The Trace Archive Steven Leonard PSG Trace Archive




















- Slides: 25

The Trace Archive Steven Leonard PSG

Trace. Archive Background Contents Database and File Structure Web pages Future Work

Background Human ramp-up 2000 Mouse Sequencing Consortium 6 th October 2000. . In fact, the incorporation of the whole genome shotgun sequencing component has led to adoption of a new, even more rapid data release policy whereby the actual raw data (that is, individual DNA sequence traces, about 500 bases long taken directly from the automated instruments) will be deposited regularly in a newly-established public databases operated by the NCBI and EBI. .

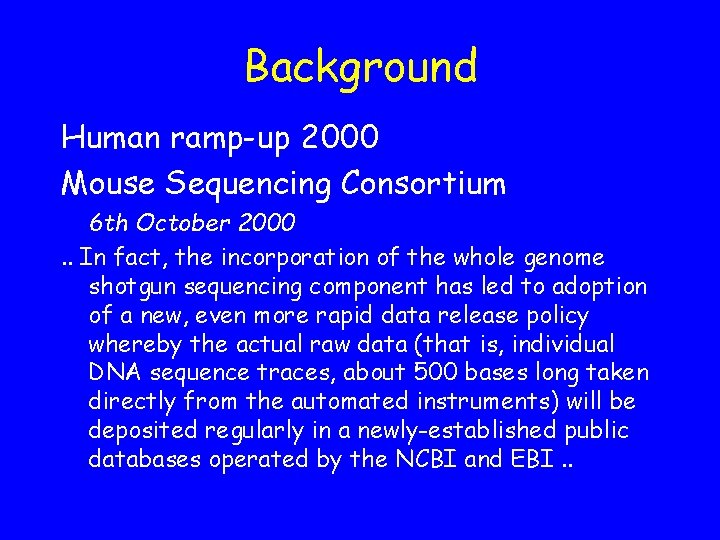
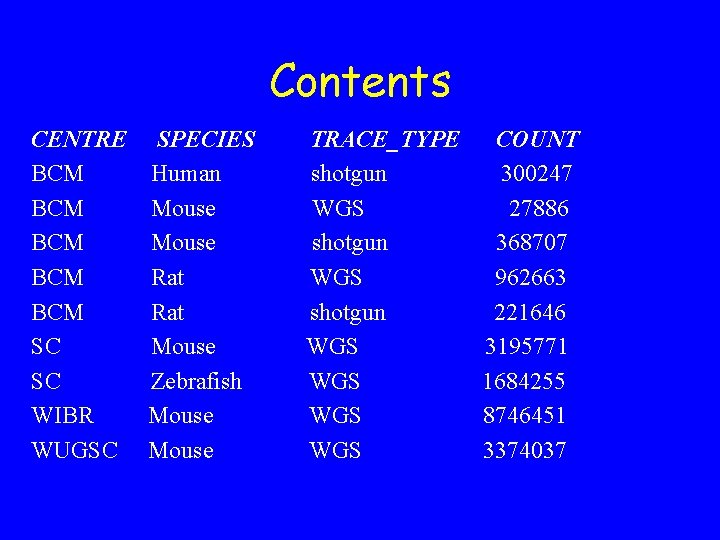
Contents CENTRE BCM BCM BCM SC SC WIBR WUGSC SPECIES Human Mouse Rat Mouse Zebrafish Mouse TRACE_TYPE COUNT shotgun 300247 WGS 27886 shotgun 368707 WGS 962663 shotgun 221646 WGS 3195771 WGS 1684255 WGS 8746451 WGS 3374037

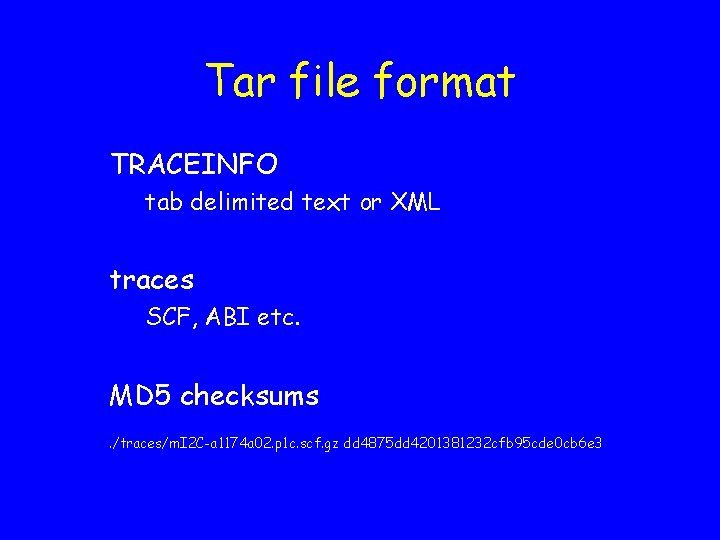
Tar file format TRACEINFO tab delimited text or XML traces SCF, ABI etc. MD 5 checksums. /traces/m. I 2 C-a 1174 a 02. p 1 c. scf. gz dd 4875 dd 4201381232 cfb 95 cde 0 cb 6 e 3

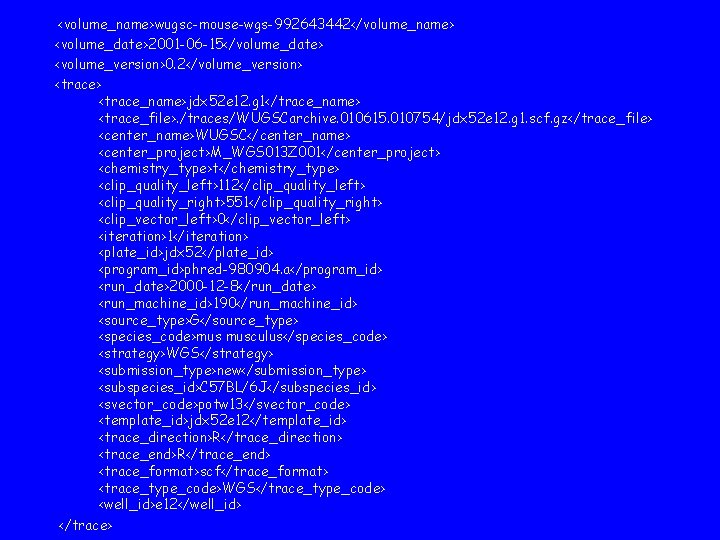
<volume_name>wugsc-mouse-wgs-992643442</volume_name> <volume_date>2001 -06 -15</volume_date> <volume_version>0. 2</volume_version> <trace_name>jdx 52 e 12. g 1</trace_name> <trace_file>. /traces/WUGSCarchive. 010615. 010754/jdx 52 e 12. g 1. scf. gz</trace_file> <center_name>WUGSC</center_name> <center_project>M_WGS 013 Z 001</center_project> <chemistry_type>t</chemistry_type> <clip_quality_left>112</clip_quality_left> <clip_quality_right>551</clip_quality_right> <clip_vector_left>0</clip_vector_left> <iteration>1</iteration> <plate_id>jdx 52</plate_id> <program_id>phred-980904. a</program_id> <run_date>2000 -12 -8</run_date> <run_machine_id>190</run_machine_id> <source_type>G</source_type> <species_code>mus musculus</species_code> <strategy>WGS</strategy> <submission_type>new</submission_type> <subspecies_id>C 57 BL/6 J</subspecies_id> <svector_code>potw 13</svector_code> <template_id>jdx 52 e 12</template_id> <trace_direction>R</trace_direction> <trace_end>R</trace_end> <trace_format>scf</trace_format> <trace_type_code>WGS</trace_type_code> <well_id>e 12</well_id> </trace>

Text trace_name trace_format trace_direction trace_end center_name center_project seq_lib_id species_code source_type strategy trace_type_code submission_type clone_id template_id run_machine_id plate_id well_id run_lane insert_size insert_stdev primer_code svector_code trace_file 19866873850757 scf F F CRA Rat. BN 2. 5. 2 L Rattus norvegicus G WGS shotgun new 19600430726162 19667033328873 S 100000035 NU 02001 XBI A 01 001 3000 0 M 13 Forward p. UC 194 C NU 02001 XBI#0984066701_A 01_00000019866873850757. pro. scf 19866873850773 scf F F CRA Rat. BN 2. 5. 2 L Rattus norvegicus G WGS shotgun new 19600430726163 19667033328874 S 100000035 NU 02001 XBI A 02 017 3000 0 M 13 Forward p. UC 194 C NU 02001 XBI#0984066701_A 02_017_00000019866873850773. pro. scf

Trace Format gzipped SCF v 3. 0 convert_trace Staden iolib-1. 8. 7 James Bonfield input SCF, ABI, etc. (CTF, ZTR) output gzipped SCF v 3. 0 (CTF, ZTR and EXP)

Database only TRACEINFO organism, centre clip L/R trace file location Index tar file with index_tar tarfile name, trace name, offset Extract files using convert_tar Staden iolib-1. 8. 7


File Structure Tar balls < 1. 5 Gbytes gzipped SCF v 3. 0 early tarballs 1 to 1 single job to extract and re-make tarball now have two jobs extract and collect

1. Extract consistency check TRACINFO, MD 5 and traces for each trace extract trace check MD 5 convert to gzipped SCF v 3. 0

2. Collect given a set of directories Calc. Size of SCF files gathers trace info verify/add defaults TRACEINFO files in original tarballs Experiment files Gather into tarballs account for duplicate traces calc. MD 5 checksums

3. Insert given a set of tarball extract TRACEINFO and parse XML indexes the tarball for each trace check for pre-existing traces get dictionary id’s populate trace_info/ancillary tables

Finally generate Fasta/Quality files generate Clip files Update FTP site Re-build SSAHA hash tables

Given a list of traces generate a single fasta files Extend this to generate a tarfile SCF FASTA QUAL TRACEINFO text or XML Need to restrict size of tarballs and cache previous results

Clipping Info Pass Fail ml 2 C-b 205 c 06. p 1 c 22 571 274 276 550 ml 2 C-b 205 c 07. p 1 c Contam CVEC: p. BACe 3. 6 ml 2 C-b 111 e 07. p 1 c Qual Where the above is "Pass|Fail", readname, start, end, GC, AT, (end-start+1) so the start and stop positions are inclusive and base counting starts at 1. I define the starting quality clip point as the start of the first 20 bp window where the integrated error rate within the window drops below 1. 00 and the ending quality clip point as the end of the last 20 bp window where the integrated error rate rises above 1. 00.

Problems Duplicate trace names 1, 000+ <10, 000 None Whitehead WUGSC/BCM Sanger (5, 000) Bad tarballs/tapes FTP errors Backup it up lost 150 Gbytes, 3, 000+ traces

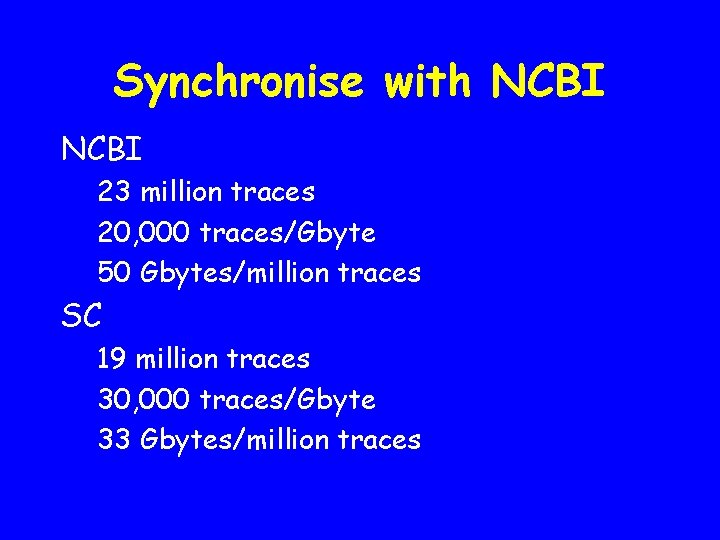
Synchronise with NCBI 23 million traces 20, 000 traces/Gbyte 50 Gbytes/million traces SC 19 million traces 30, 000 traces/Gbyte 33 Gbytes/million traces

Acknowledgements Sanger Richard Durbin, Jim Mullikin Andy Smith, Simon Mercer Tony Cox + Web team James Cuff + SSG Santhi Sivadasan, Martin. Widlake NCBI Vladimir Alekseyev, Eugene Yaschenko, Deanna Church

Future Work Other organisms C. briggsae Dicty Tetraodon Xenopus EST’s submit to EMBL from the archive True Trace Server Asp Gap 4, etc.

==> Netscape URL http: //trace. ensembl. org/ search on trace_name trace/list of traces add quality values add trace (need Java)

==> Netscape SSAHA Server 14 GBytes to build, 4 -5 Gbytes to run preload file in browser for search passed reads organism specific modified headers

==> Netscape FTP site • fasta/qual/traceinfo – fixed • clip – updated – Duplicate/Updated

==> NETSCAPE Chromosome 1