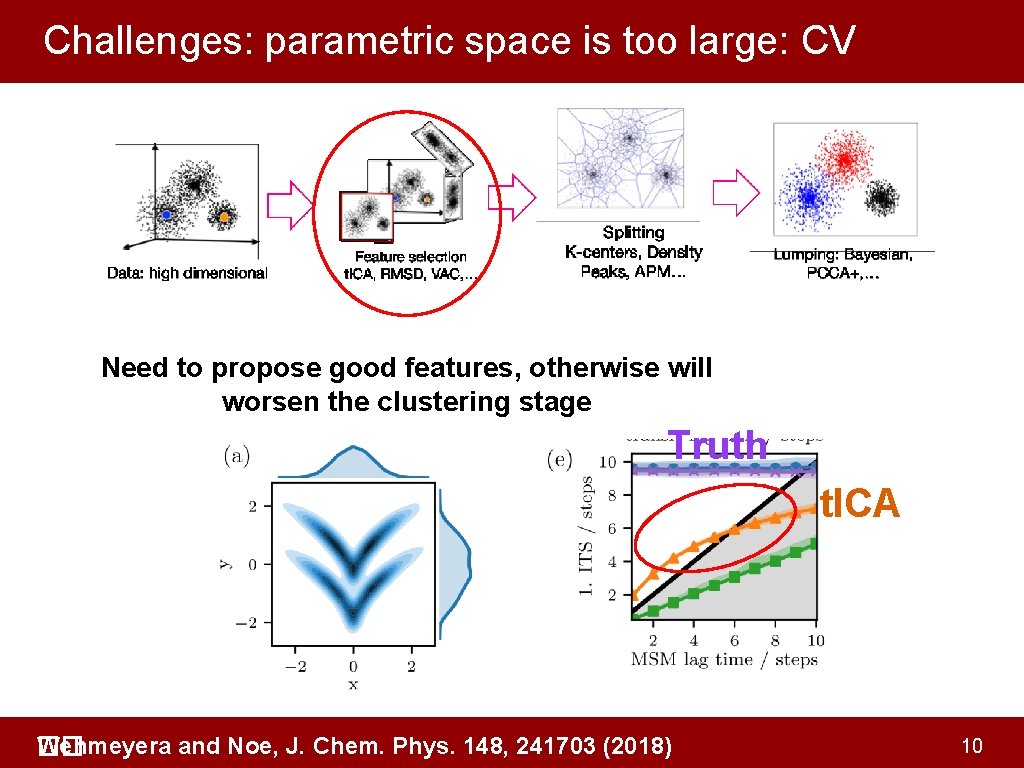
Challenges: parametric space is too large: CV Need to propose good features, otherwise will worsen the clustering stage Truth t. ICA Wehmeyera and Noe, J. Chem. Phys. 148, 241703 (2018) �� 10

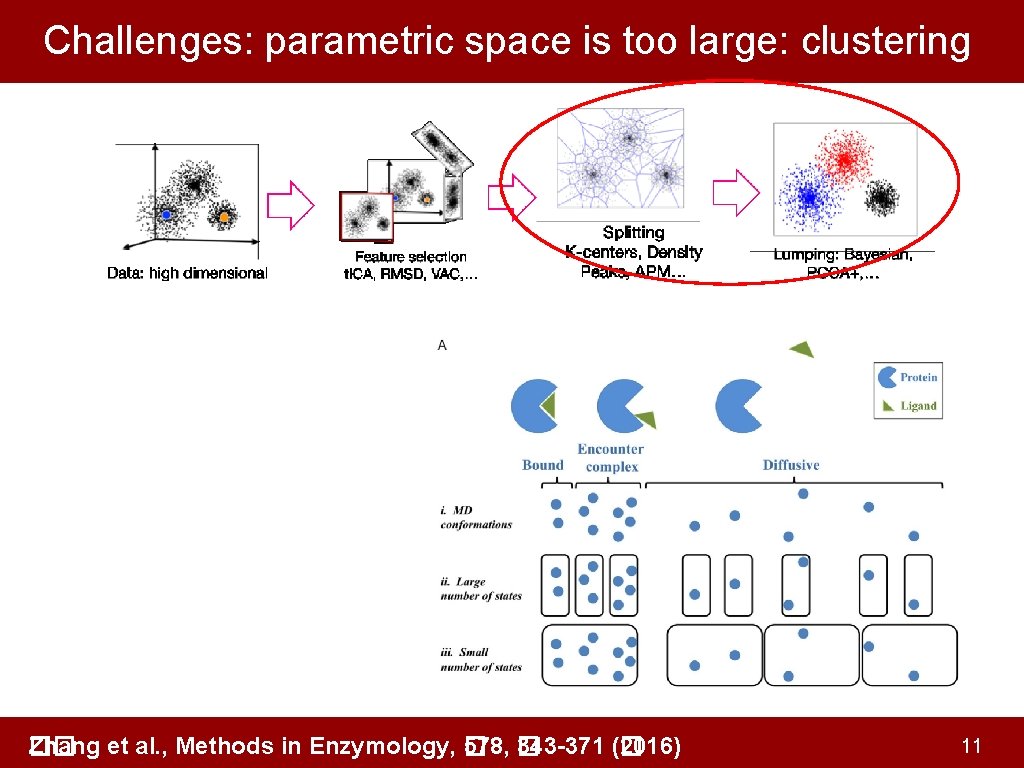
Challenges: parametric space is too large: clustering Zhang et al. , Methods in Enzymology, � �� 578, � 343 -371 (� 2016) 11

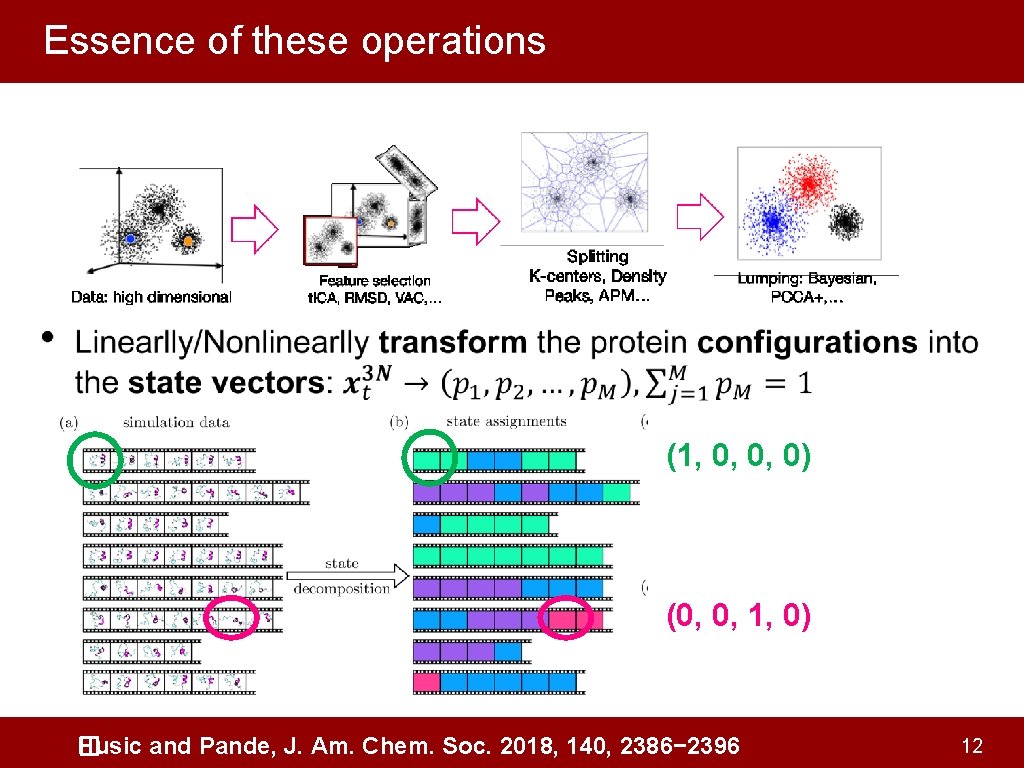
Essence of these operations • (1, 0, 0, 0) (0, 0, 1, 0) Husic and Pande, J. Am. Chem. Soc. 2018, 140, 2386− 2396 � 12

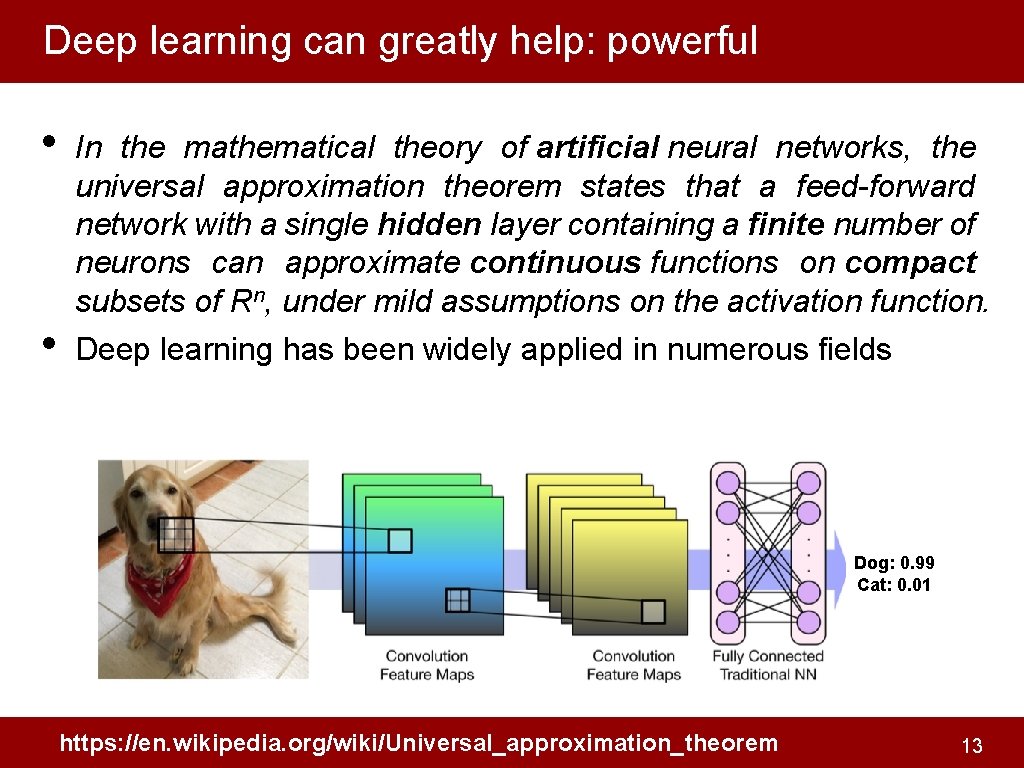
Deep learning can greatly help: powerful • • In the mathematical theory of artificial neural networks, the universal approximation theorem states that a feed-forward network with a single hidden layer containing a finite number of neurons can approximate continuous functions on compact subsets of Rn, under mild assumptions on the activation function. Deep learning has been widely applied in numerous fields Dog: 0. 99 Cat: 0. 01 https: //en. wikipedia. org/wiki/Universal_approximation_theorem 13

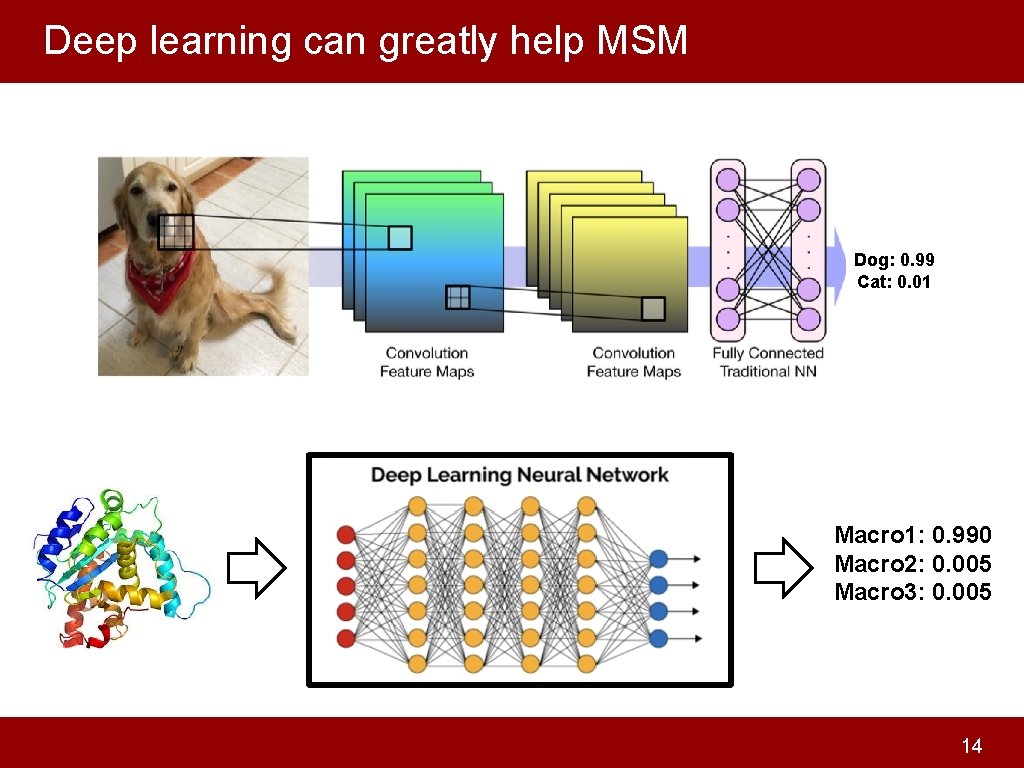
Deep learning can greatly help MSM Dog: 0. 99 Cat: 0. 01 Macro 1: 0. 990 Macro 2: 0. 005 Macro 3: 0. 005 14

Outline • General protocol of building MSM • Challenges with MSM • VAMPnets • Time-lagged auto-encoder 15

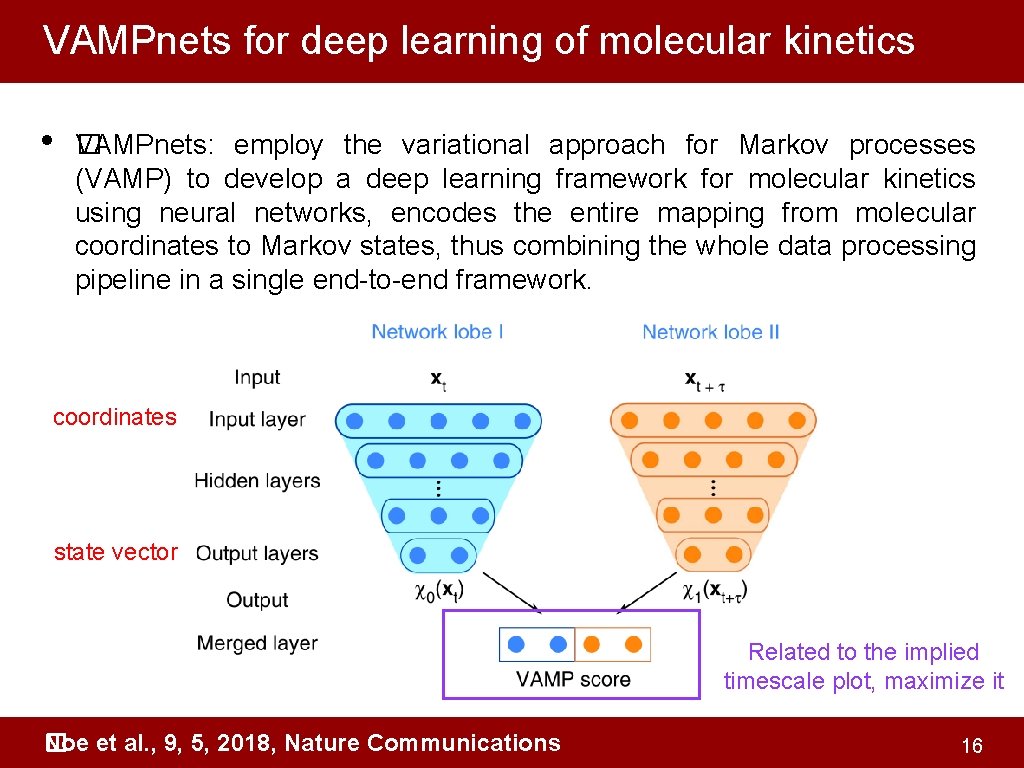
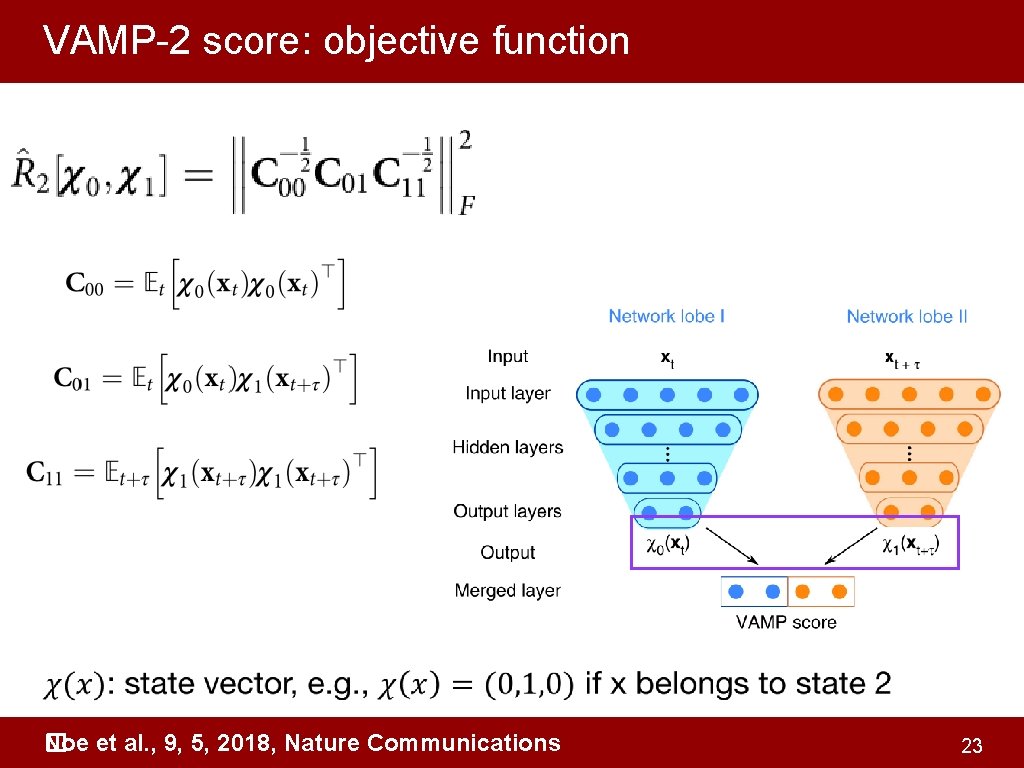
VAMPnets for deep learning of molecular kinetics • �AMPnets: employ the variational approach for Markov processes V (VAMP) to develop a deep learning framework for molecular kinetics using neural networks, encodes the entire mapping from molecular coordinates to Markov states, thus combining the whole data processing pipeline in a single end-to-end framework. coordinates state vector Related to the implied timescale plot, maximize it Noe et al. , 9, 5, 2018, Nature Communications � 16

Understanding VAMPnets • The basic structure of neural network • What is VAMP score 17

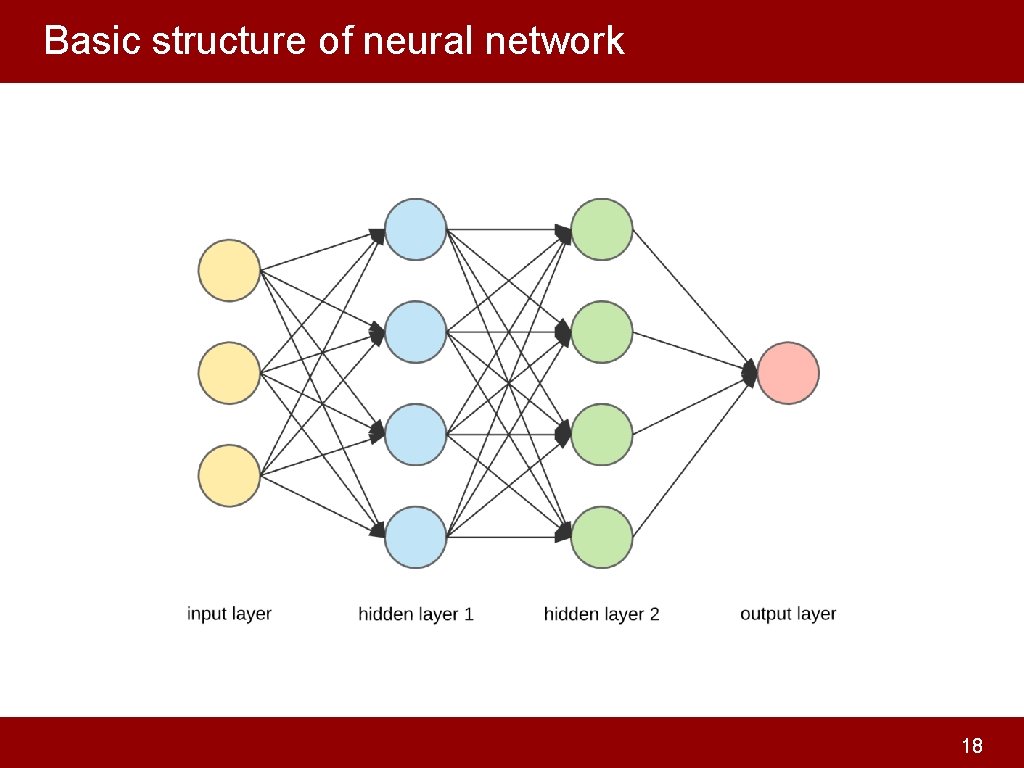
Basic structure of neural network 18

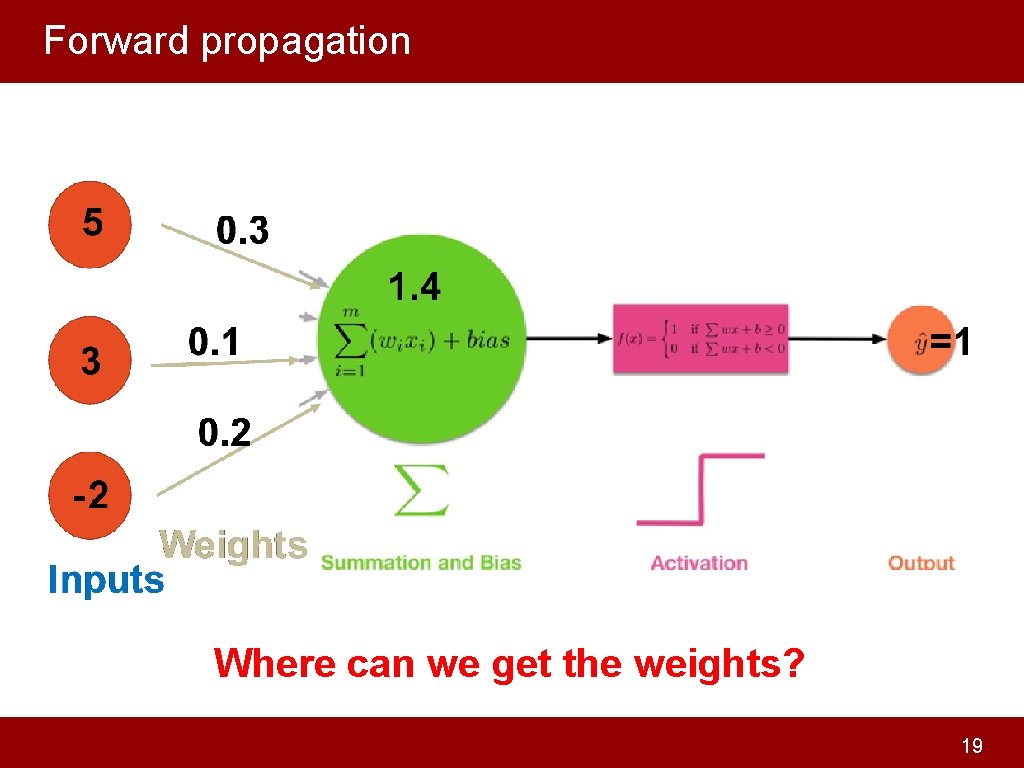
Forward propagation Where can we get the weights? 19

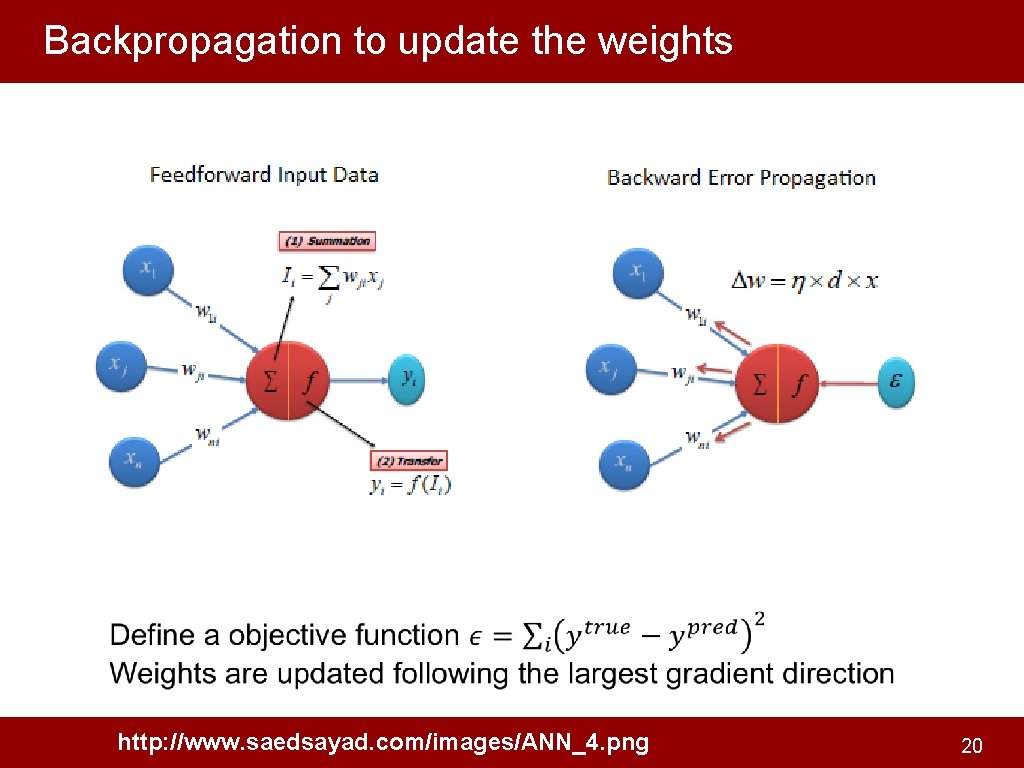
Backpropagation to update the weights http: //www. saedsayad. com/images/ANN_4. png 20

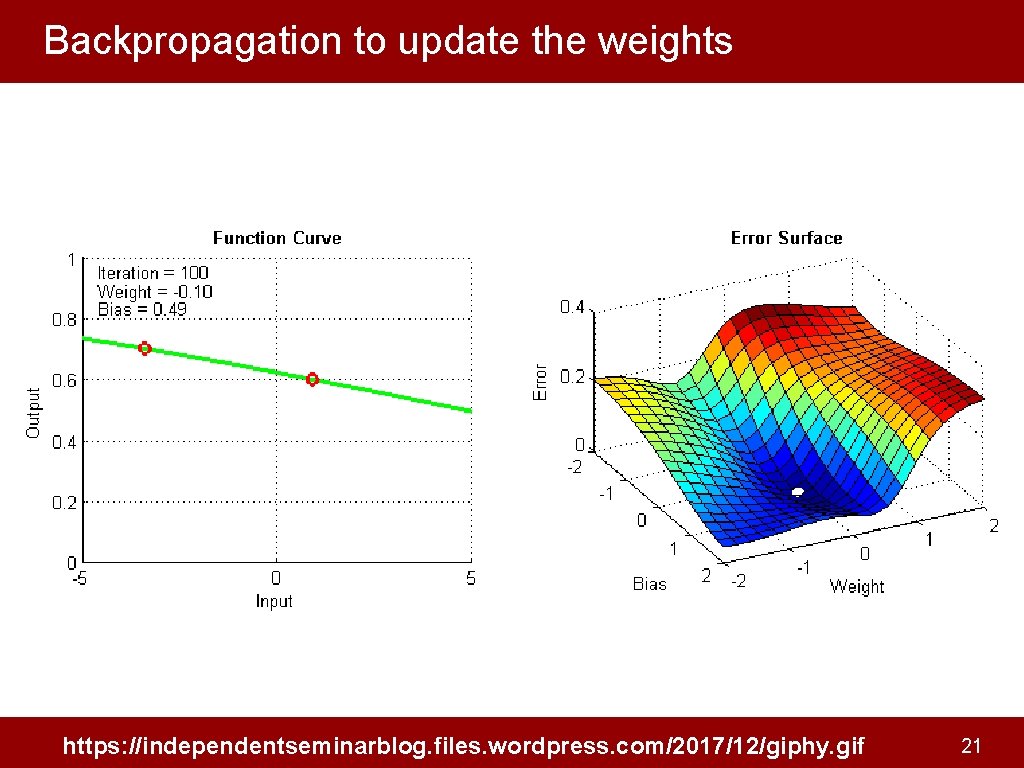
Backpropagation to update the weights https: //independentseminarblog. files. wordpress. com/2017/12/giphy. gif 21

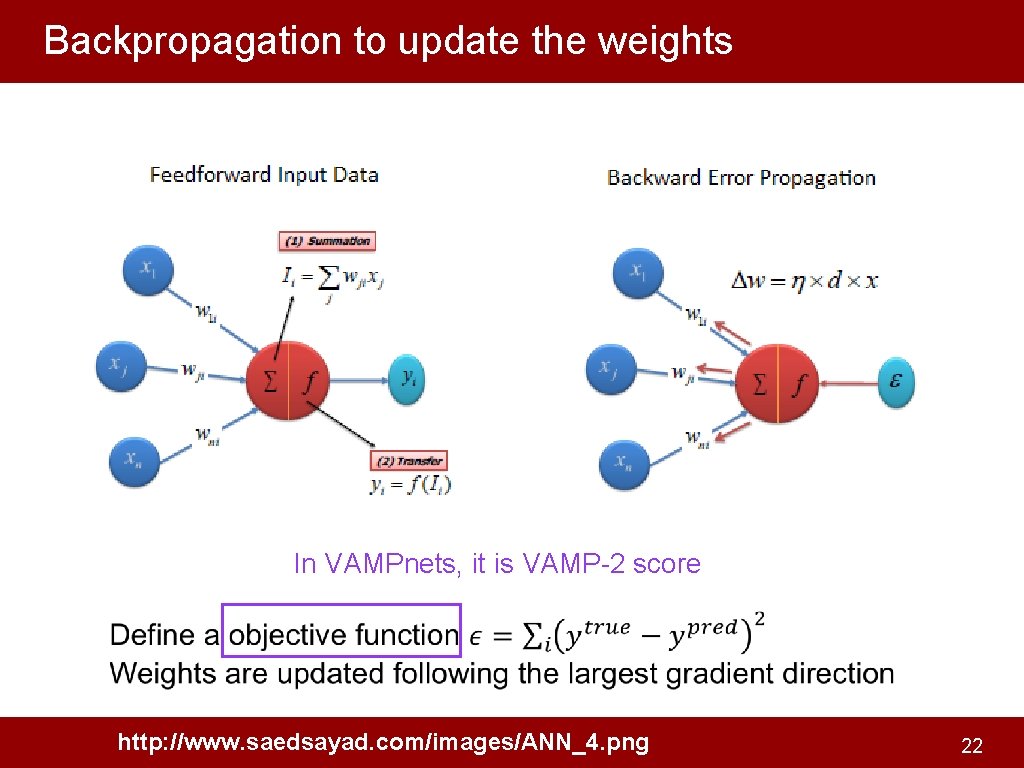
Backpropagation to update the weights In VAMPnets, it is VAMP-2 score http: //www. saedsayad. com/images/ANN_4. png 22

VAMP-2 score: objective function Noe et al. , 9, 5, 2018, Nature Communications � 23

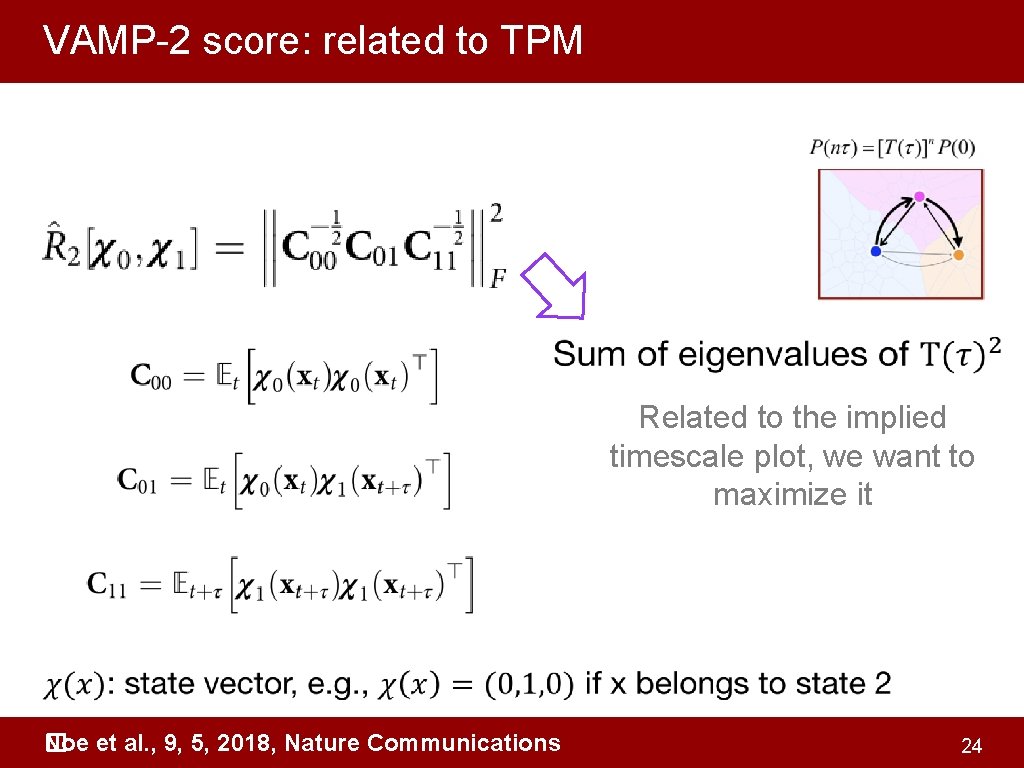
VAMP-2 score: related to TPM Related to the implied timescale plot, we want to maximize it Noe et al. , 9, 5, 2018, Nature Communications � 24

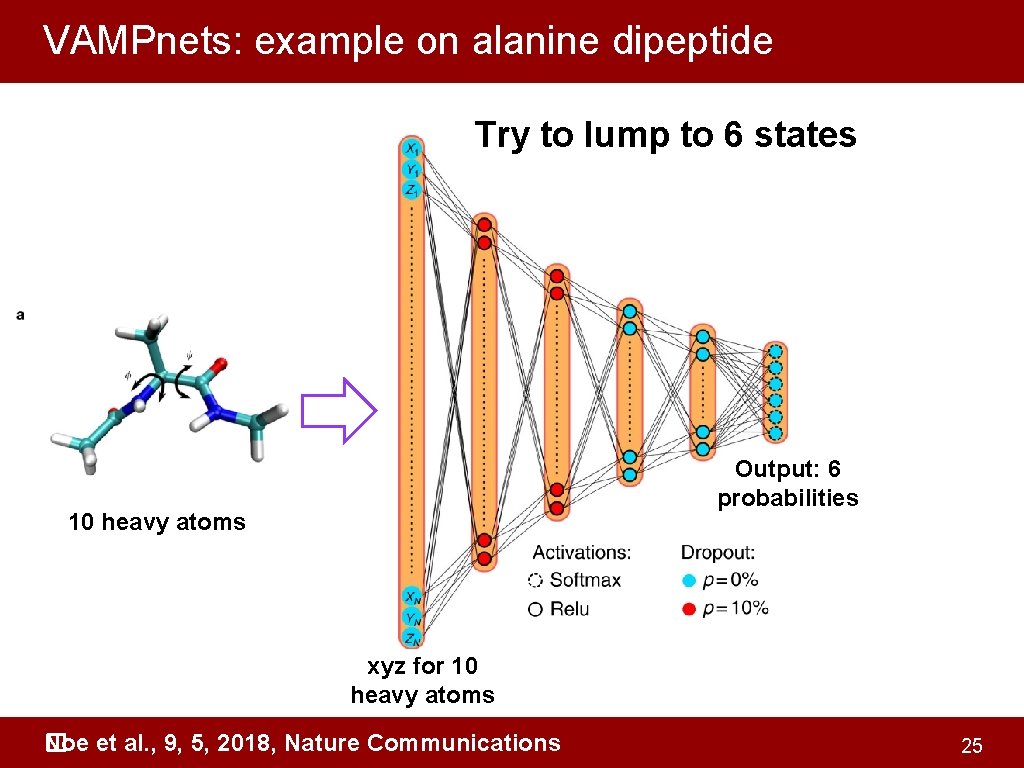
VAMPnets: example on alanine dipeptide Try to lump to 6 states Output: 6 probabilities 10 heavy atoms xyz for 10 heavy atoms Noe et al. , 9, 5, 2018, Nature Communications � 25

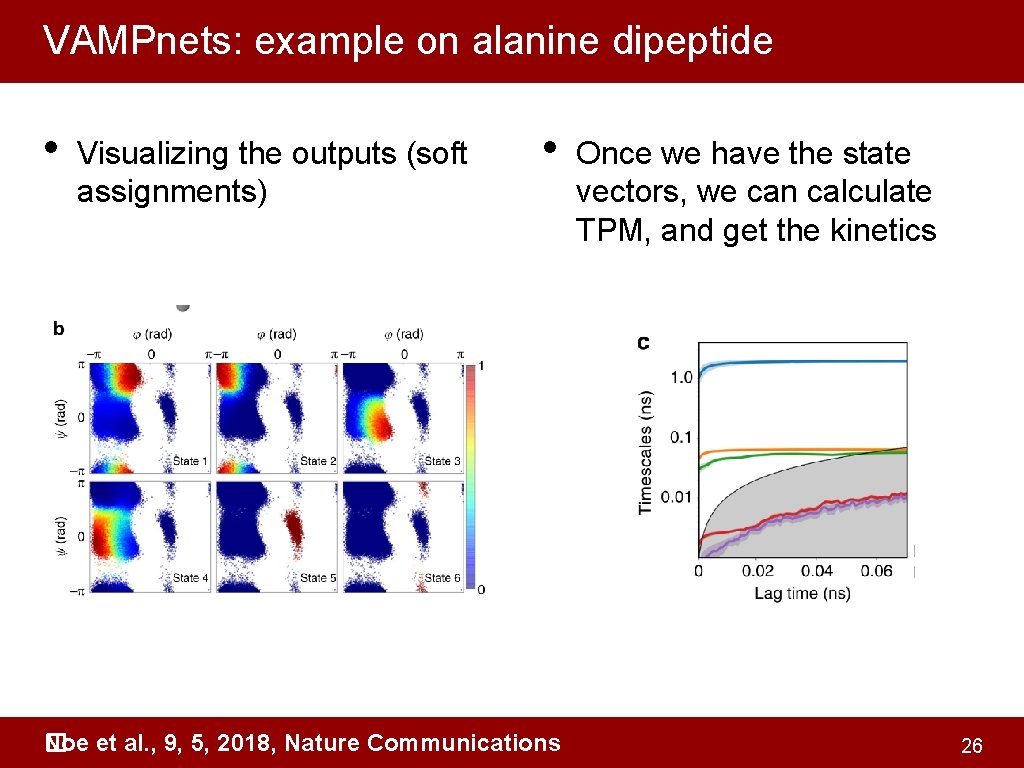
VAMPnets: example on alanine dipeptide • Visualizing the outputs (soft assignments) • Noe et al. , 9, 5, 2018, Nature Communications � Once we have the state vectors, we can calculate TPM, and get the kinetics 26

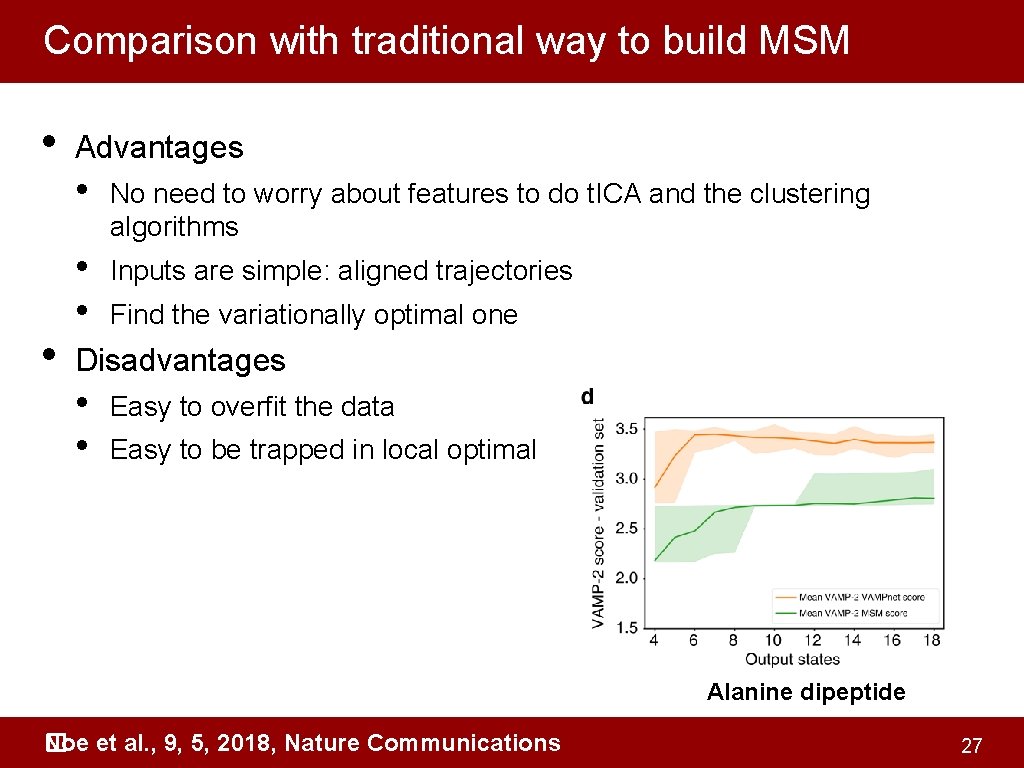
Comparison with traditional way to build MSM • • Advantages • No need to worry about features to do t. ICA and the clustering algorithms • • Inputs are simple: aligned trajectories Find the variationally optimal one Disadvantages • • Easy to overfit the data Easy to be trapped in local optimal Alanine dipeptide Noe et al. , 9, 5, 2018, Nature Communications � 27

Outline • General protocol of building MSM • Challenges with MSM • VAMPnets • Time-lagged auto-encoder 28

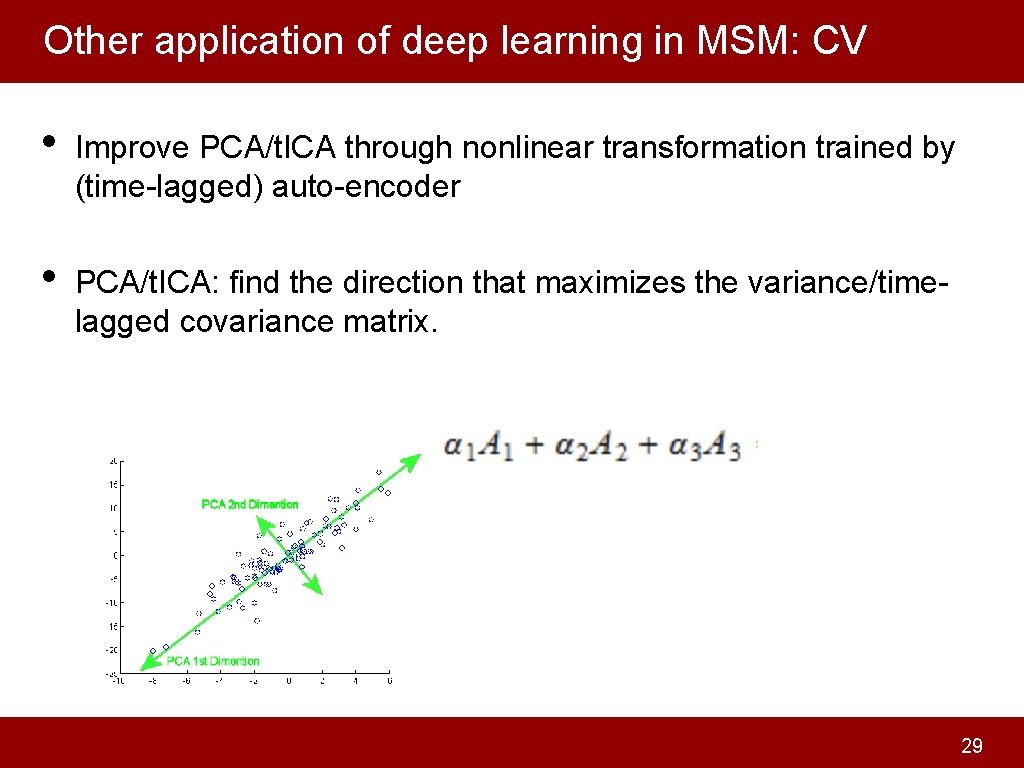
Other application of deep learning in MSM: CV • Improve PCA/t. ICA through nonlinear transformation trained by (time-lagged) auto-encoder • PCA/t. ICA: find the direction that maximizes the variance/timelagged covariance matrix. 29

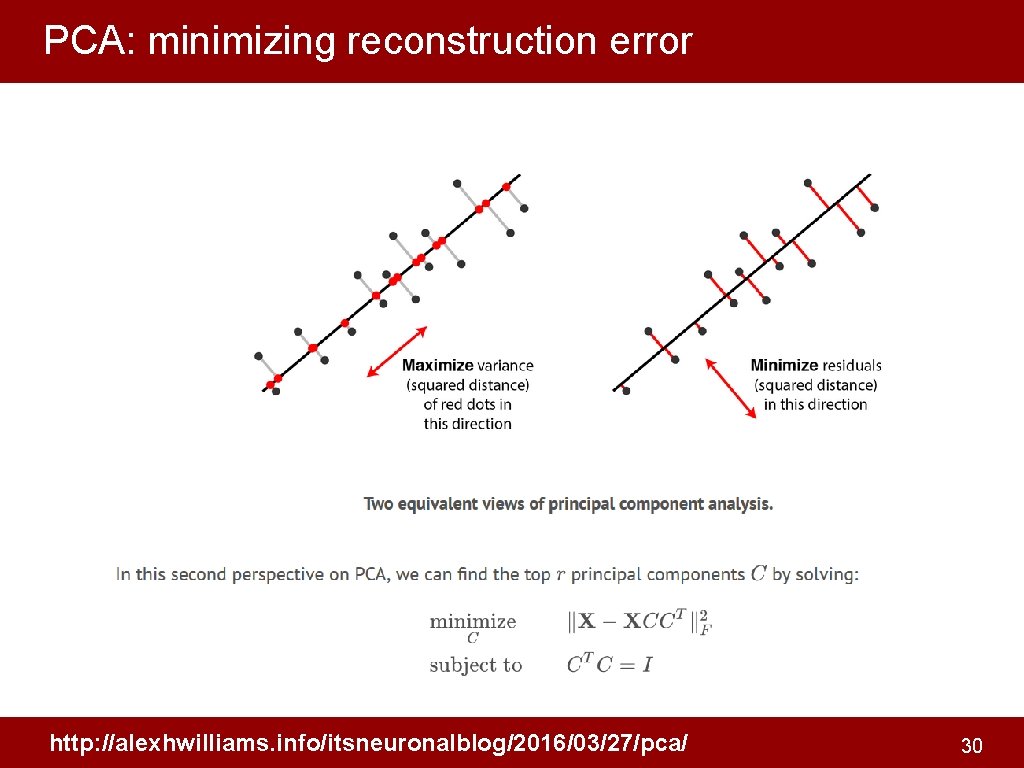
PCA: minimizing reconstruction error http: //alexhwilliams. info/itsneuronalblog/2016/03/27/pca/ 30

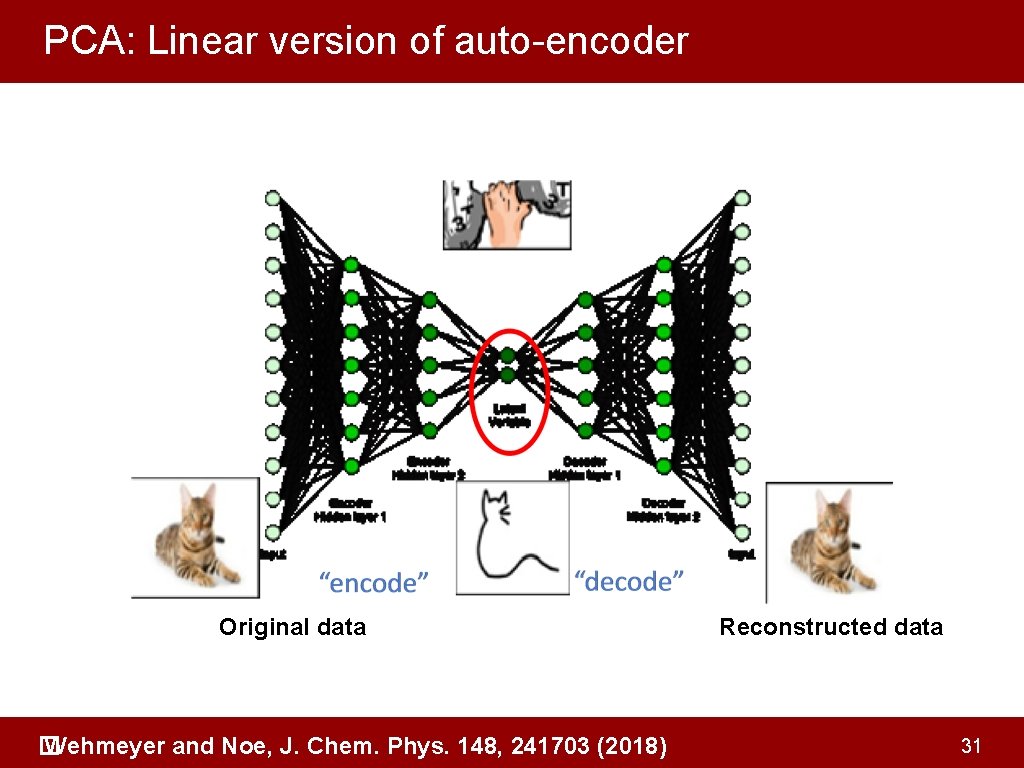
PCA: Linear version of auto-encoder Original data Wehmeyer and Noe, J. Chem. Phys. 148, 241703 (2018) � Reconstructed data 31

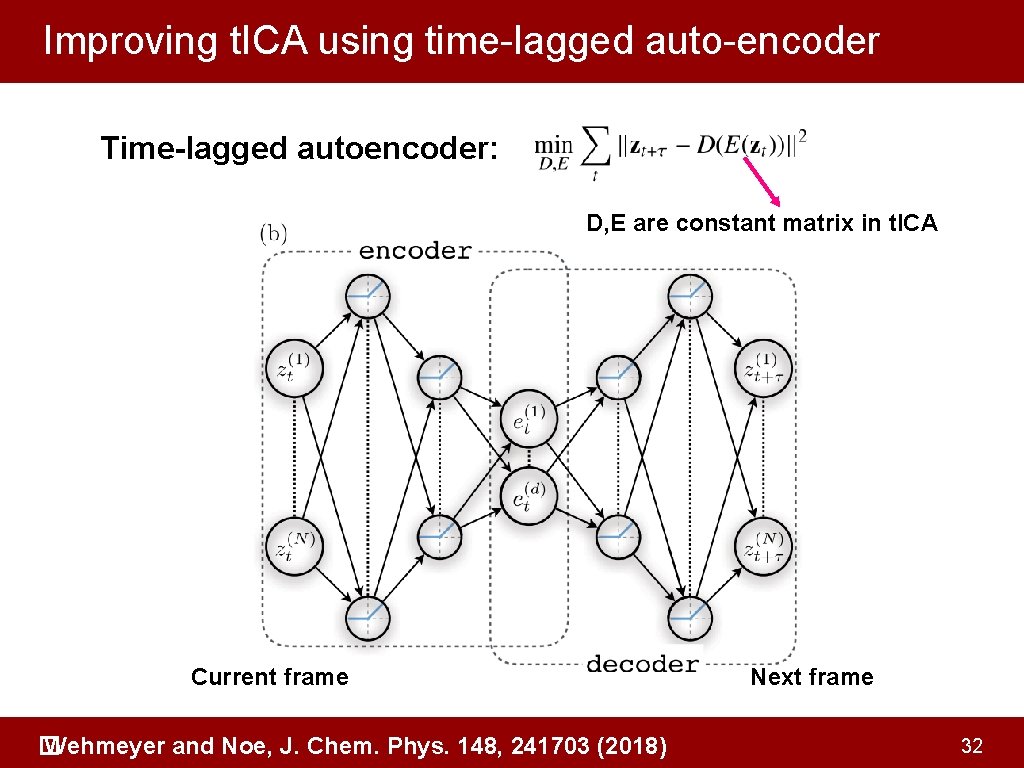
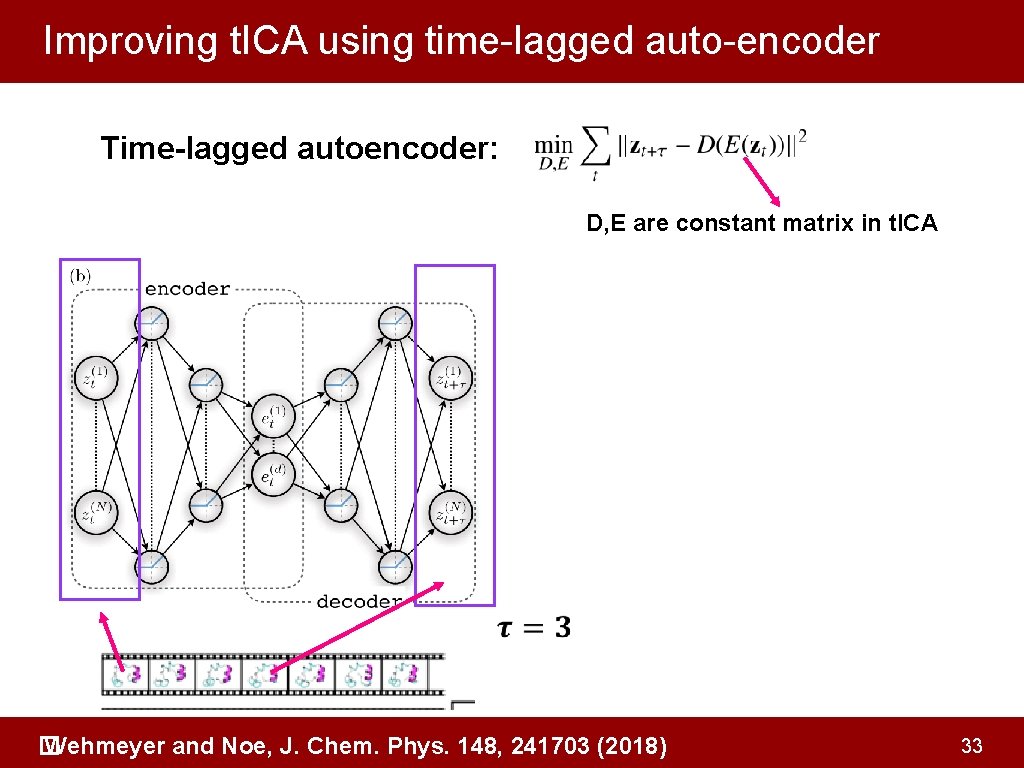
Improving t. ICA using time-lagged auto-encoder Time-lagged autoencoder: D, E are constant matrix in t. ICA Current frame Wehmeyer and Noe, J. Chem. Phys. 148, 241703 (2018) � Next frame 32

Improving t. ICA using time-lagged auto-encoder Time-lagged autoencoder: D, E are constant matrix in t. ICA Wehmeyer and Noe, J. Chem. Phys. 148, 241703 (2018) � 33

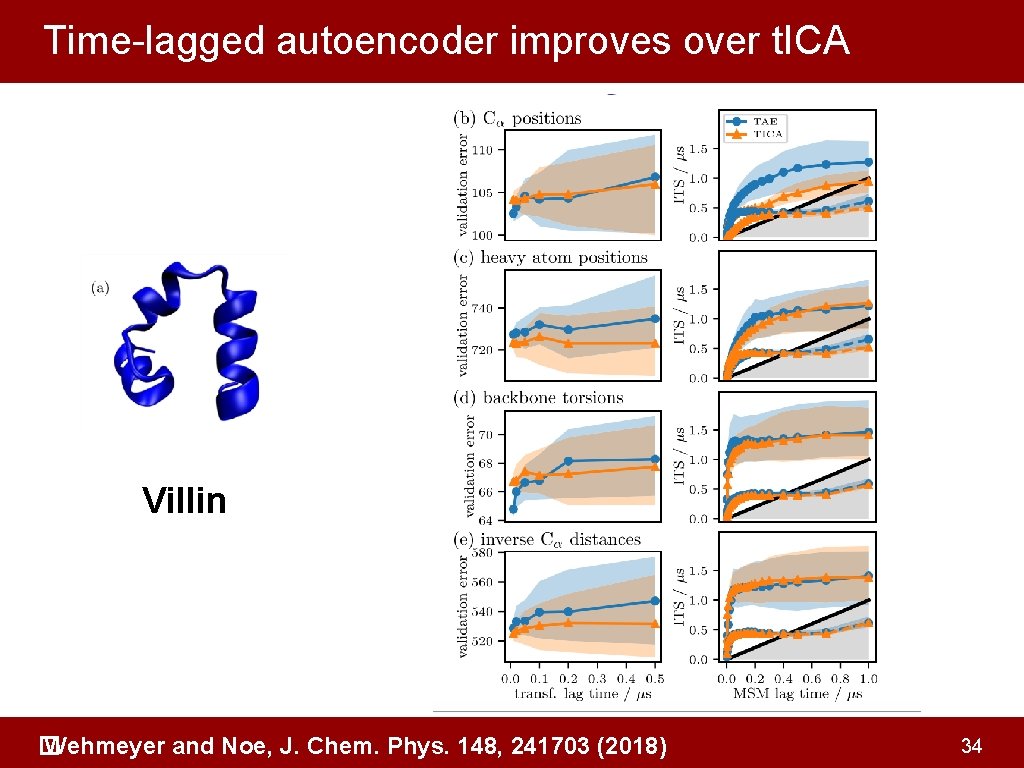
Time-lagged autoencoder improves over t. ICA Villin Wehmeyer and Noe, J. Chem. Phys. 148, 241703 (2018) � 34

Summary • Deep learning improves MSM in reducing the number of prior knowledge • However, deep learning may overfit the data when our sampling is not enough 35