The General Linear Model GLM in SPM 5



- Slides: 28

The General Linear Model (GLM) in SPM 5 Klaas Enno Stephan Wellcome Trust Centre for Neuroimaging University College London With many thanks to my colleagues in the FIL methods group, particularly Stefan Kiebel, for useful slides SPM Short Course, May 2007 Wellcome Trust Centre for Neuroimaging

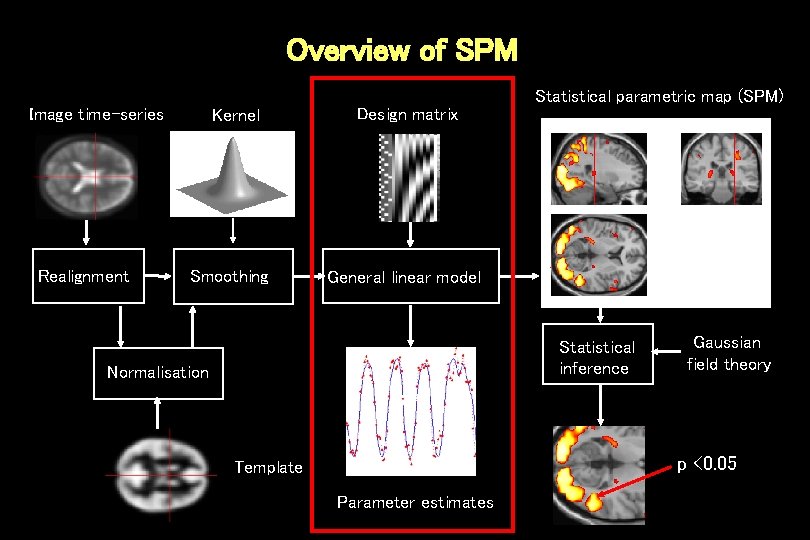
Overview of SPM Image time-series Realignment Kernel Smoothing Design matrix Statistical parametric map (SPM) General linear model Statistical inference Normalisation Gaussian field theory p <0. 05 Template Parameter estimates

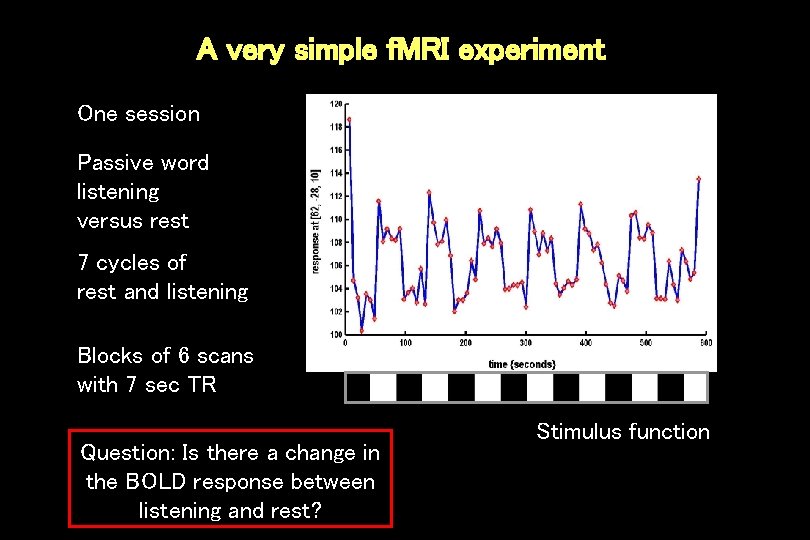
A very simple f. MRI experiment One session Passive word listening versus rest 7 cycles of rest and listening Blocks of 6 scans with 7 sec TR Question: Is there a change in the BOLD response between listening and rest? Stimulus function

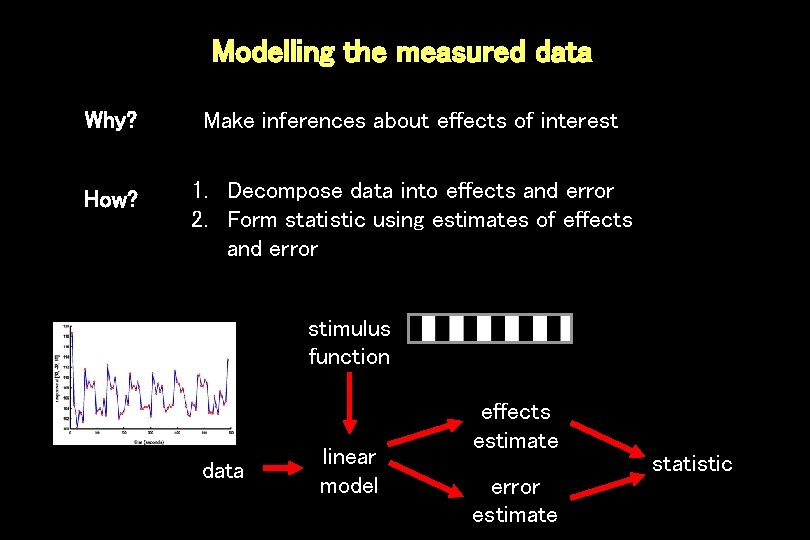
Modelling the measured data Why? Make inferences about effects of interest How? 1. Decompose data into effects and error 2. Form statistic using estimates of effects and error stimulus function data linear model effects estimate error estimate statistic

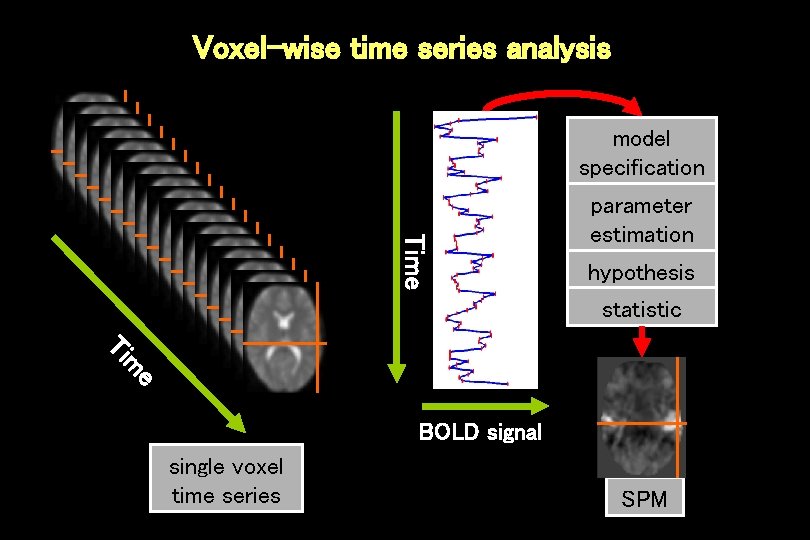
Voxel-wise time series analysis model specification Time parameter estimation hypothesis statistic e m Ti BOLD signal single voxel time series SPM

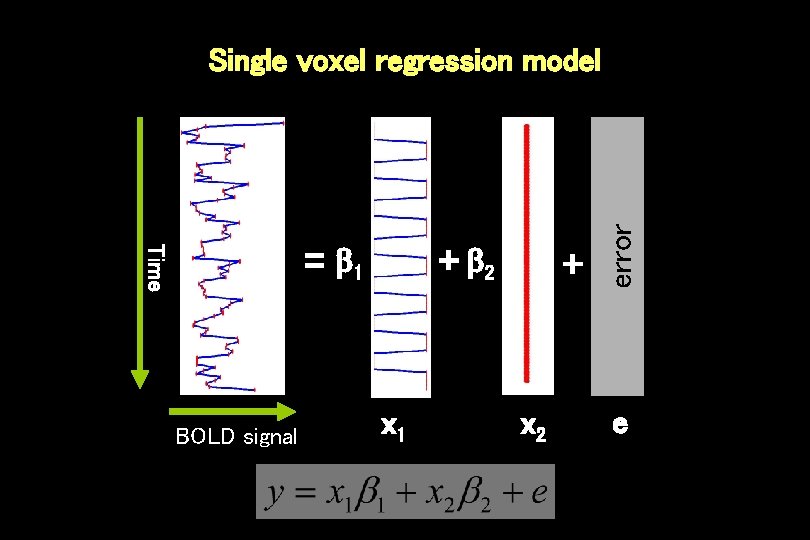
Time = 1 BOLD signal + 2 x 1 + x 2 error Single voxel regression model e

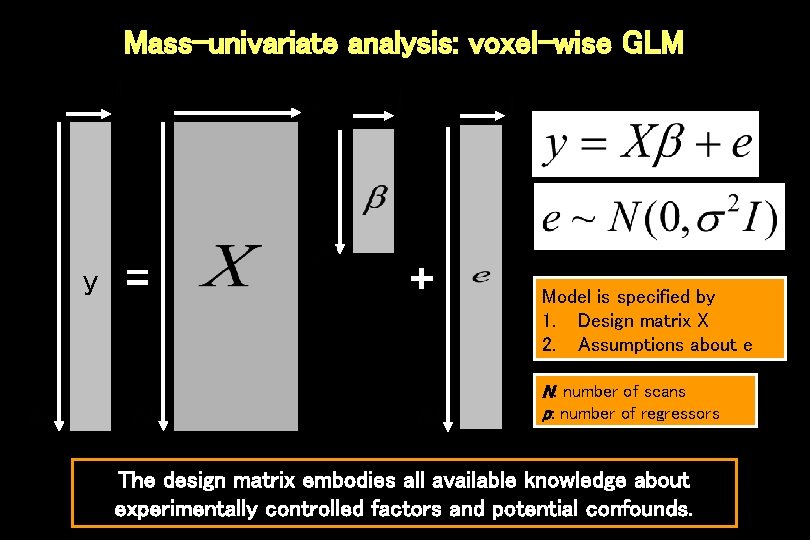
Mass-univariate analysis: voxel-wise GLM y = + Model is specified by 1. Design matrix X 2. Assumptions about e N: number of scans p: number of regressors The design matrix embodies all available knowledge about experimentally controlled factors and potential confounds.

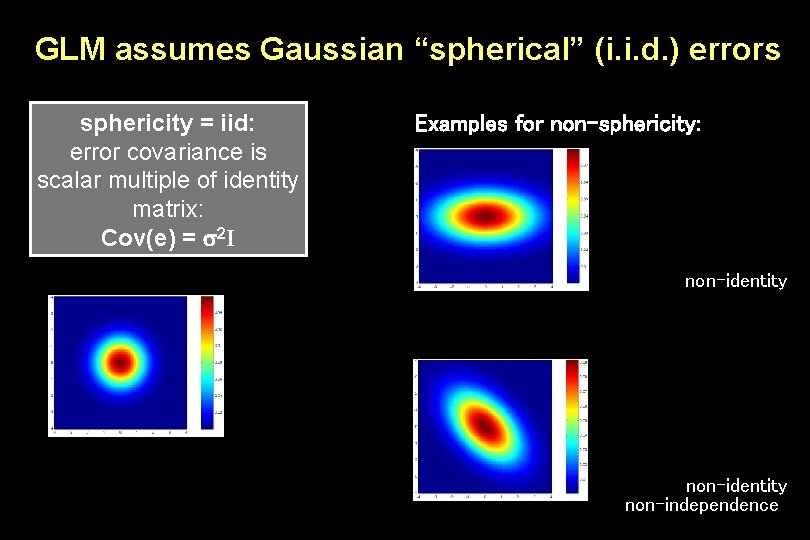
GLM assumes Gaussian “spherical” (i. i. d. ) errors sphericity = iid: error covariance is scalar multiple of identity matrix: Cov(e) = 2 I Examples for non-sphericity: non-identity non-independence

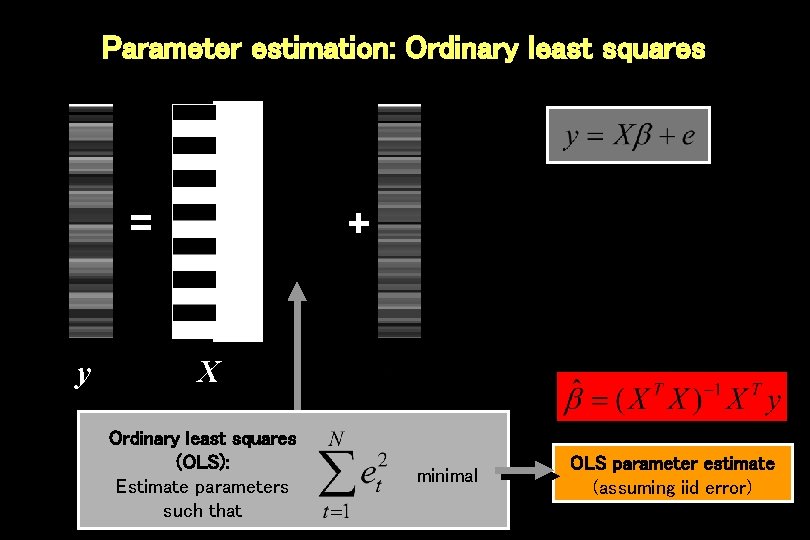
Parameter estimation: Ordinary least squares = y + X Ordinary least squares (OLS): Estimate parameters such that minimal OLS parameter estimate (assuming iid error)

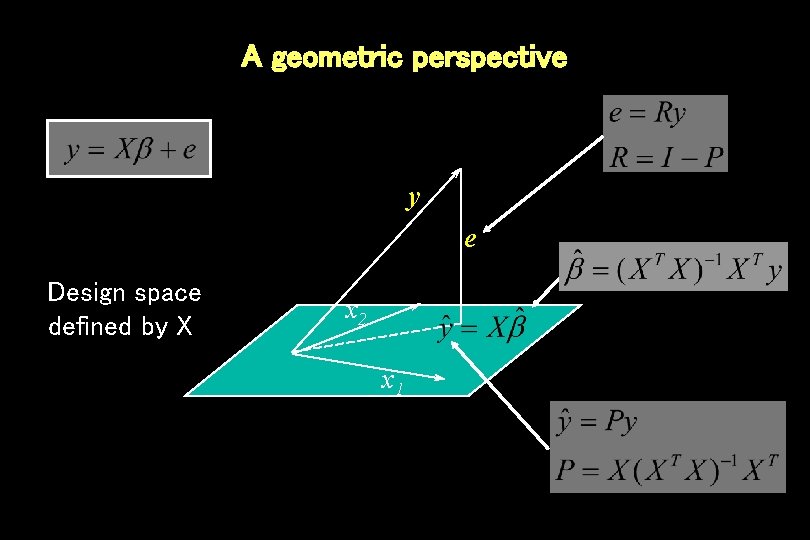
A geometric perspective y e Design space defined by X x 2 x 1

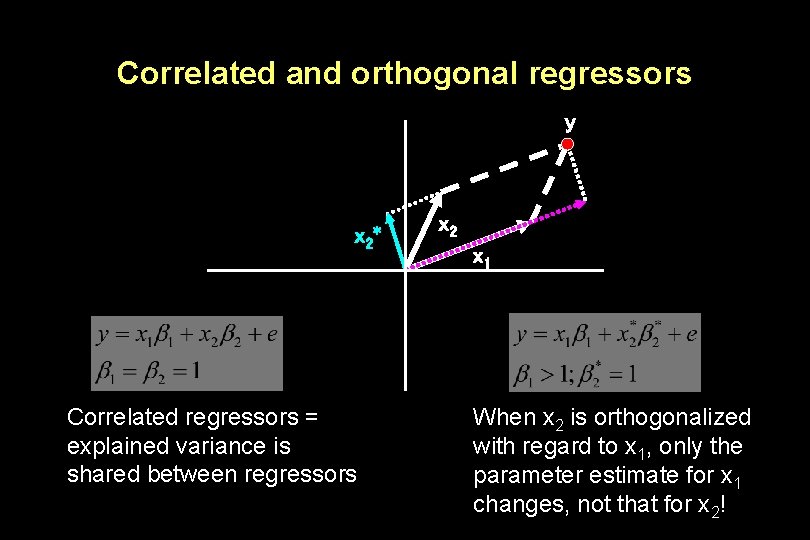
Correlated and orthogonal regressors y x 2* Correlated regressors = explained variance is shared between regressors x 2 x 1 When x 2 is orthogonalized with regard to x 1, only the parameter estimate for x 1 changes, not that for x 2!

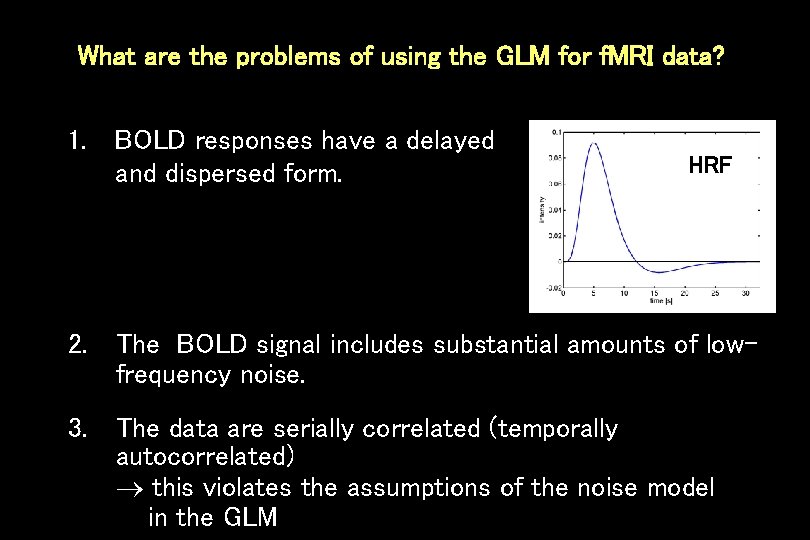
What are the problems of using the GLM for f. MRI data? 1. BOLD responses have a delayed and dispersed form. HRF 2. The BOLD signal includes substantial amounts of lowfrequency noise. 3. The data are serially correlated (temporally autocorrelated) this violates the assumptions of the noise model in the GLM

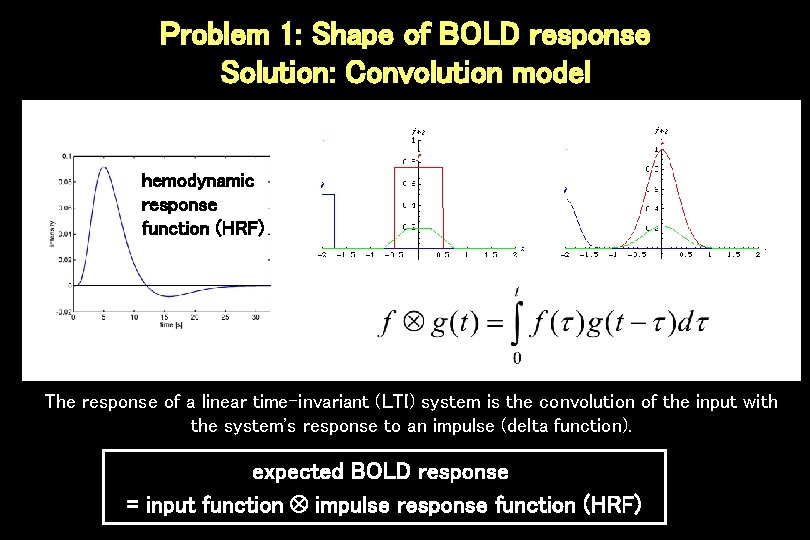
Problem 1: Shape of BOLD response Solution: Convolution model hemodynamic response function (HRF) The response of a linear time-invariant (LTI) system is the convolution of the input with the system's response to an impulse (delta function). expected BOLD response = input function impulse response function (HRF)

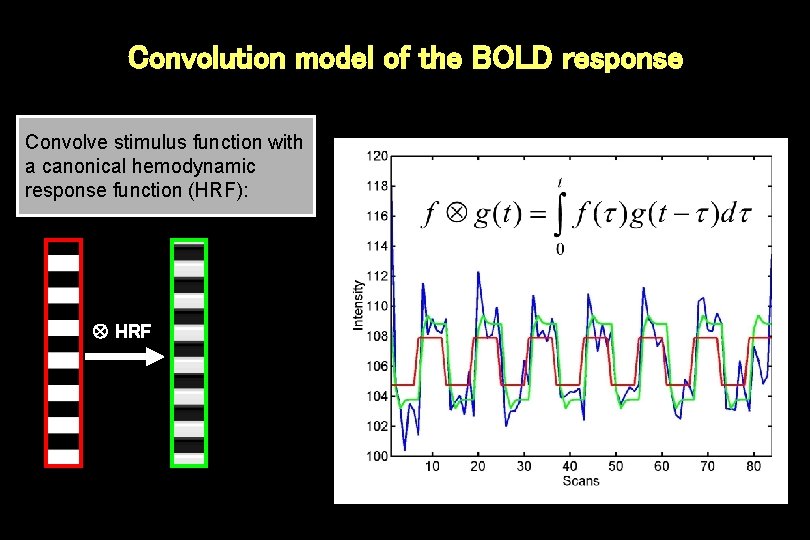
Convolution model of the BOLD response Convolve stimulus function with a canonical hemodynamic response function (HRF): HRF

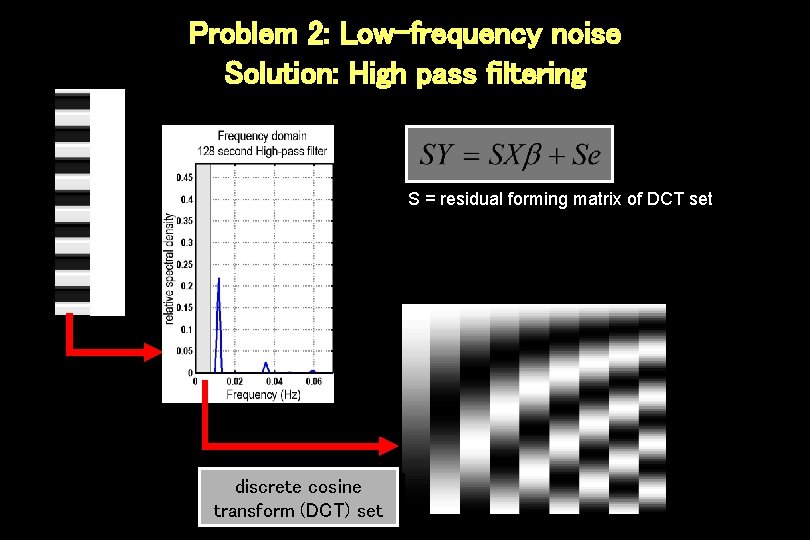
Problem 2: Low-frequency noise Solution: High pass filtering S = residual forming matrix of DCT set discrete cosine transform (DCT) set

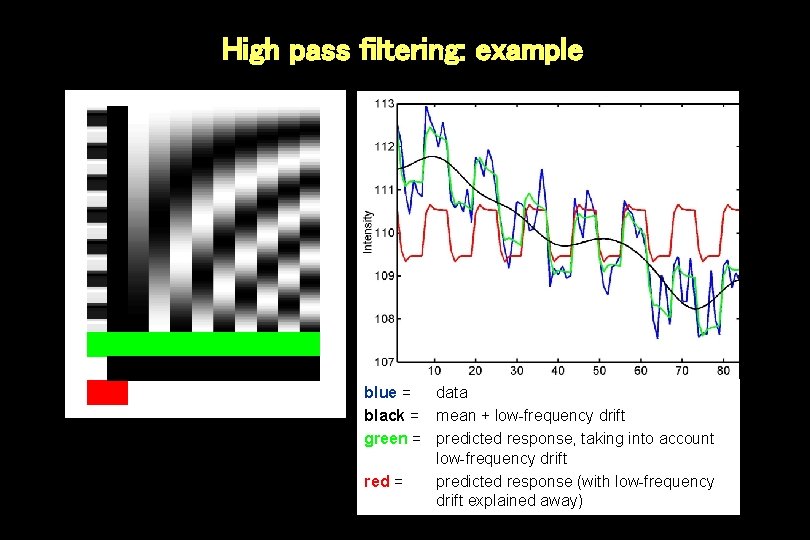
High pass filtering: example blue = data black = mean + low-frequency drift green = predicted response, taking into account low-frequency drift red = predicted response (with low-frequency drift explained away)

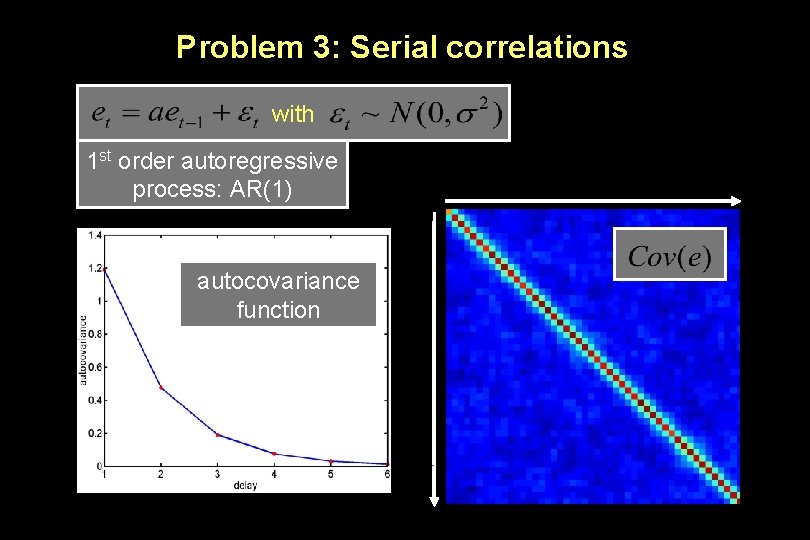
Problem 3: Serial correlations with 1 st order autoregressive process: AR(1) autocovariance function

Dealing with serial correlations • Pre-colouring: impose some known autocorrelation structure on the data (filtering with matrix W) and use Satterthwaite correction for df’s. • Pre-whitening: 1. Use an enhanced noise model with hyperparameters for multiple error covariance components. 2. Use estimated autocorrelation to specify matrix W and use weighted least squares (WLS) estimation.

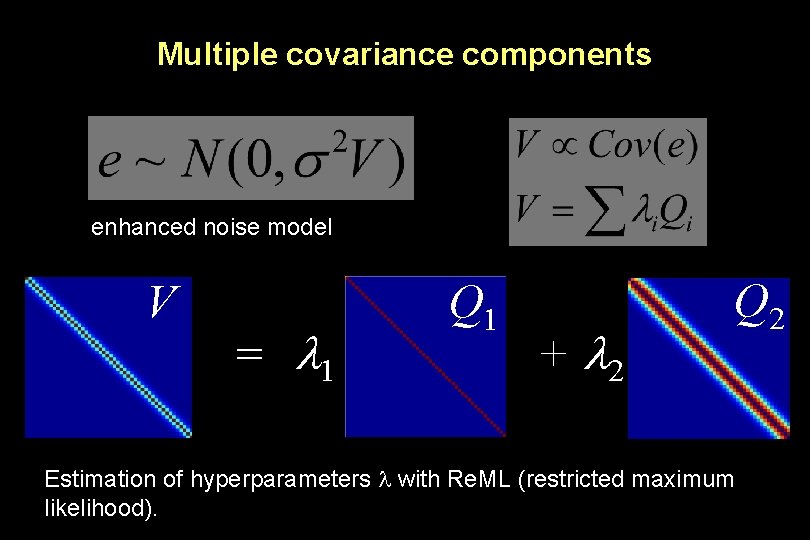
Multiple covariance components enhanced noise model V = 1 Q 1 + 2 Q 2 Estimation of hyperparameters with Re. ML (restricted maximum likelihood).

Hyperparameter estimation Three hyperparameters: 1, 2, • Assumption: error covariance pattern is identical within voxels (of same tissue class), variance is not voxel-specific estimates of , but single estimate of for all voxels • Estimation of in SPM proceeds by pooling over all voxels that survive an F-test at p<0. 001 based on OLS estimates (“first pass” during estimation in SPM)

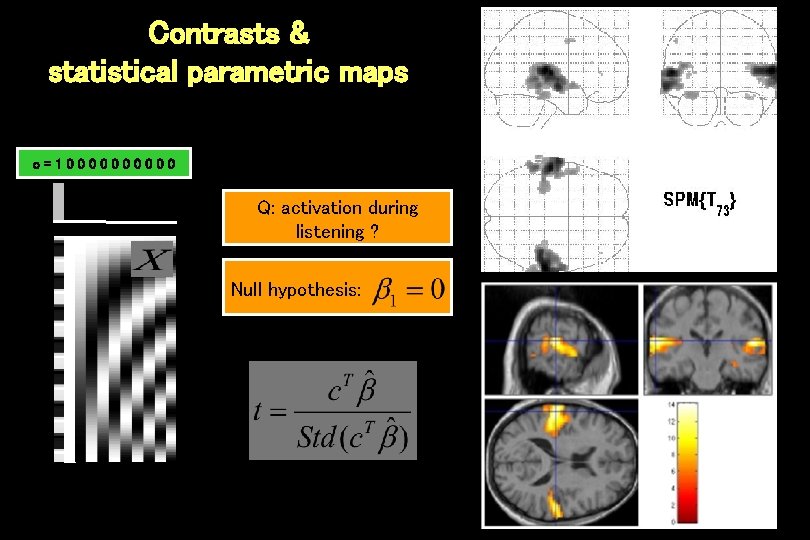
Contrasts & statistical parametric maps c=100000 Q: activation during listening ? Null hypothesis:

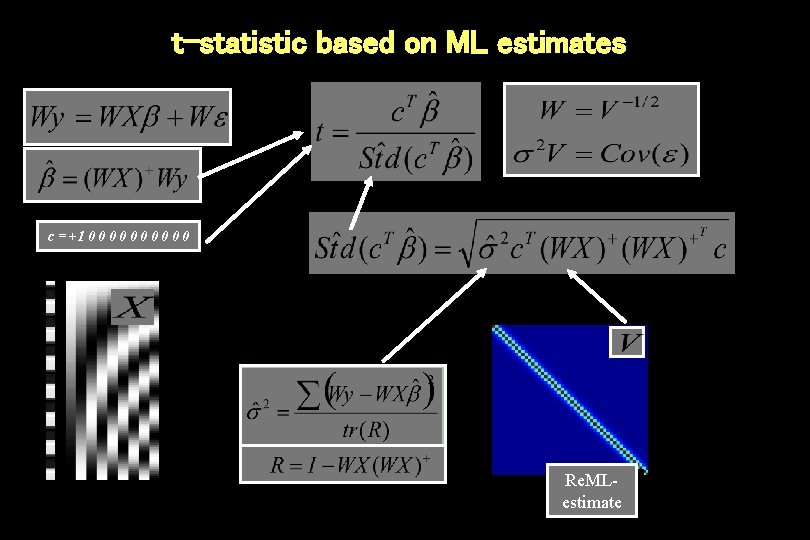
t-statistic based on ML estimates c = +1 0 0 0 0 0 Re. MLestimate

Physiological confounds • head movements • arterial pulsations • breathing • eye blinks • adaptation affects, fatigue, fluctuations in concentration, etc.

Outlook: further challenges • correction for multiple comparisons • variability in the HRF across voxels • slice timing • limitations of frequentist statistics Bayesian analyses • GLM ignores interactions among voxels models of effective connectivity

Summary • Mass-univariate approach: same GLM for each voxel • GLM includes all known experimental effects and confounds • Convolution with a canonical HRF • High-pass filtering to account for low-frequency drifts • Estimation of multiple variance components (e. g. to account for serial correlations) • Parametric statistics


Correction for multiple comparisons • Mass-univariate approach: We apply the GLM to each of a huge number of voxels (usually > 100, 000). • Threshold of p<0. 05 more than 5000 voxels significant by chance! • Massive problem with multiple comparisons! • Solution: Gaussian random field theory (Will’s talk)

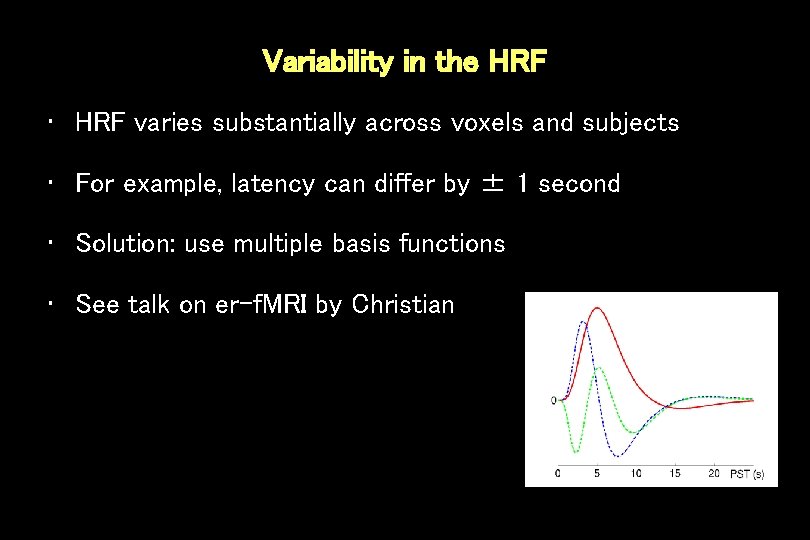
Variability in the HRF • HRF varies substantially across voxels and subjects • For example, latency can differ by ± 1 second • Solution: use multiple basis functions • See talk on er-f. MRI by Christian