Part 1 Working with RNASeq Data RNAseq overview












- Slides: 79

Part 1: Working with RNA-Seq Data

RNA-seq: overview Genome. …TCTGAAACAATGCTTCAATCTAACTTATCATTGGGA…. 2

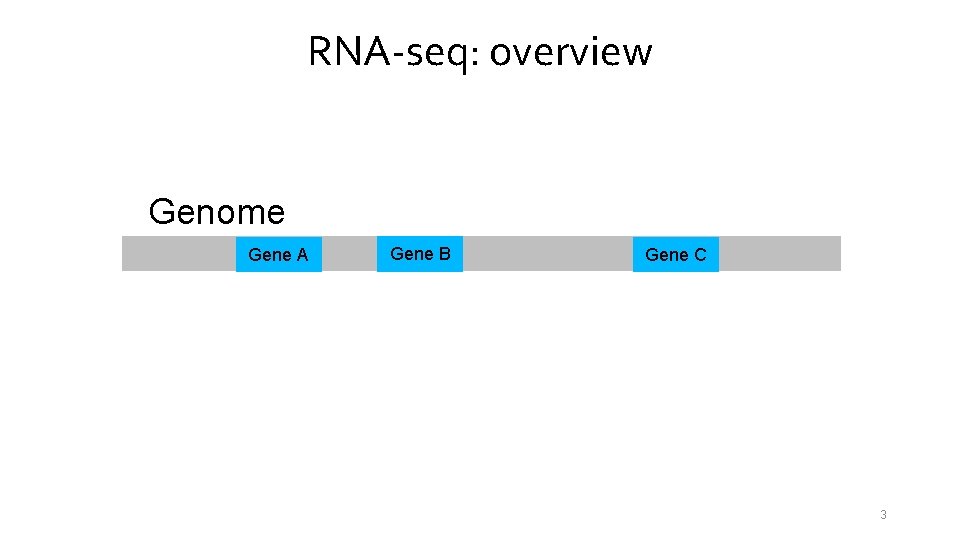
RNA-seq: overview Genome Gene A Gene B Gene C 3

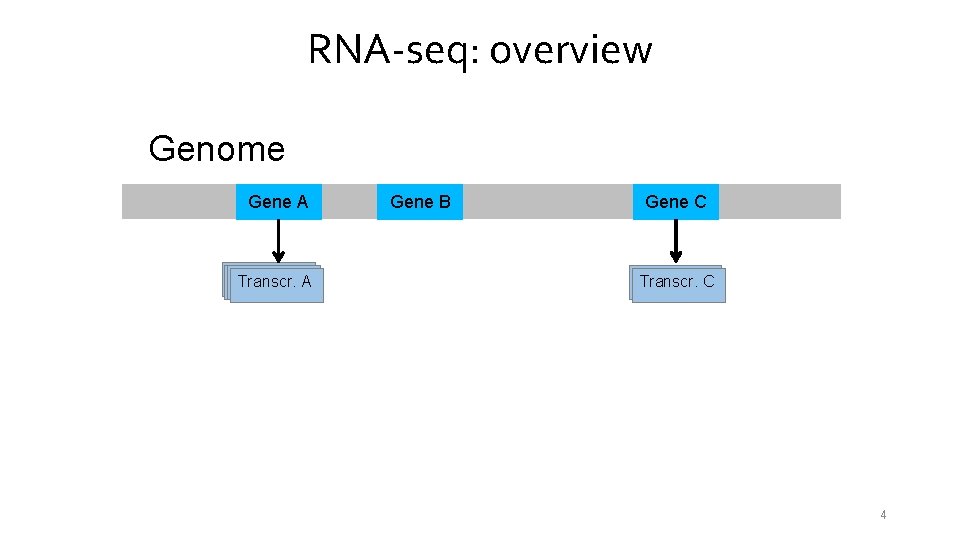
RNA-seq: overview Genome Gene A Transcr. A A Gene B Gene C Transcr. A C 4

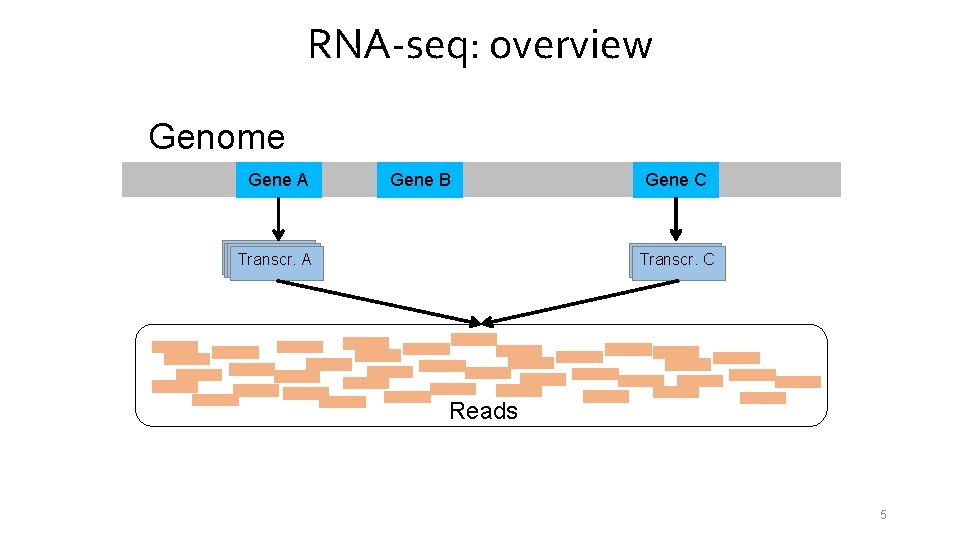
RNA-seq: overview Genome Gene A Gene B Transcr. A A Gene C Transcr. A C Reads 5

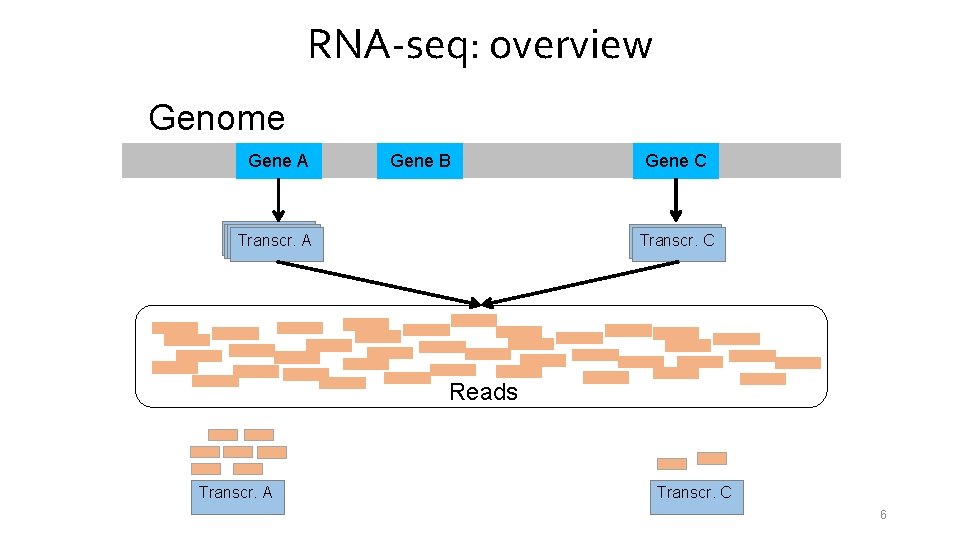
RNA-seq: overview Genome Gene A Gene B Transcr. A A Gene C Transcr. A C Reads Transcr. A Transcr. C 6

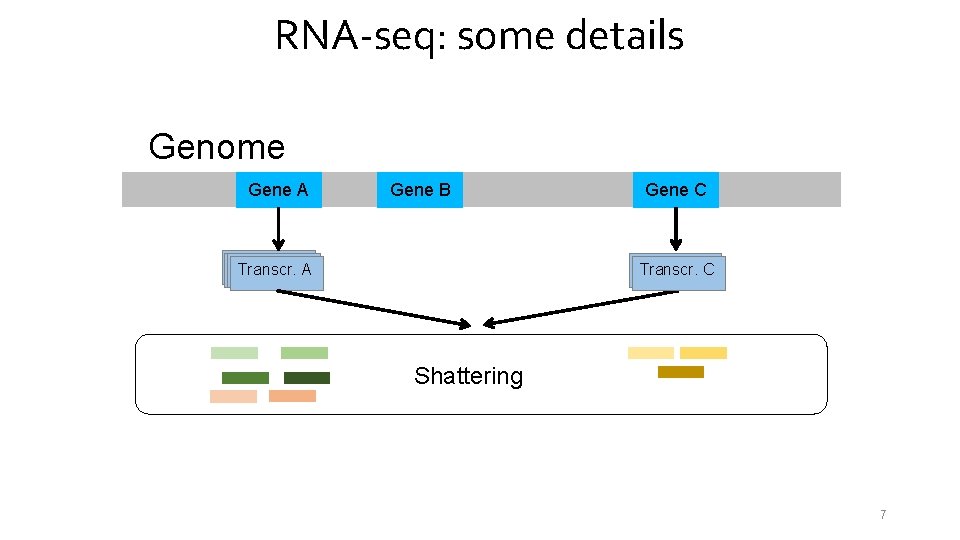
RNA-seq: some details Genome Gene A Gene B Transcr. A Gene C Transcr. C C Shattering 7

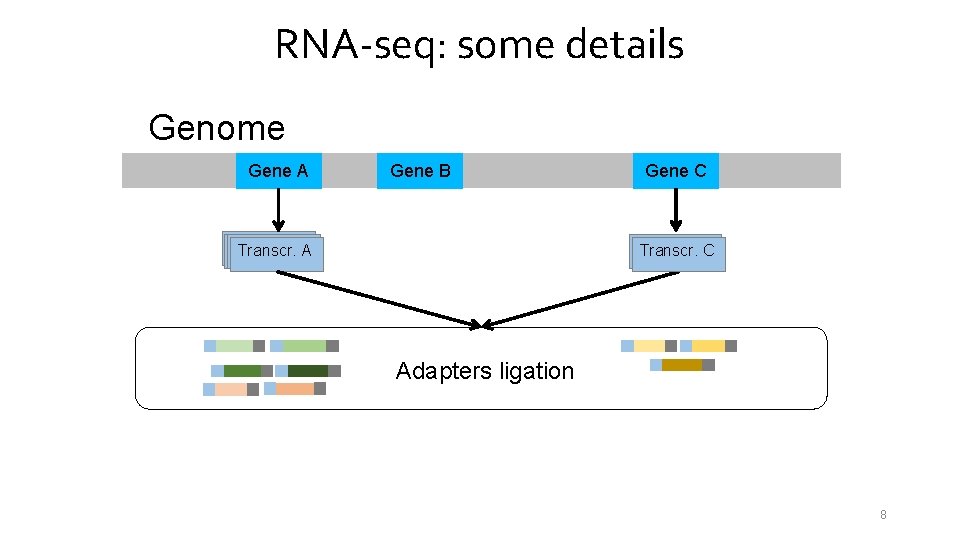
RNA-seq: some details Genome Gene A Gene B Transcr. A Gene C Transcr. C Adapters ligation 8

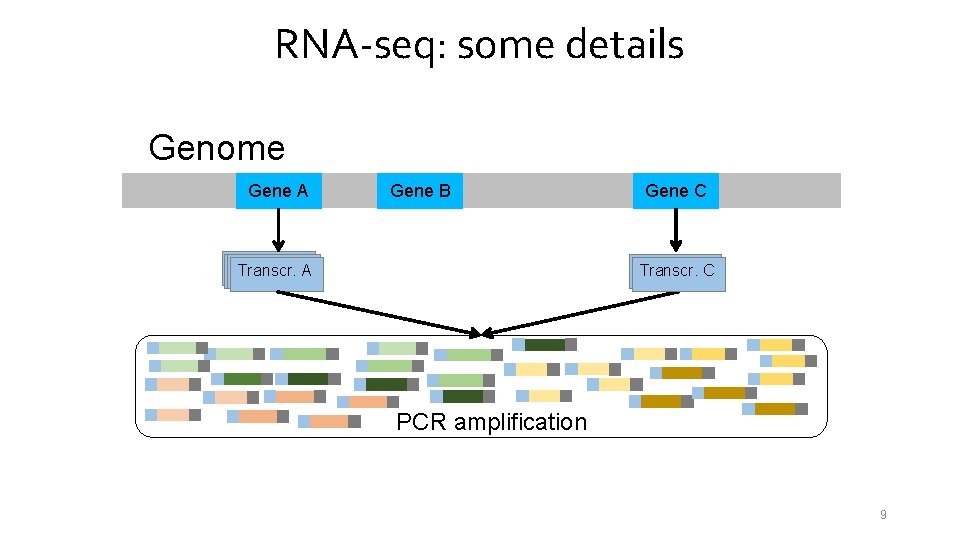
RNA-seq: some details Genome Gene A Gene B Transcr. A Gene C Transcr. C PCR amplification 9

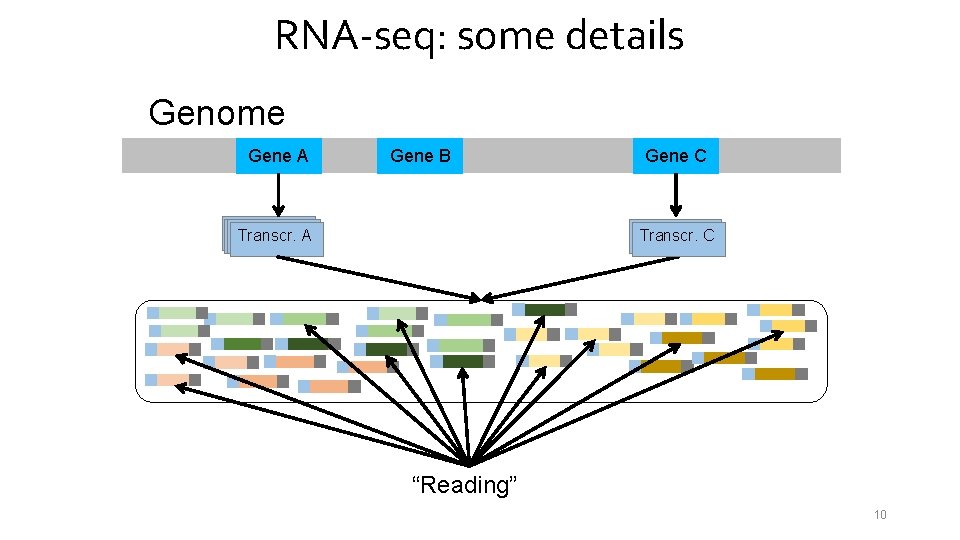
RNA-seq: some details Genome Gene A Gene B Transcr. A Gene C Transcr. C “Reading” 10

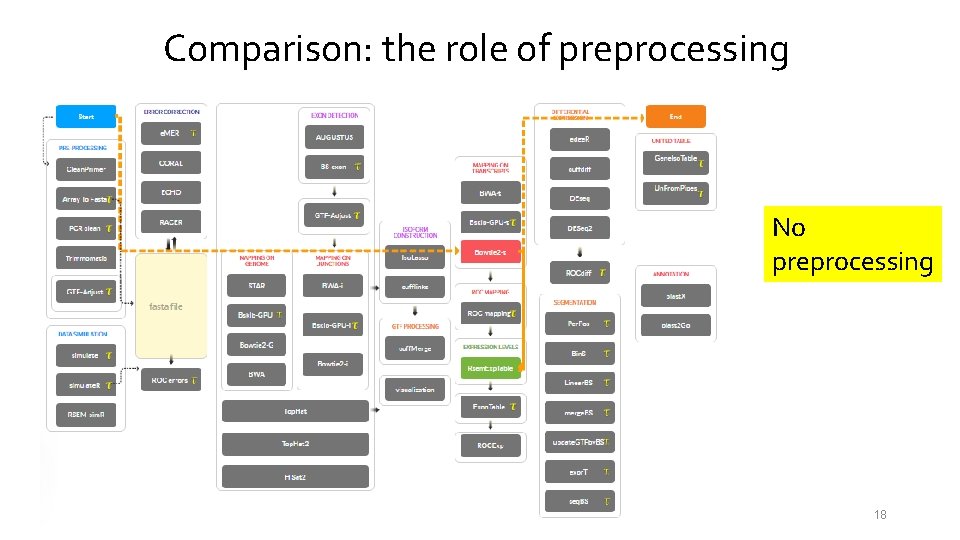
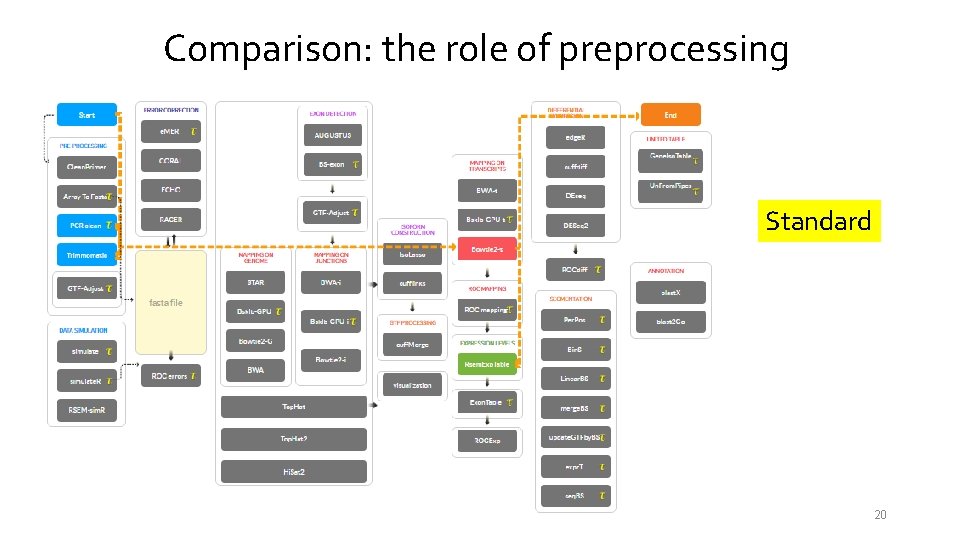
RNA-seq: per-sample processing Preprocessing: • Adapters removal plus additional trimming • Removing PCR duplicates Mapping • Mapping on the set of known transcripts • Mapping on genome (and potential identification of novel transcripts) • Combined strategy Quantification of expression levels 11

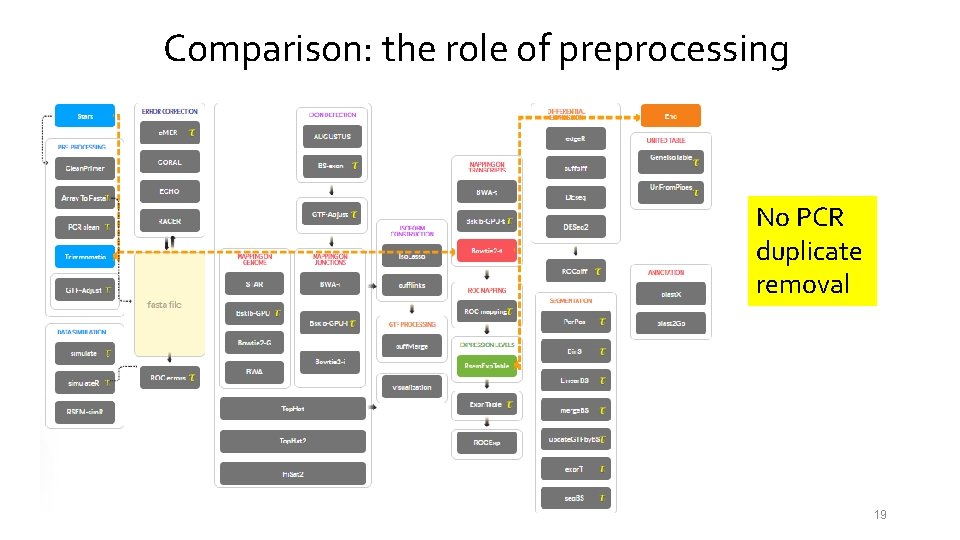
RNA-seq: Comments PCR removal should be used with caution to avoid removing natural duplicates (valuable links: http: //www. cureffi. org/2012/12/11/how-pcr-duplicates-arise-in-next-generation-sequencing/ https: //www. ncbi. nlm. nih. gov/pmc/articles/PMC 4965708/ - DNA-seq and variant calling https: //www. ncbi. nlm. nih. gov/pmc/articles/PMC 4597324/ - RNA-seq, Ch. IP-seq data https: //www. ncbi. nlm. nih. gov/pmc/articles/PMC 3871669/ - trimming 12

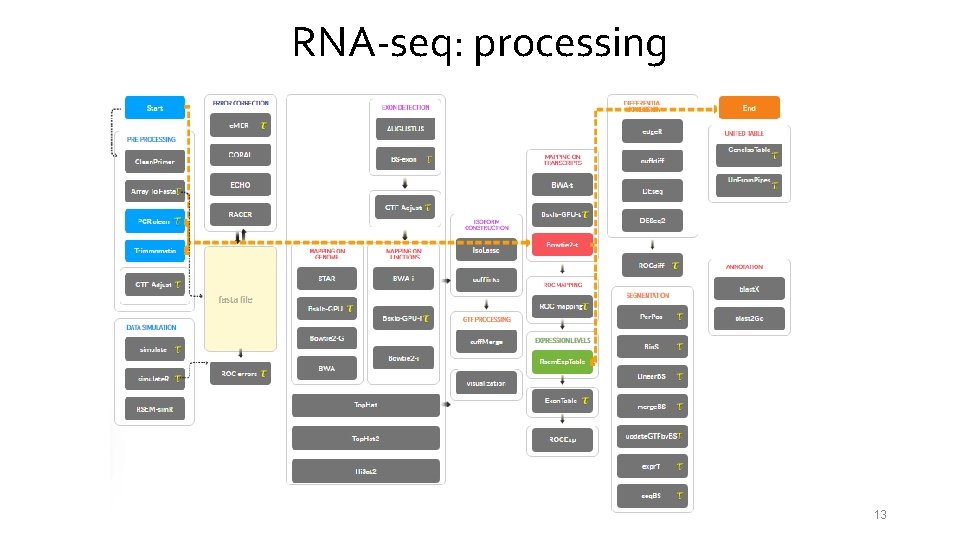
RNA-seq: processing 13

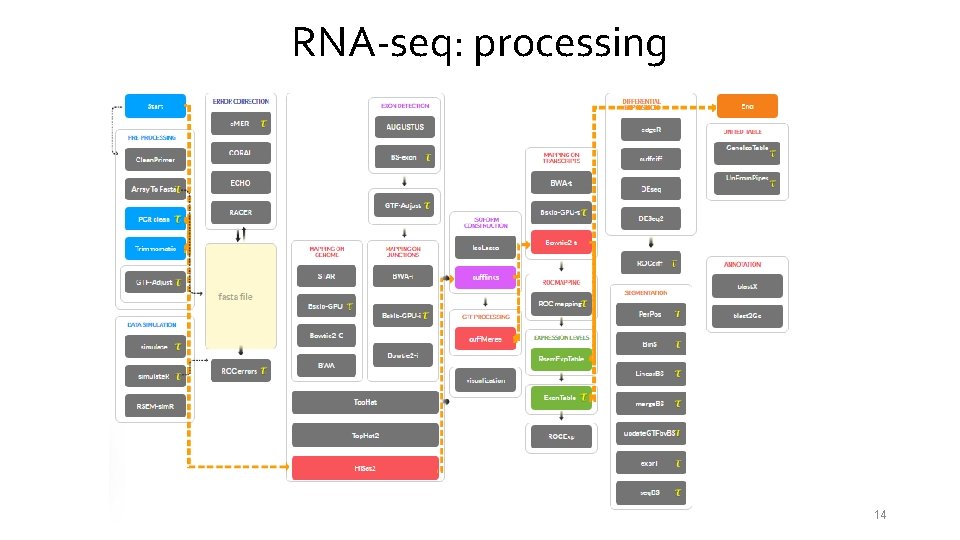
RNA-seq: processing 14

RNA-seq: expression level quantification Standard measures • read counts (raw, expected) • FPKM – fragments per kilo base per million mapped reads: Number of reads mapped on the gene / ((total number of mapped reads – in millions) x (gene length – in kilobases)) • TPM – transcripts per million For one sample TPMg = C x FPKMg, where C is selected in such a way that sum of all TPMg is one million. But constants C are different for different samples. 15

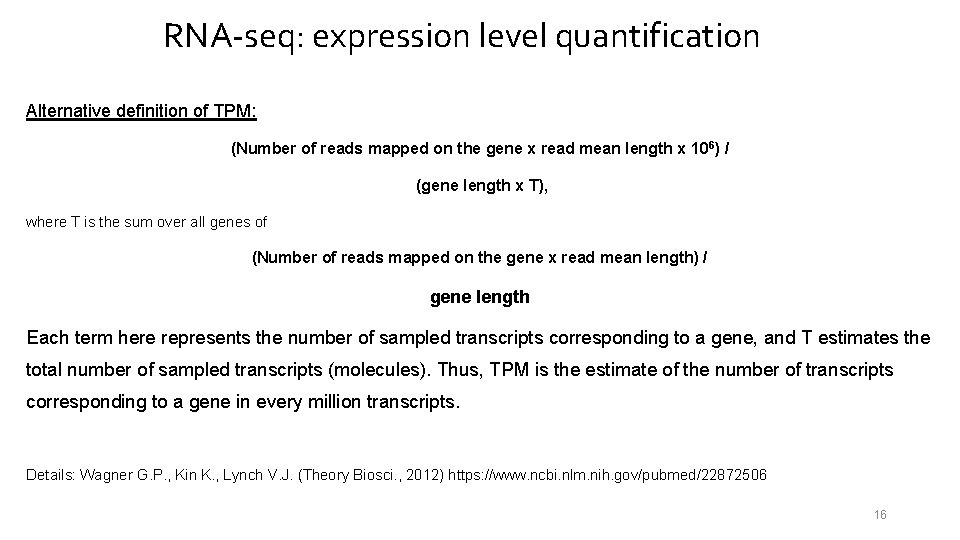
RNA-seq: expression level quantification Alternative definition of TPM: (Number of reads mapped on the gene x read mean length x 106) / (gene length x T), where T is the sum over all genes of (Number of reads mapped on the gene x read mean length) / gene length Each term here represents the number of sampled transcripts corresponding to a gene, and T estimates the total number of sampled transcripts (molecules). Thus, TPM is the estimate of the number of transcripts corresponding to a gene in every million transcripts. Details: Wagner G. P. , Kin K. , Lynch V. J. (Theory Biosci. , 2012) https: //www. ncbi. nlm. nih. gov/pubmed/22872506 16

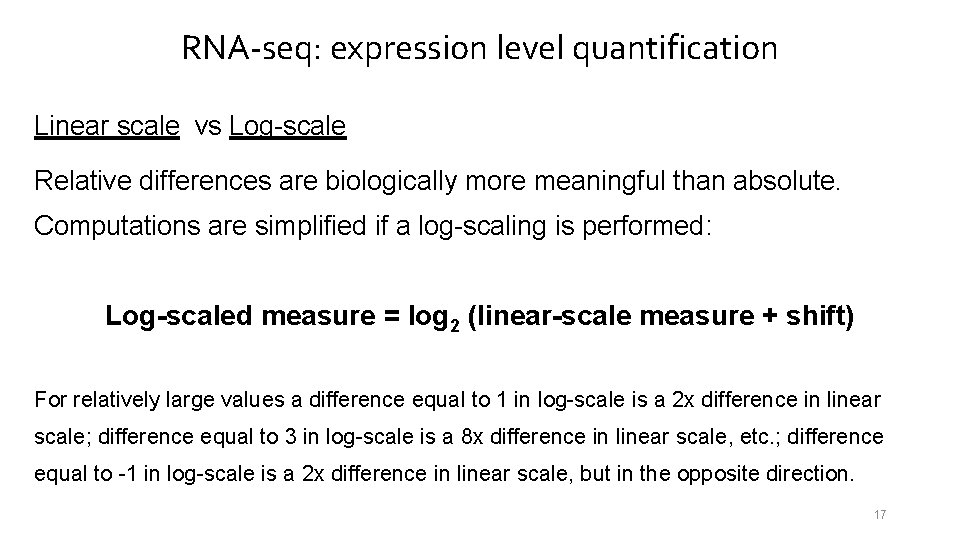
RNA-seq: expression level quantification Linear scale vs Log-scale Relative differences are biologically more meaningful than absolute. Computations are simplified if a log-scaling is performed: Log-scaled measure = log 2 (linear-scale measure + shift) For relatively large values a difference equal to 1 in log-scale is a 2 x difference in linear scale; difference equal to 3 in log-scale is a 8 x difference in linear scale, etc. ; difference equal to -1 in log-scale is a 2 x difference in linear scale, but in the opposite direction. 17

Comparison: the role of preprocessing No preprocessing 18

Comparison: the role of preprocessing No PCR duplicate removal 19

Comparison: the role of preprocessing Standard 20

Comparison: the role of preprocessing (output) 21

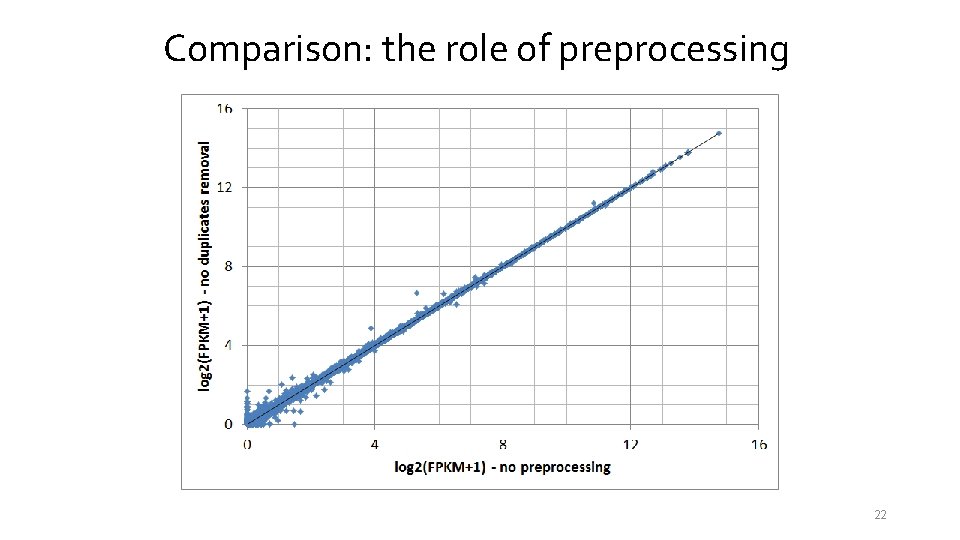
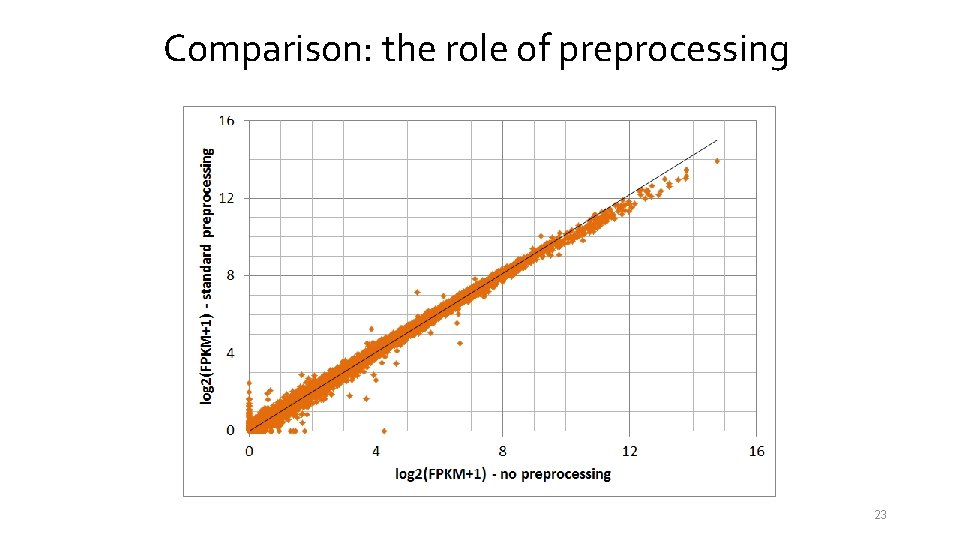
Comparison: the role of preprocessing 22

Comparison: the role of preprocessing 23

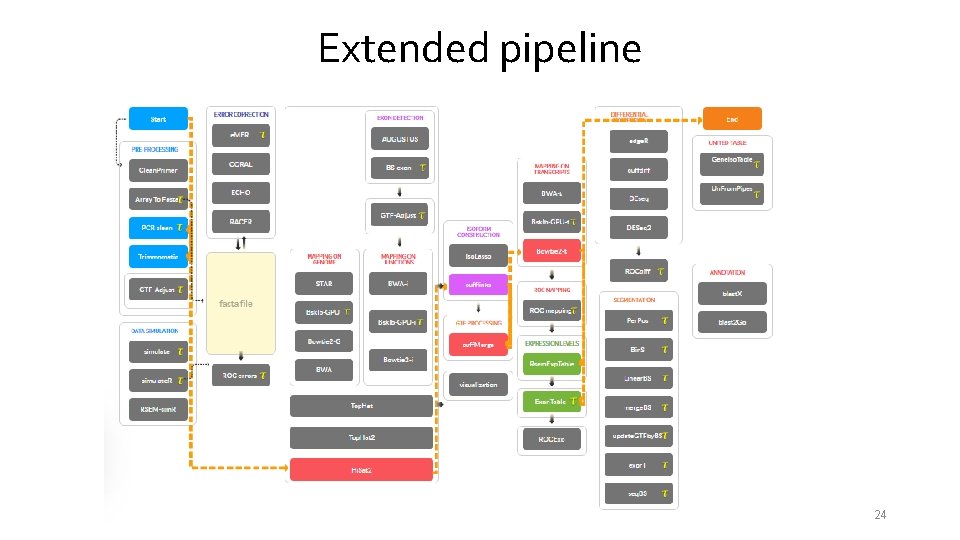
Extended pipeline 24

Extended pipeline 25

BREAK 26

Part 2: Differential expression and pathway / gene set enrichment analysis

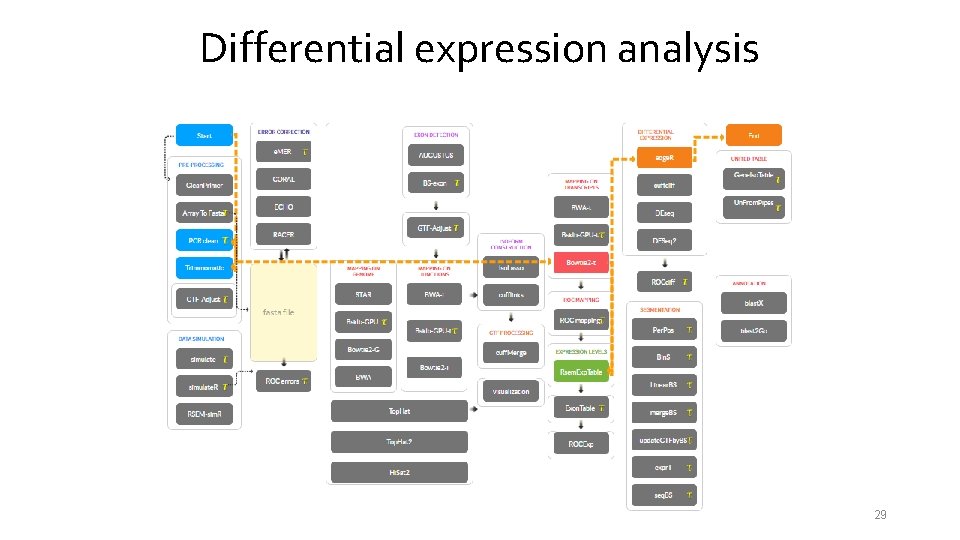
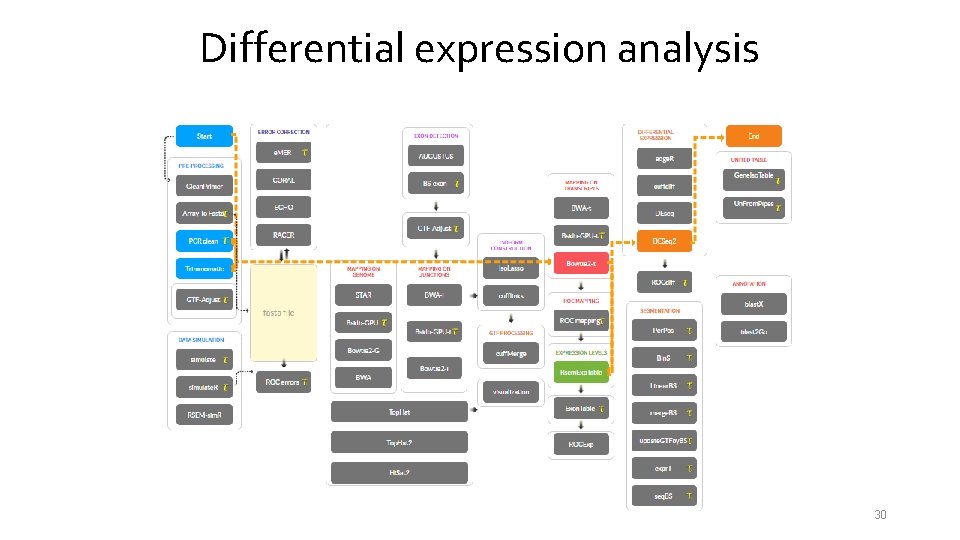
Differential expression analysis Quantities related to the degree of differential expression: • Difference between mean expression levels – fold change (please, pay attention to scale); • Statistical significance – p-value, adjusted p-value (e. g. , FDR) • Expression level magnitude (caution with lowexpressed genes from the analysis). 28

Differential expression analysis 29

Differential expression analysis 30

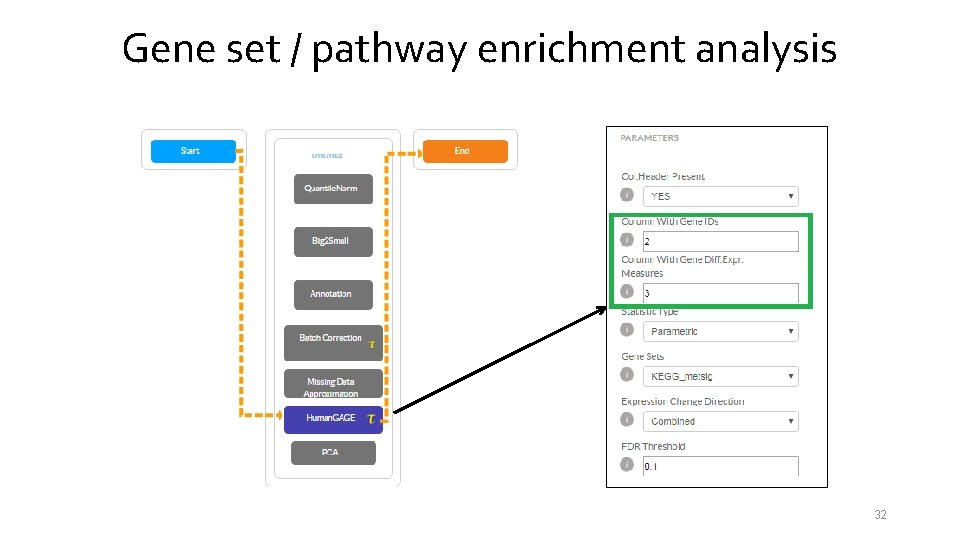
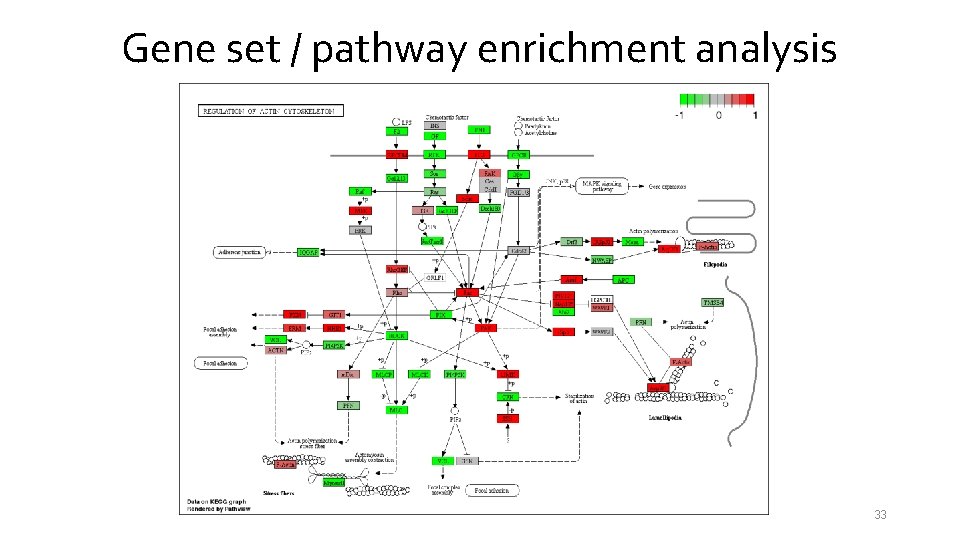
Gene set / pathway enrichment analysis Possible options: • Use only lists (thresholding required): one of the standard tools here is The Database for Annotation, Visualization and Integrated Discovery – DAVID (https: //david. ncifcrf. gov/home. jsp, https: //davidd. ncifcrf. gov/). • Take into consideration degrees of differential expression; • Additionally take into consideration pathway topology. 31

Gene set / pathway enrichment analysis 32

Gene set / pathway enrichment analysis 33

BREAK 34

Part 3: Unsupervised analysis

Unsupervised analysis: PCA 36

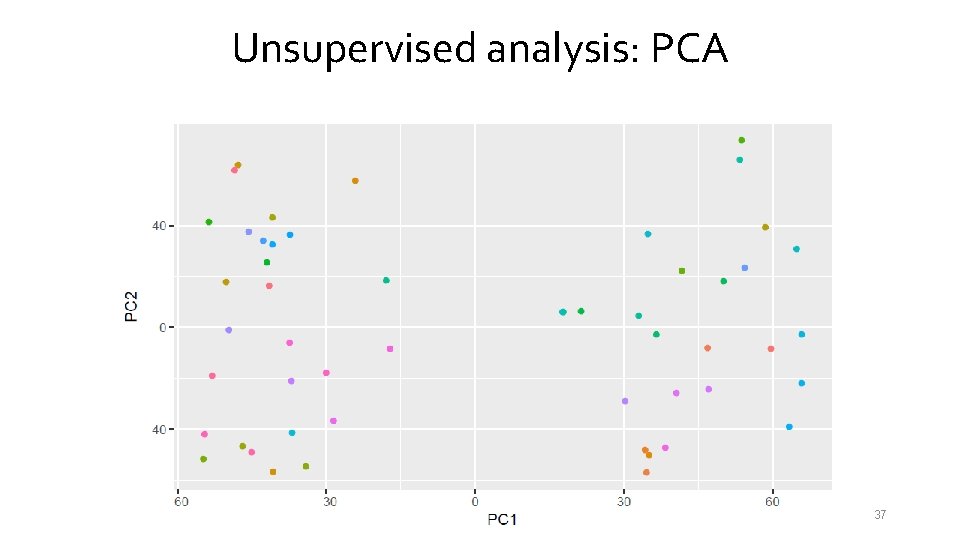
Unsupervised analysis: PCA 37

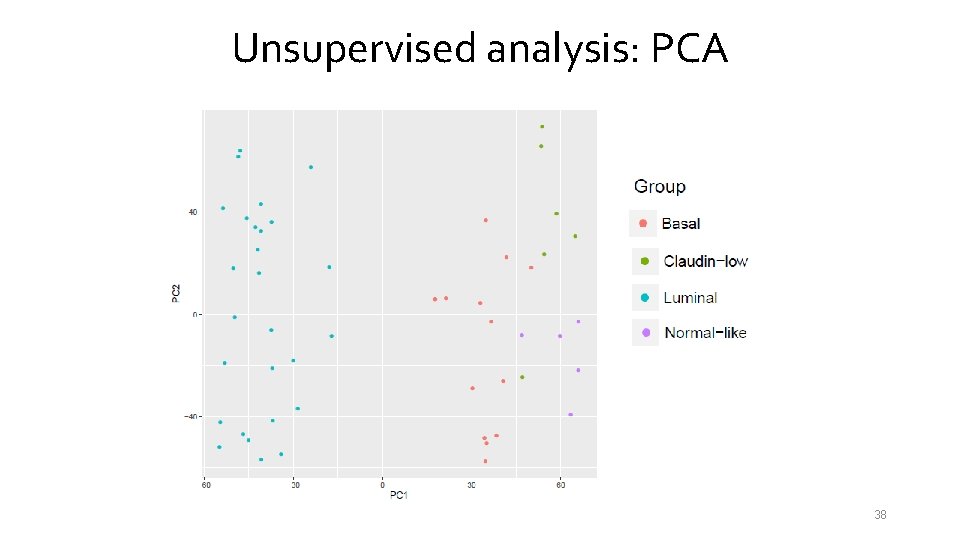
Unsupervised analysis: PCA 38

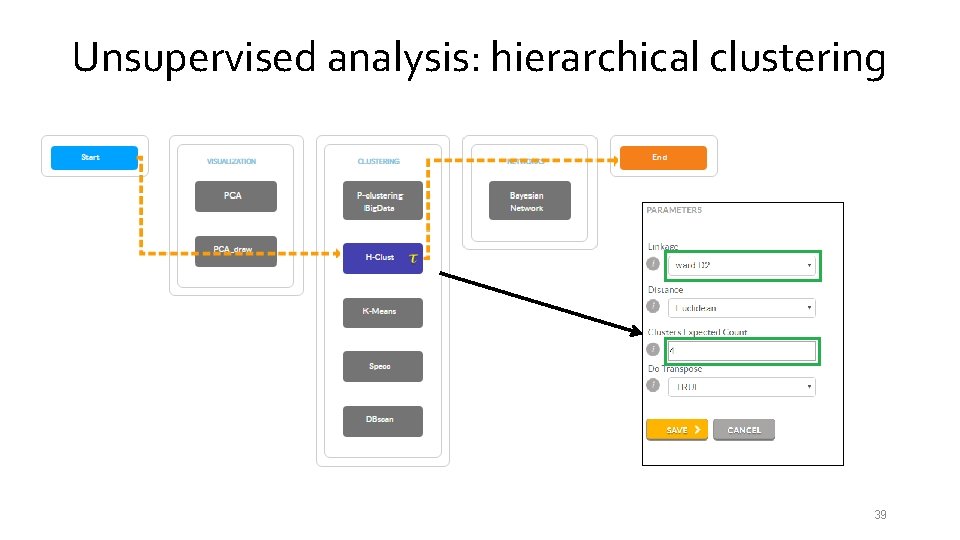
Unsupervised analysis: hierarchical clustering 39

Unsupervised analysis: hierarchical clustering 40

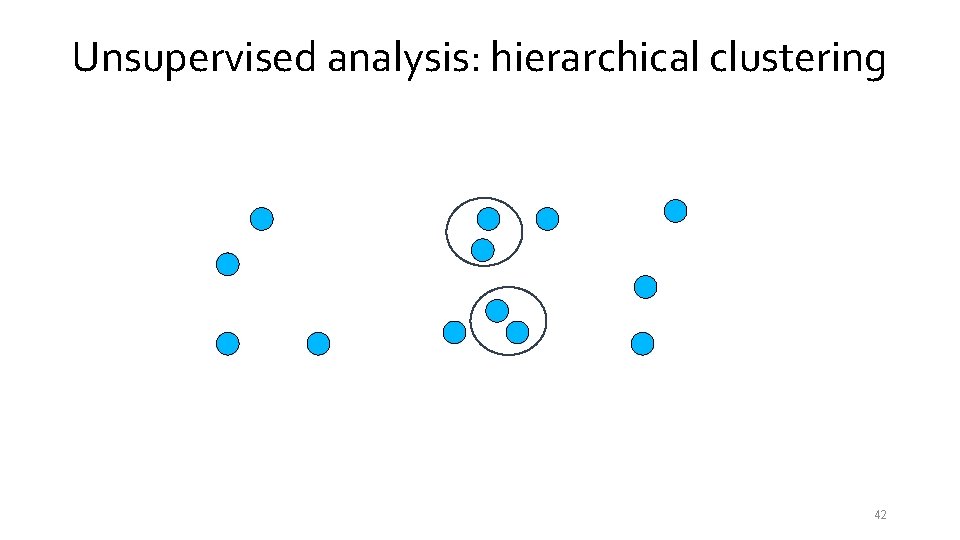
Unsupervised analysis: hierarchical clustering 41

Unsupervised analysis: hierarchical clustering 42

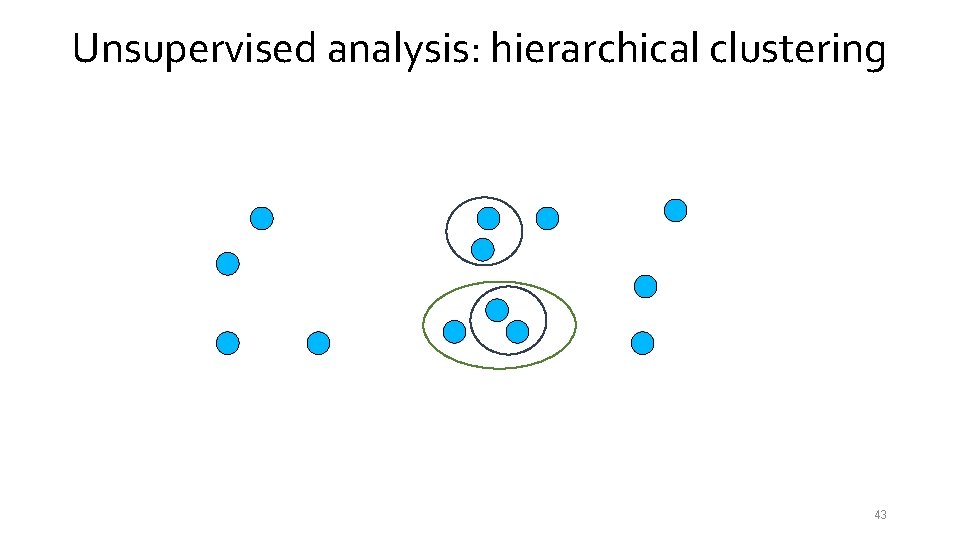
Unsupervised analysis: hierarchical clustering 43

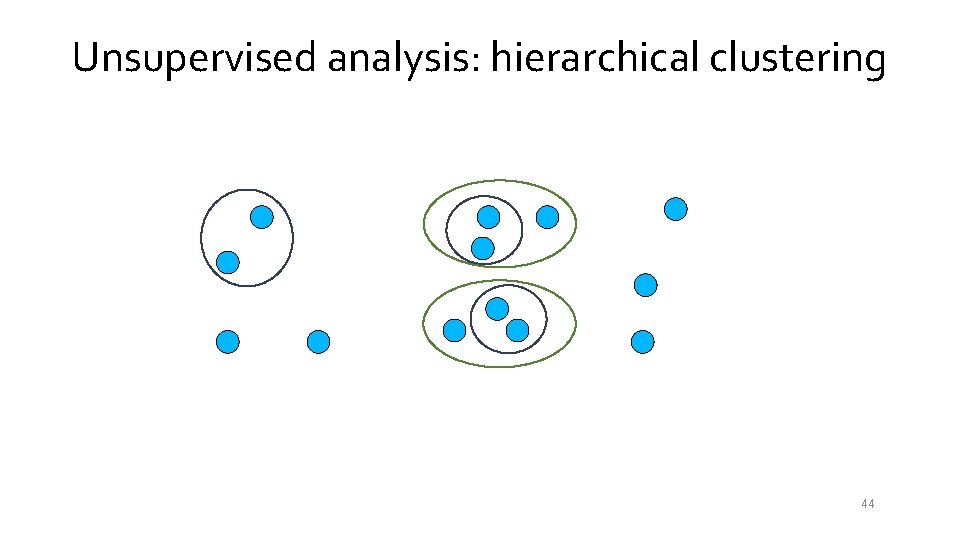
Unsupervised analysis: hierarchical clustering 44

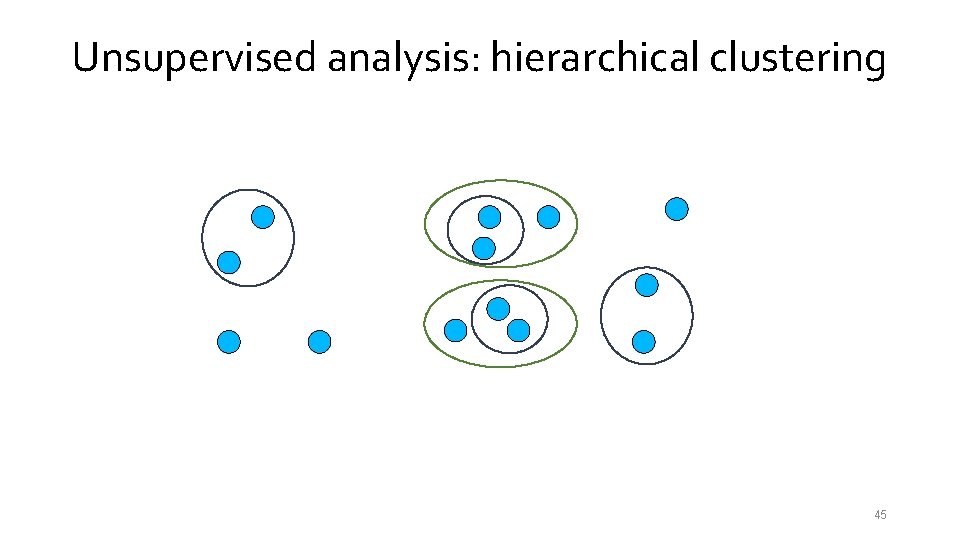
Unsupervised analysis: hierarchical clustering 45

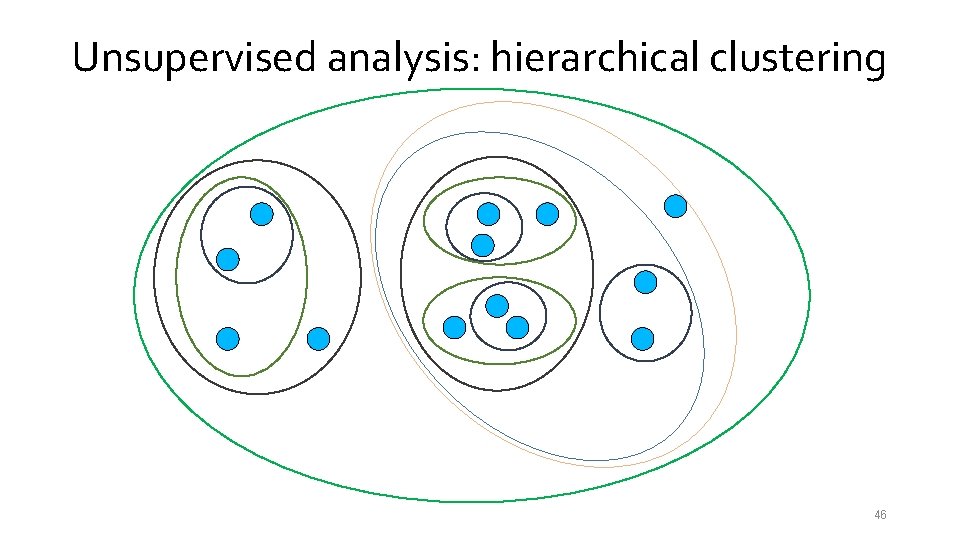
Unsupervised analysis: hierarchical clustering 46

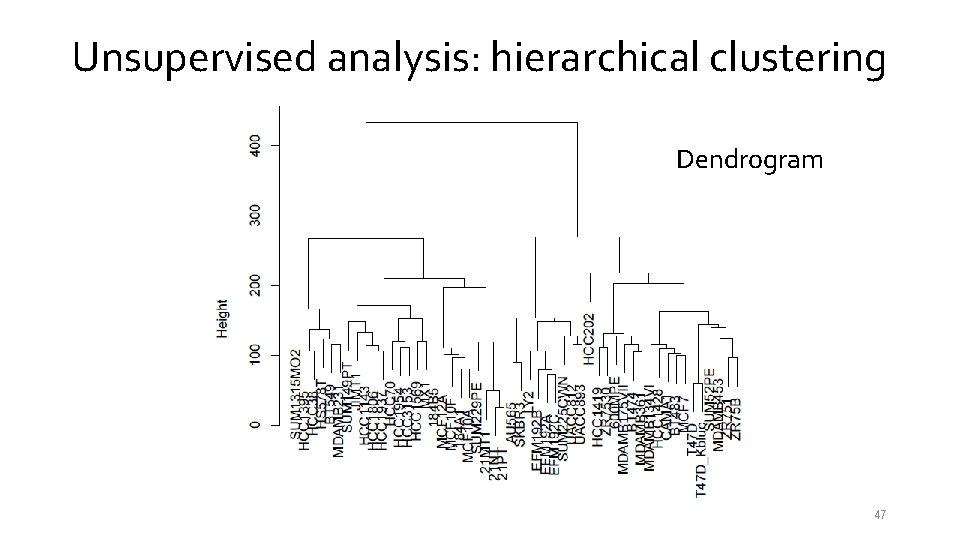
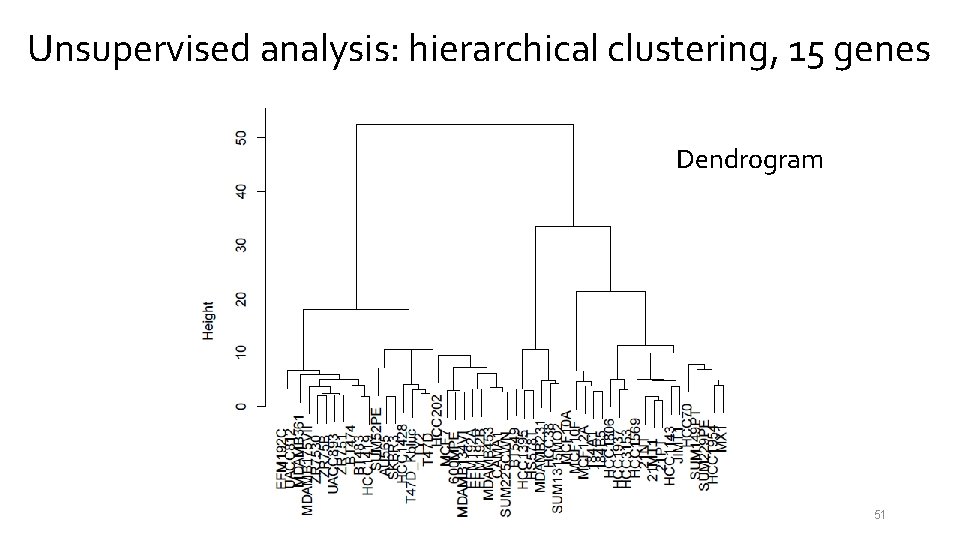
Unsupervised analysis: hierarchical clustering Dendrogram 47

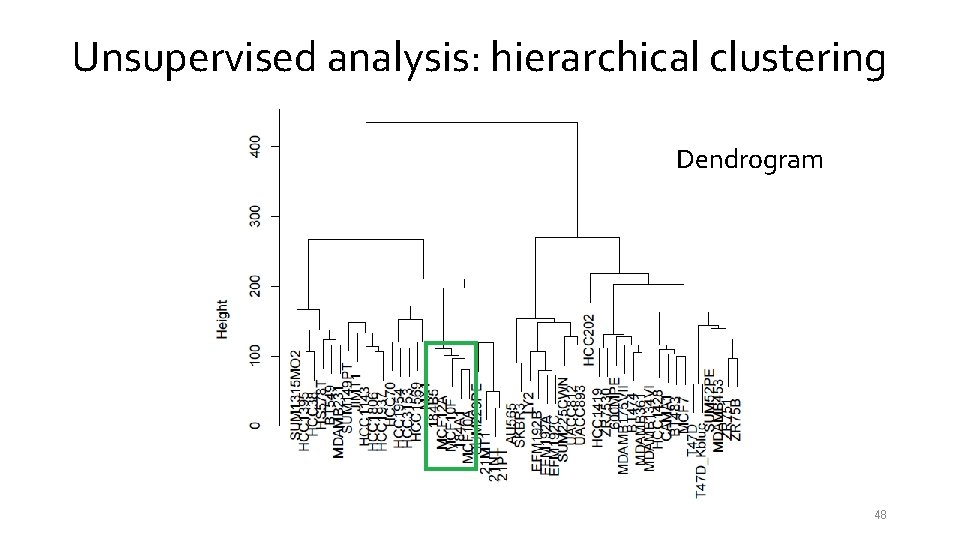
Unsupervised analysis: hierarchical clustering Dendrogram 48

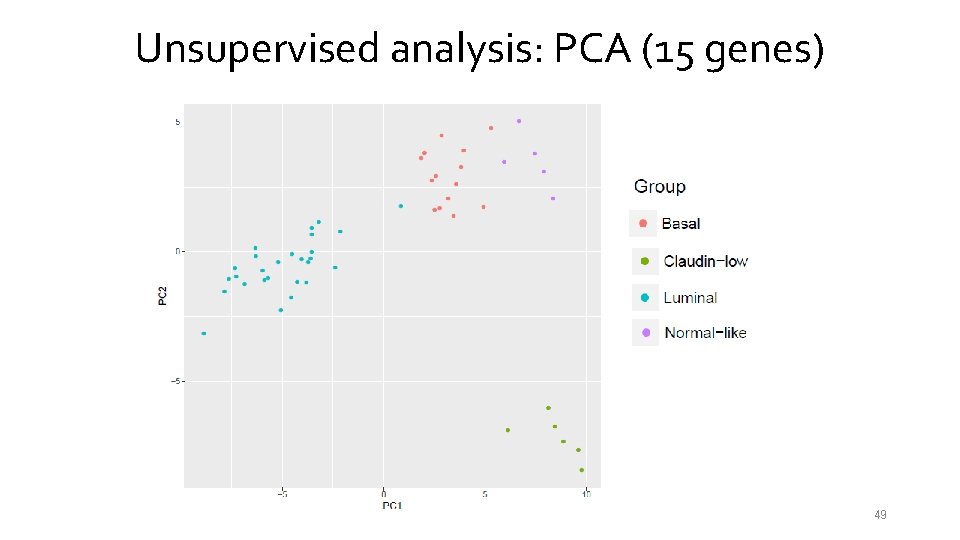
Unsupervised analysis: PCA (15 genes) 49

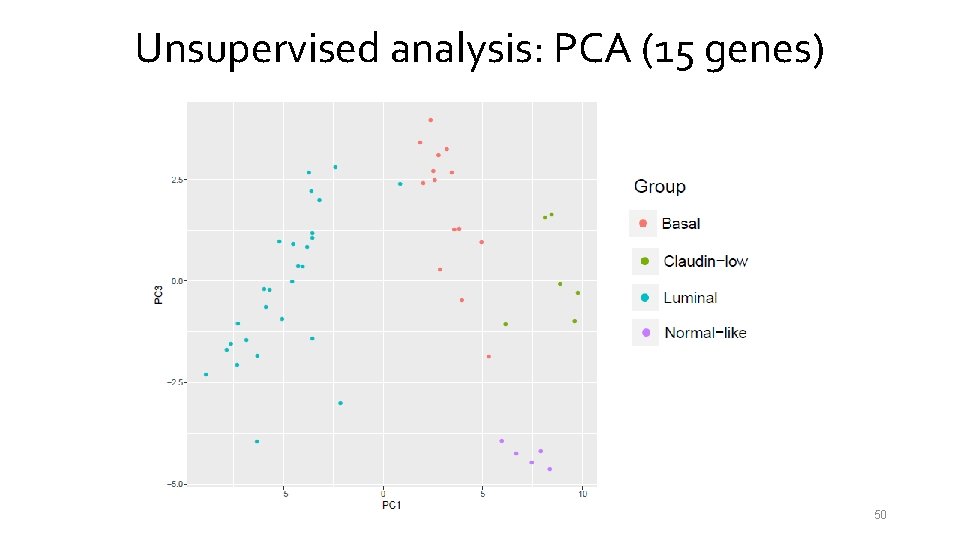
Unsupervised analysis: PCA (15 genes) 50

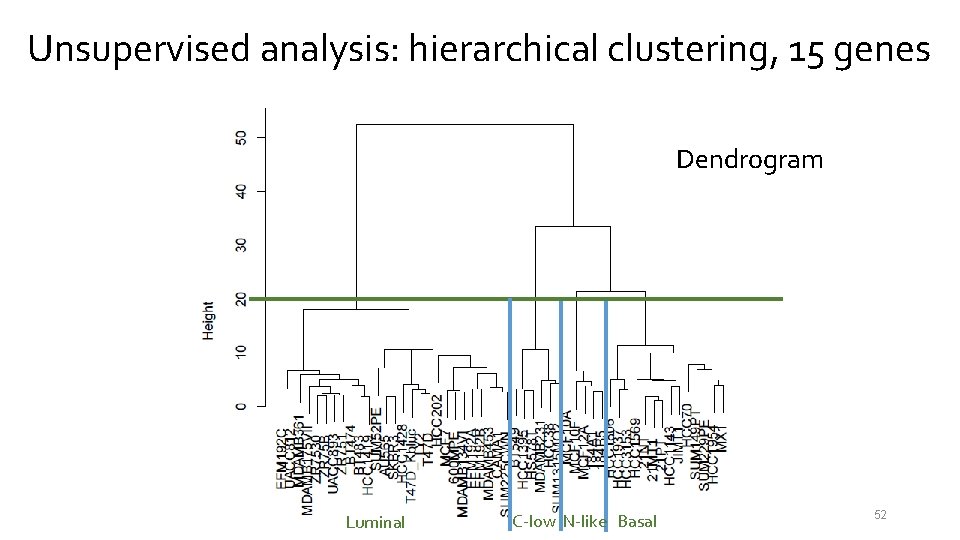
Unsupervised analysis: hierarchical clustering, 15 genes Dendrogram 51

Unsupervised analysis: hierarchical clustering, 15 genes Dendrogram Luminal C-low N-like Basal 52

Gene annotation: ENSG to Gene Symbols plus GO 53

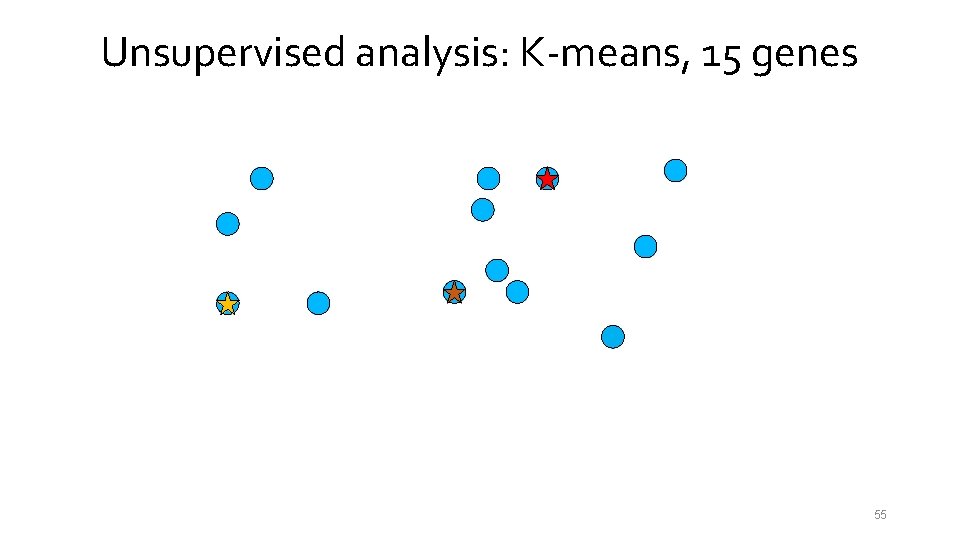
Unsupervised analysis: K-means, 15 genes 54

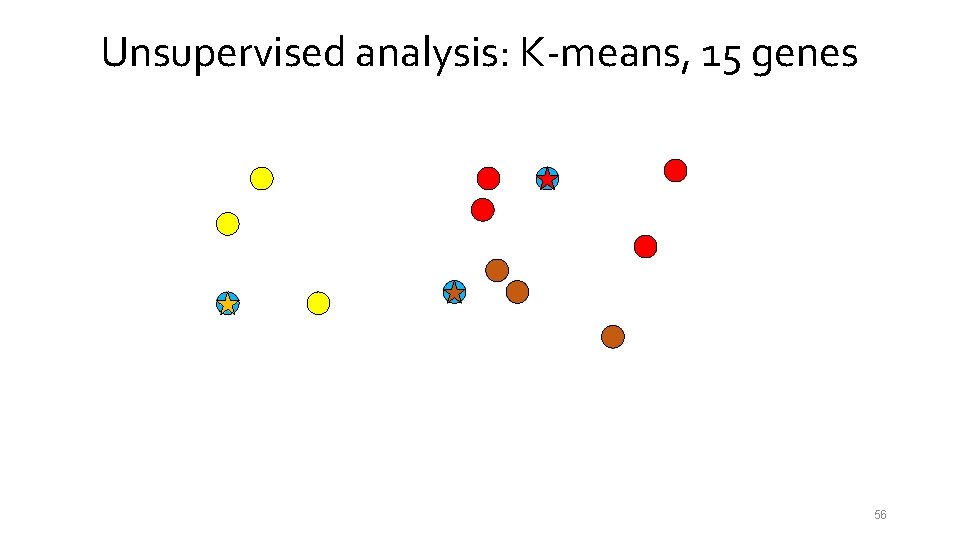
Unsupervised analysis: K-means, 15 genes 55

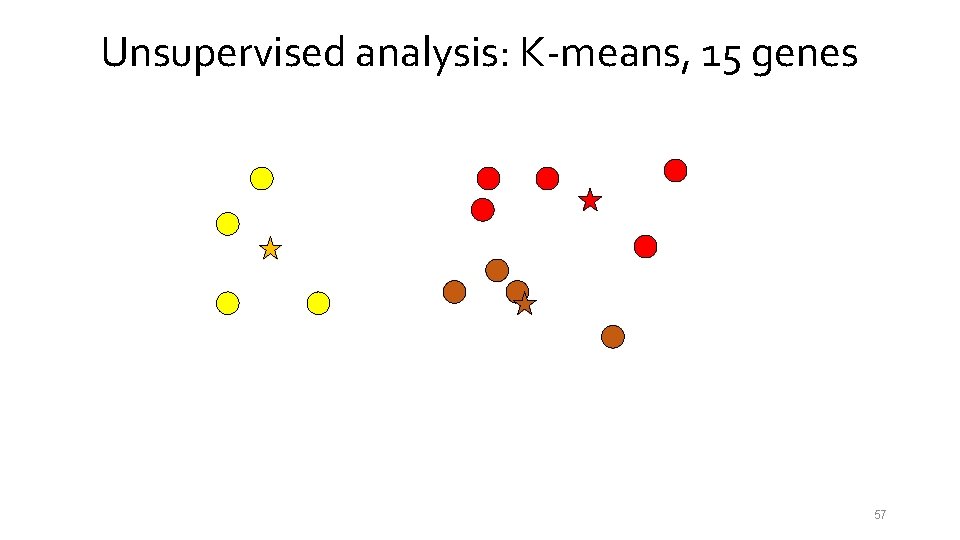
Unsupervised analysis: K-means, 15 genes 56

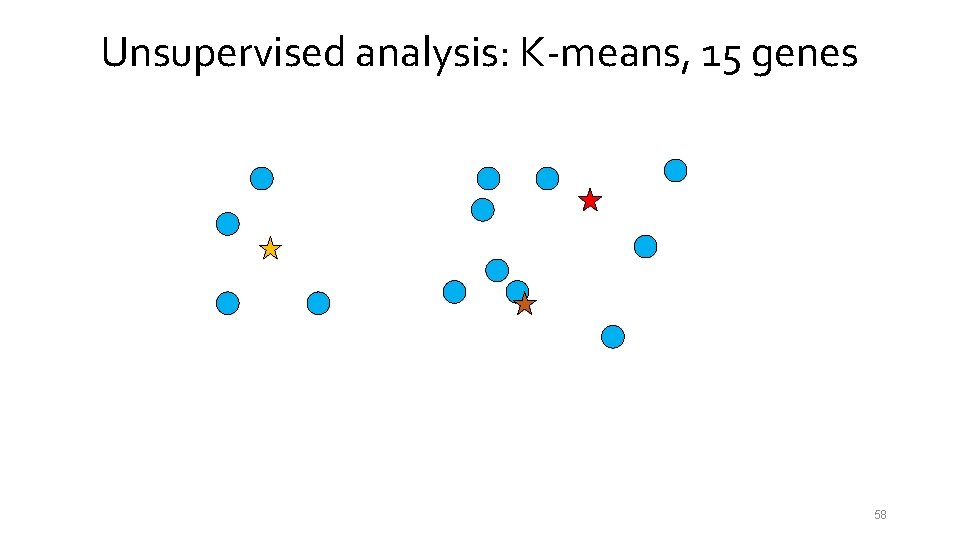
Unsupervised analysis: K-means, 15 genes 57

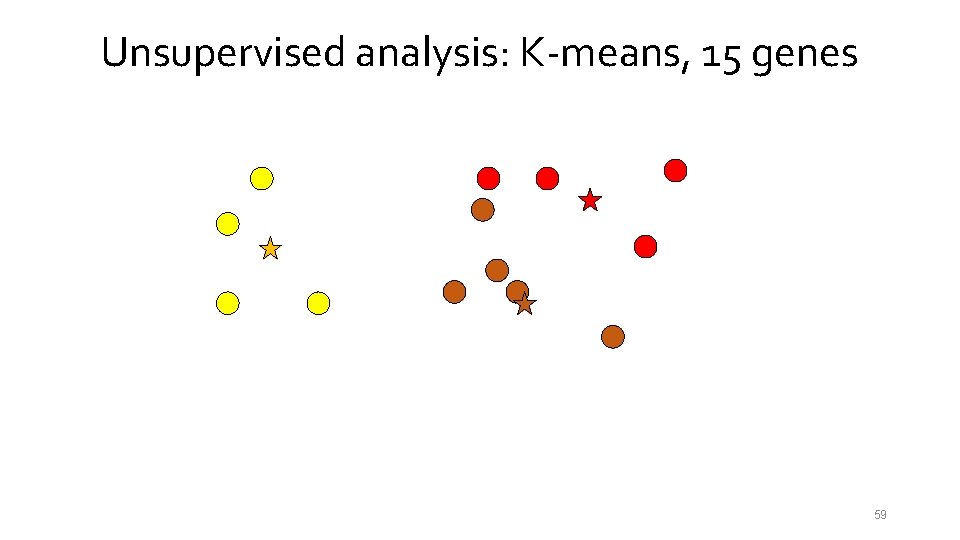
Unsupervised analysis: K-means, 15 genes 58

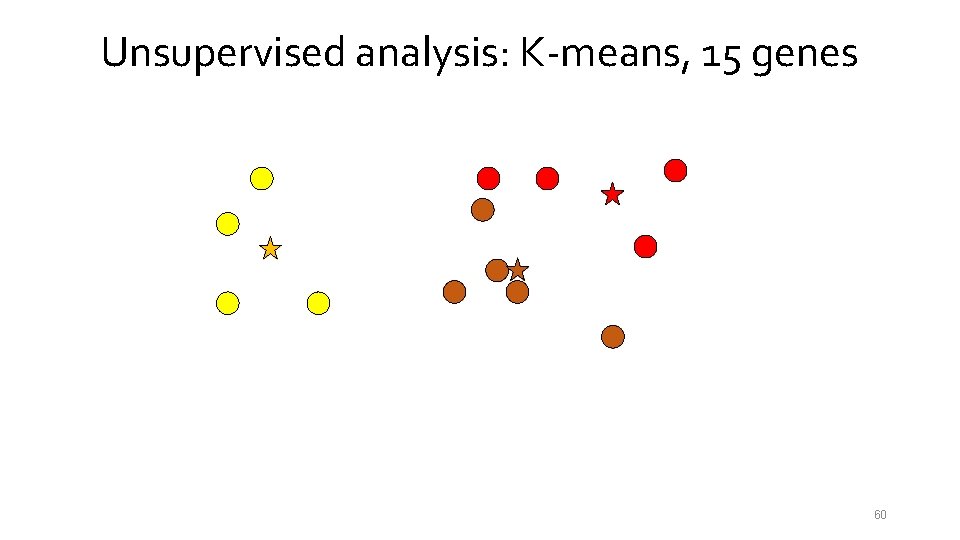
Unsupervised analysis: K-means, 15 genes 59

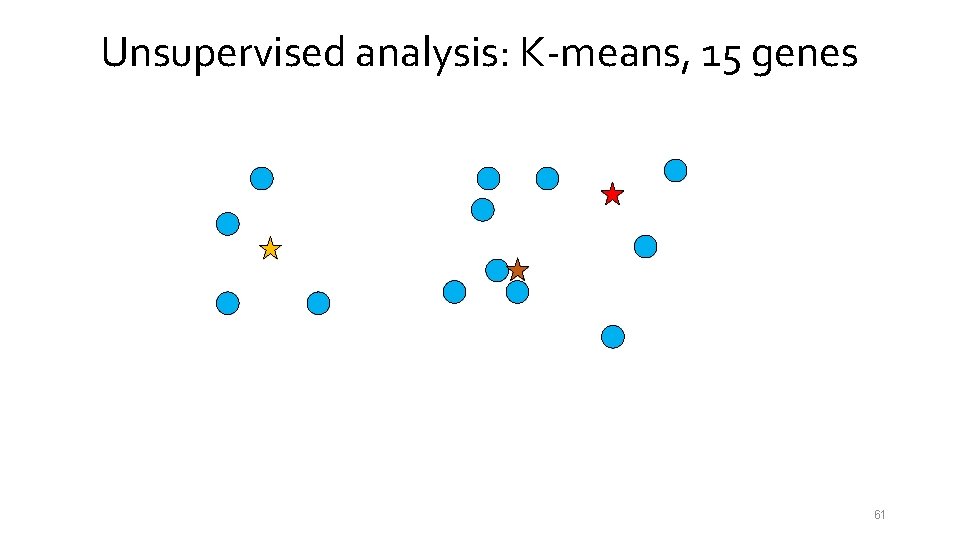
Unsupervised analysis: K-means, 15 genes 60

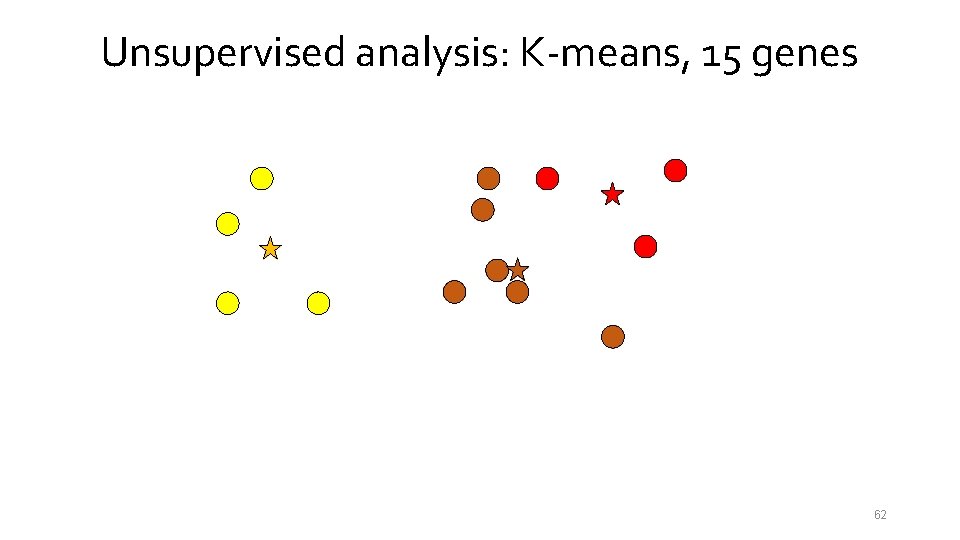
Unsupervised analysis: K-means, 15 genes 61

Unsupervised analysis: K-means, 15 genes 62

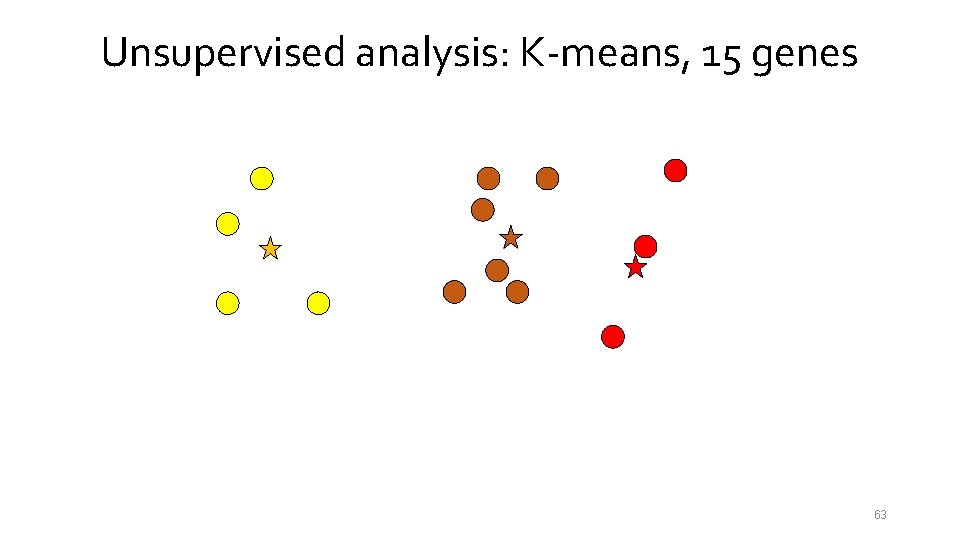
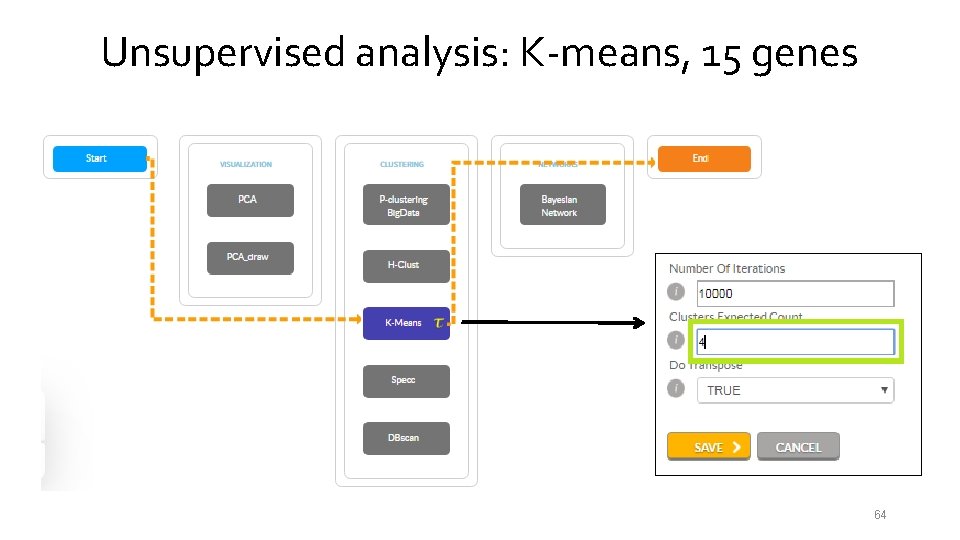
Unsupervised analysis: K-means, 15 genes 63

Unsupervised analysis: K-means, 15 genes 64

Unsupervised analysis: K-means, 15 genes “The SUM 52 PE cell line was derived from a pleural effusion and was found to be negative for ER and PR expression, however the original primary tumor from this patient was positive for both hormone receptors”. Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010; 32(1 -2): 35 -48. Ethier SP, Kokeny KE, Ridings JW, Dilts CA. erb. B family receptor expression and growth regulation in a newly isolated human breast cancer cell line. Cancer Res. 1996; 56(4): 899 -907. 65

BREAK 66

Part 4: Supervised analysis: classification

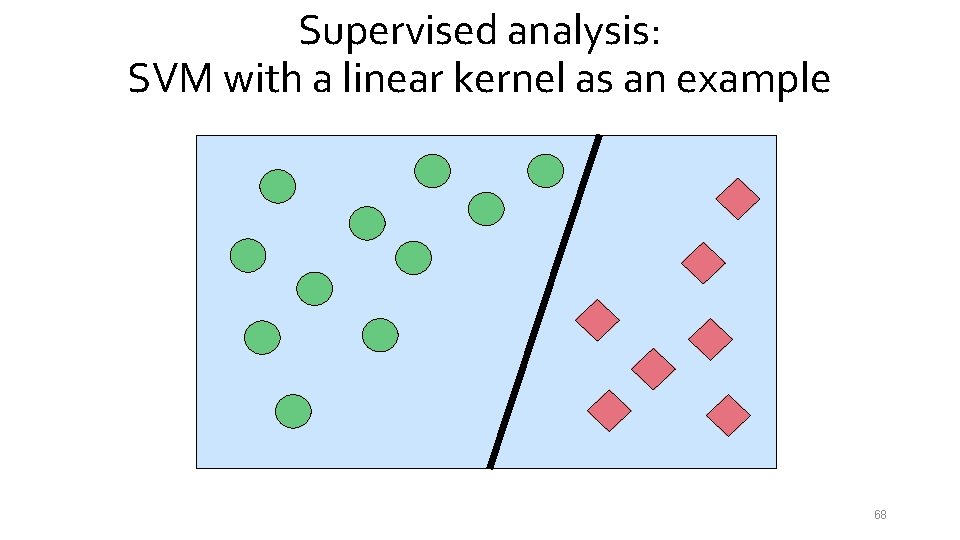
Supervised analysis: SVM with a linear kernel as an example 68

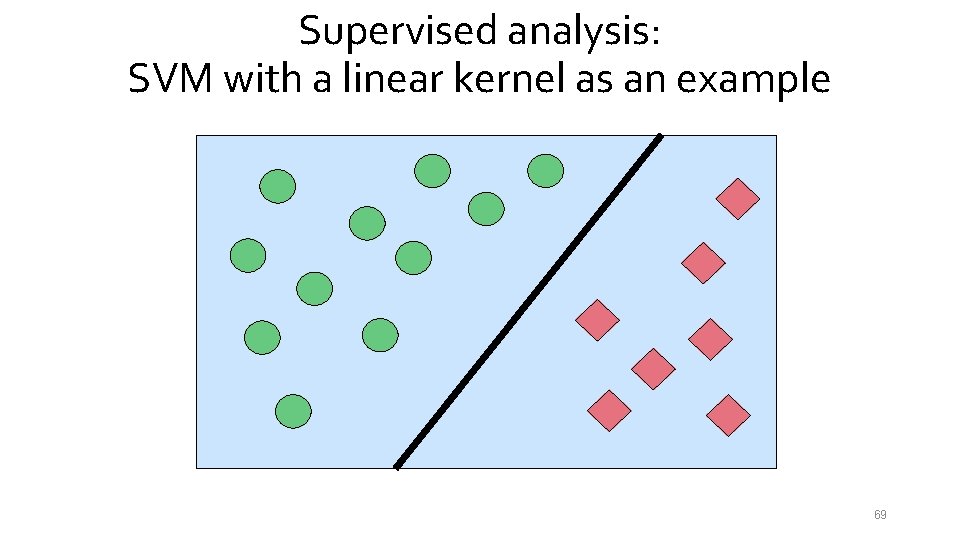
Supervised analysis: SVM with a linear kernel as an example 69

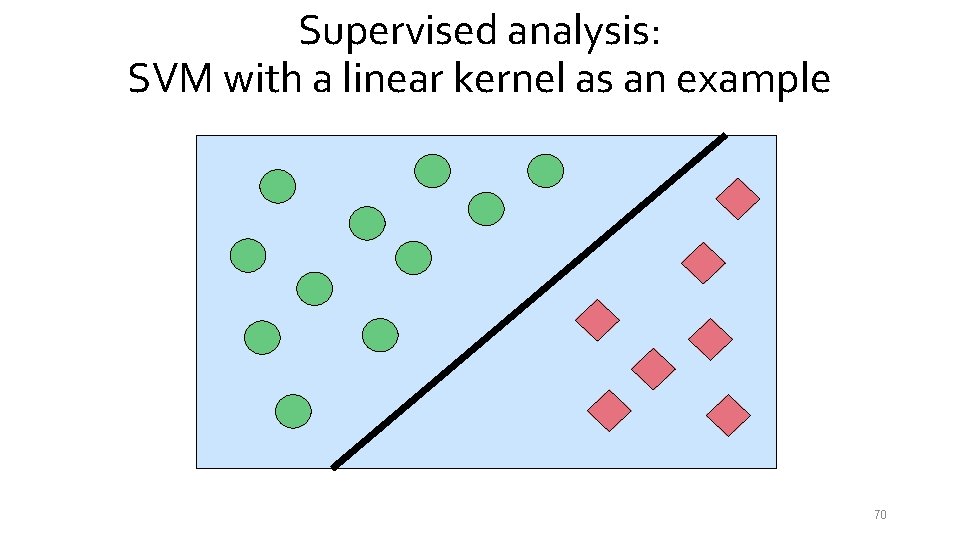
Supervised analysis: SVM with a linear kernel as an example 70

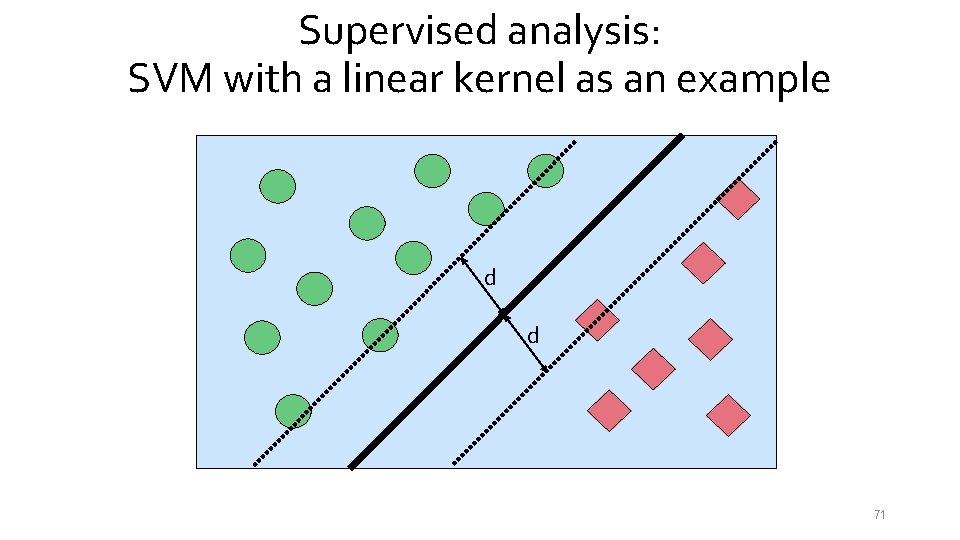
Supervised analysis: SVM with a linear kernel as an example d d 71

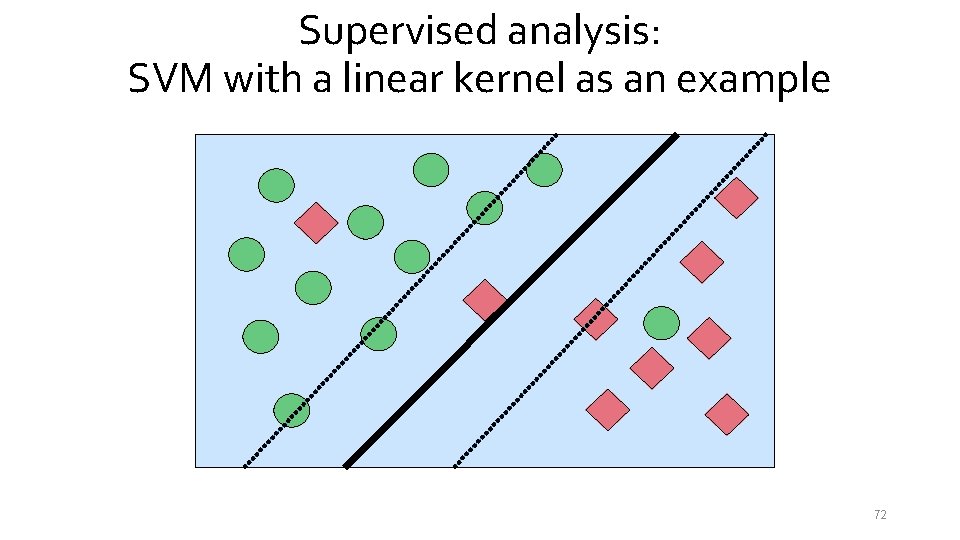
Supervised analysis: SVM with a linear kernel as an example 72

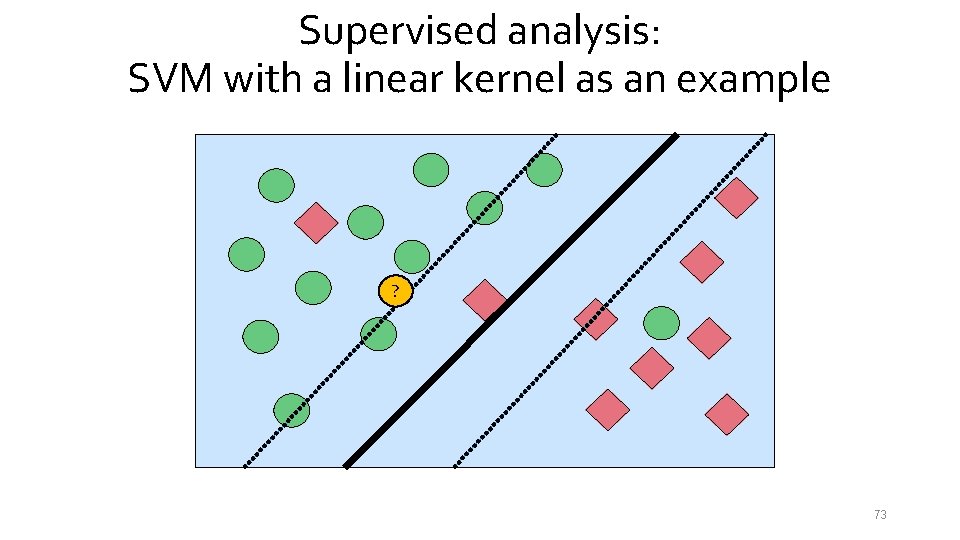
Supervised analysis: SVM with a linear kernel as an example ? 73

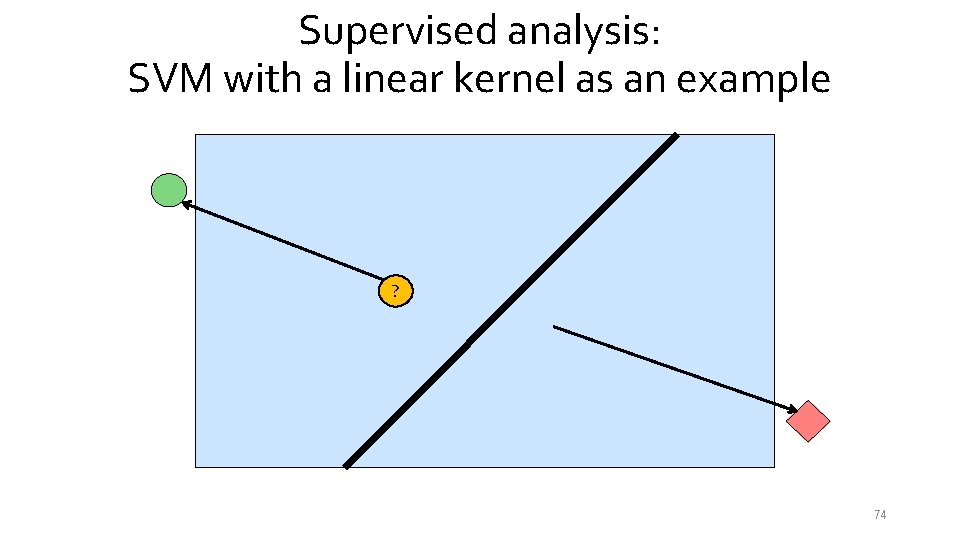
Supervised analysis: SVM with a linear kernel as an example ? 74

Supervised analysis: available methods • Linear Discriminant Analysis (LDA) • Quadratic Discriminant Analysis (QDA) • Random Forest • Support Vector Machine (SVM) • Naïve Bayes 75

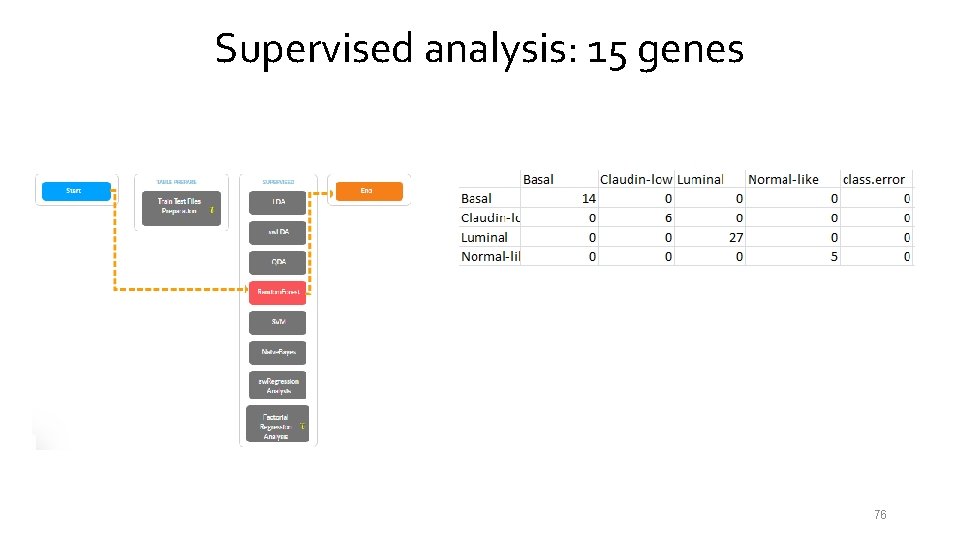
Supervised analysis: 15 genes 76

BREAK 77

BREAK HANDSON Separation of TCGA and breast cancer PDX samples 78

BREAK HANDSON Analysis of a subset of breast cancer PDX samples 79