NMR Spectroscopy Question and Answer Session Judith KleinSeetharaman












- Slides: 49

NMR Spectroscopy Question and Answer Session Judith Klein-Seetharaman Department of Structural Biology jks 33@pitt. edu Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session

What did we do? • NMR instrumentation • NMR sample preparation considerations – Water signals – Water suppression – Detergents • NMR setup – downstairs – – zg zgpr selabs hsqc • Spectral processing: Topspin Software 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 2

What did we do then? • Converting raw spectral formats to other formats using NMRpipe – Example: HIV protease relaxation data • Assignment of signals with one strategy – – – HNCOCACBG HNCOCA HNCACB HNCACO • Example: Ubiquitin assignment with NMRview. J 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 3

What would be the next step? • For determining a structure with NMR spectroscopy, after assignment, we would… – Collect constraints: • • NOE Dipolar coupling Scalar coupling constants (gives dihedral angles) Solvent exchange – Calculate a structure based on these constraints 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 4

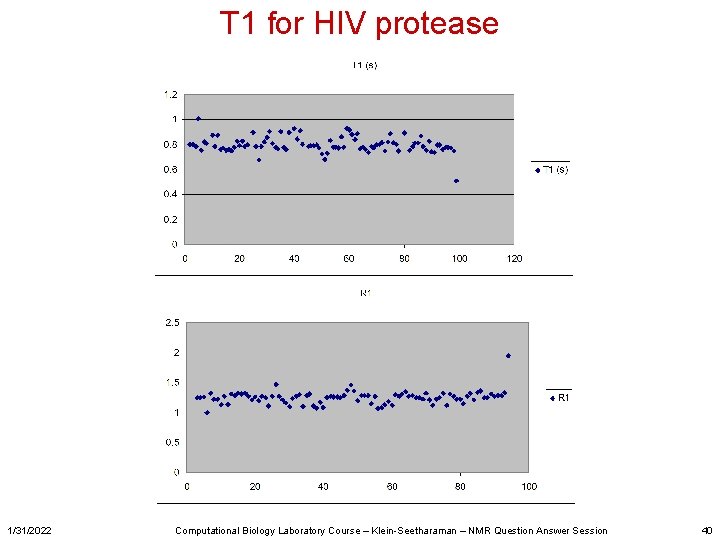
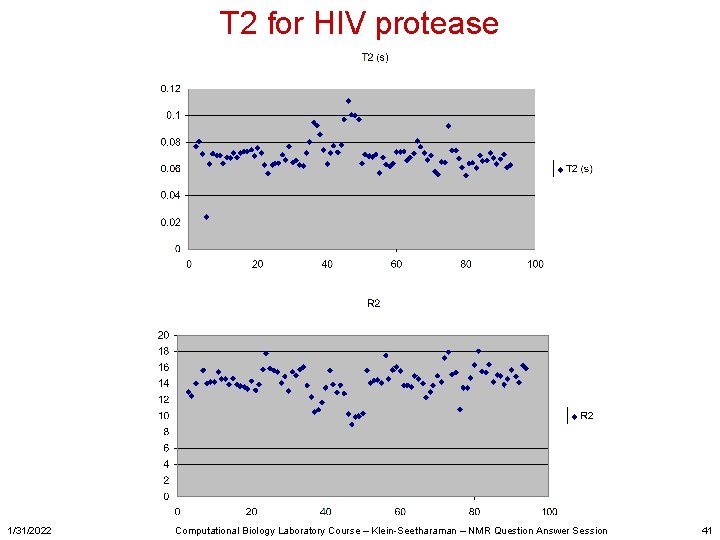
What are other things one can do with NMR? • For study of dynamics – Measure and analyze T 1, T 2, Het. NOE data – We went through T 1 analysis in computer lab (conversion of raw data with NMRpipe, analysis with NMRview. J) – T 2 was your homework • Other studies might include chemical shift perturbations • Temperature titrations • … 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 5

Homework Assignments 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 6

Homework Part 1 • NMR instrumentation • NMR sample preparation considerations – Water signals – Water suppression – Detergents • NMR setup – downstairs – – zg zgpr selabs hsqc • Spectral processing: Topspin Software 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 7

Experiments Run • zg = records all 1 H's in your sample • zgpr = same as zg but with water suppression • selabs = the way we set it up, it records only NH region • hsqcfpgpf 3 gphwg = in short: hsqc = records all 1 H's attached to 15 N 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 8

• click on Topspin icon and open the program 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 9

• click on guest, expand MB 3_Jan 2007 directory • This is the directory that contains all of the data that was acquired in class. 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 10

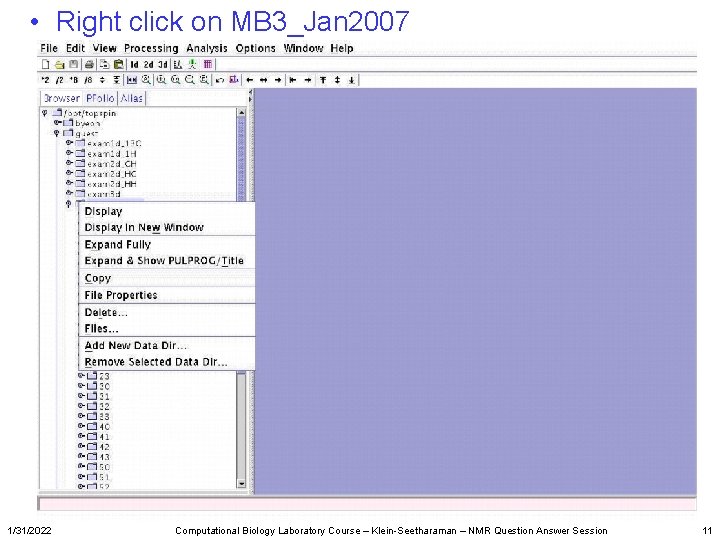
• Right click on MB 3_Jan 2007 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 11

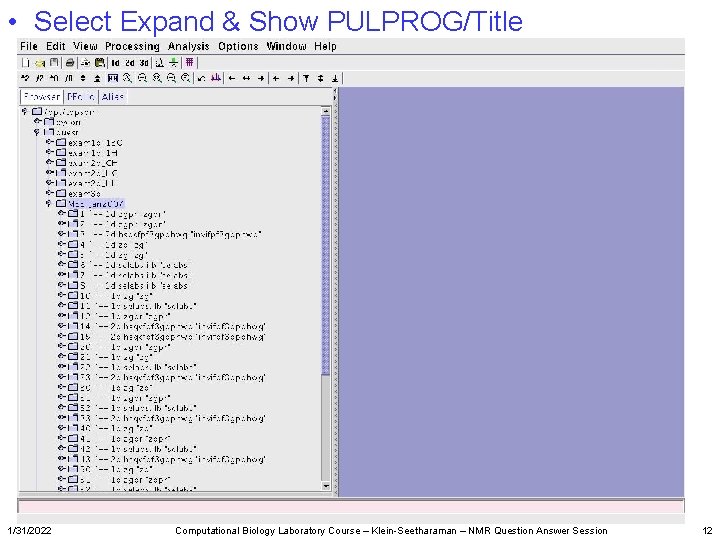
• Select Expand & Show PULPROG/Title 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 12

Samples • • • 1/31/2022 Sample IDs Experiment IDs B 1 -7 F 10 -15 A 20 -23 D 30 -33 E 40 -43 H 50 -53 B 60, 61 F 70, 71 F 100 -103 B 110 -113 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 13

• • 1 = zgpr of Sample B 2 = zgpr of Sample B 3 = hsqc of Sample B 4 = zg of Sample B 5 = zg of Sample B 6 = selabs of Sample B 7 = selabs of Sample B • • • 10 = zg of Sample F 11 = selabs of Sample F 12 = zgpr of Sample F 13 = hsqc of sample F 14 = hsqc of sample F 15 = hsqc of sample F • • • ----- experiments done with MB 3 students: 20 = zgpr of Sample A 21 = zg of sample A 22 = selabs of sample A 23 = hsqc of sample A • • 30 = zg of Sample D 31 = zgpr of Sample D 32 = selabs of Sample D 33 = hsqc of Sample D 1/31/2022 Samples • • 40 = zg of sample E 41 = zgpr of sample E 42 = selabs of Sample E 43 = hsqc of Sample E • • 50 = zg of Sample H 51 = zgpr of Sample H 52 = selabs of sample H 53 = hsqc of sample H • • 60 = zg of sample B 61 = zgpr of sample B • • 70 = zg of sample F 71 = zgpr of sample F • • • ---------experiments done with comp bio students 100 = zg of sample F 101 = zgpr of sample F 102 = selabs of sample F 103 = hsqc of sample F • • 110 = zg of sample B 111 = zg of sample B 112 = selabs of sample B 113 = hsqc of sample B Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 14

Solving the Mystery of the Samples • Drag the experiment that you want to analyze into the main purple window area. • If you do not see a spectrum, you need to type efp • This will process the data so that you can see the spectrum. • In the case of the hsqc it will also ask you for an experiment number where to store the processed data, you can use the default or enter a number, • e. g. 2. Another alternative is to type “rser 1” that will retrieve the first slice of the 2 d hsqc experiment also. 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 15

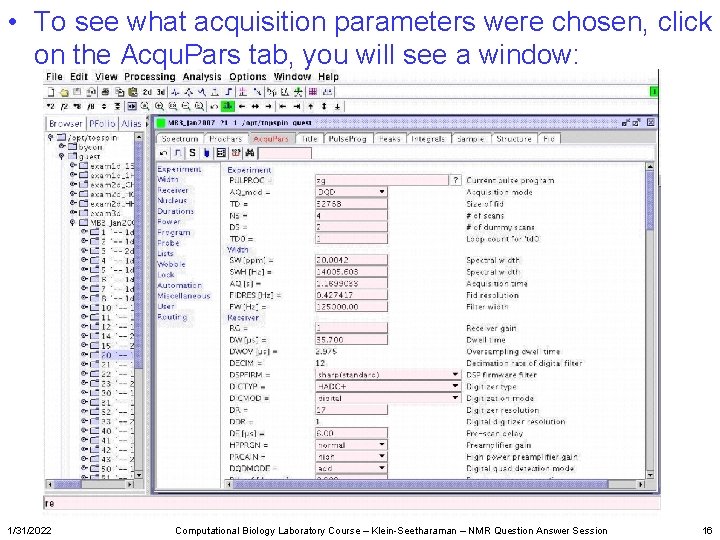
• To see what acquisition parameters were chosen, click on the Acqu. Pars tab, you will see a window: 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 16

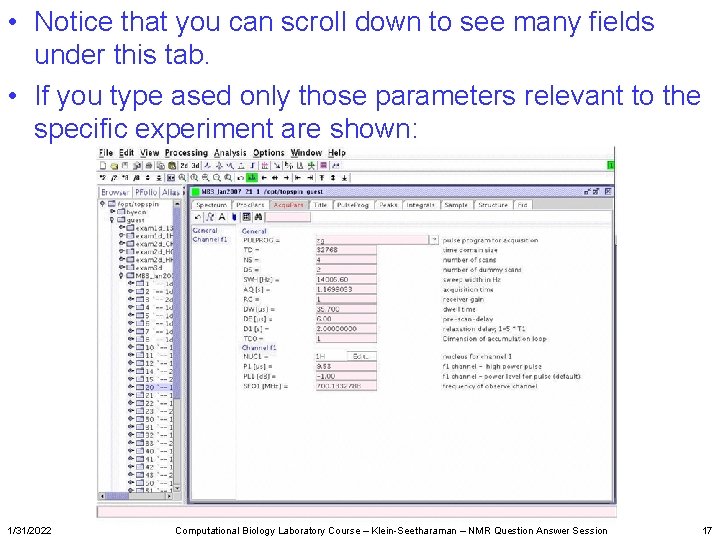
• Notice that you can scroll down to see many fields under this tab. • If you type ased only those parameters relevant to the specific experiment are shown: 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 17

Some tips • If you want to move more quickly from experiment to experiment, type re and then the experiment number, e. g. • re 21 gives the experiment stored in directory 21 • To save a picture of a spectrum select File, Export and then give it a file name extension indicating which format you would like (e. g. . jpg) • Note that you may have to phase your spectrum. 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 18

Phasing 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 19

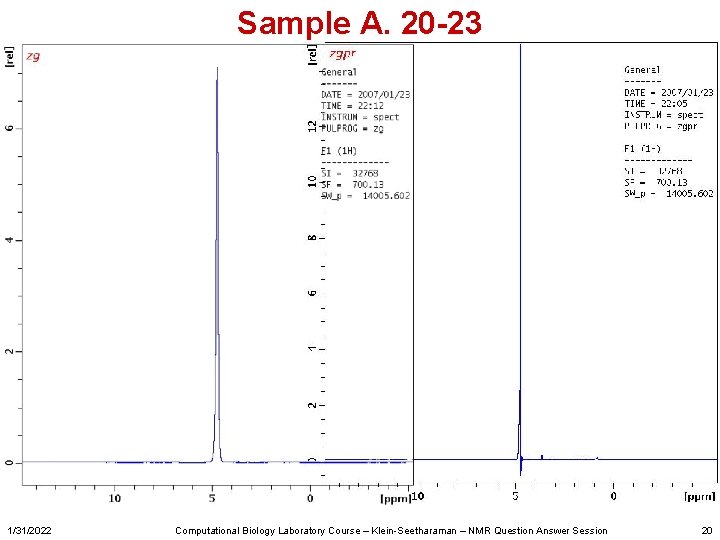
Sample A. 20 -23 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 20

1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 21

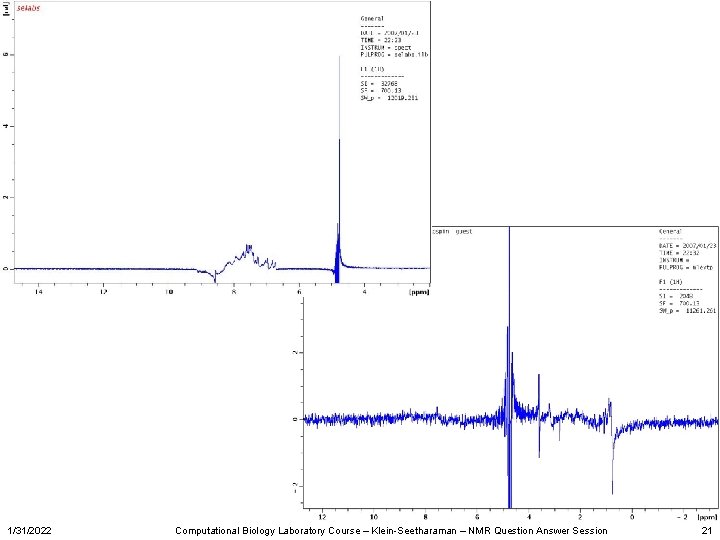
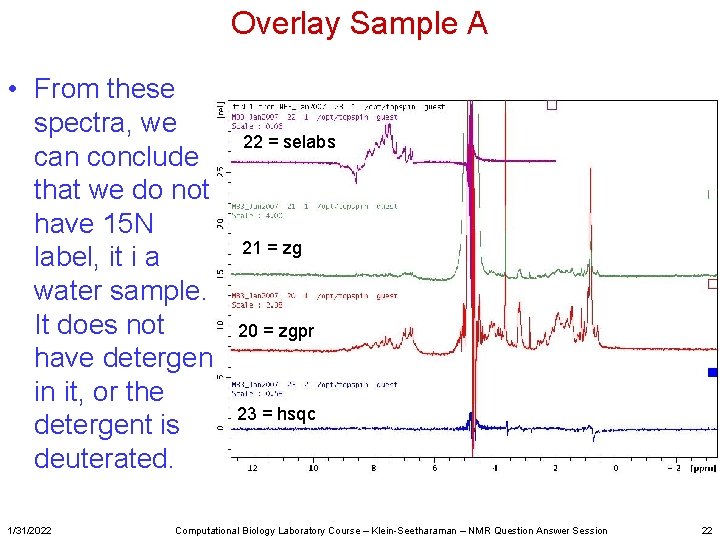
Overlay Sample A • From these spectra, we can conclude that we do not have 15 N label, it i a water sample. It does not have detergent in it, or the detergent is deuterated. 1/31/2022 22 = selabs 21 = zg 20 = zgpr 23 = hsqc Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 22

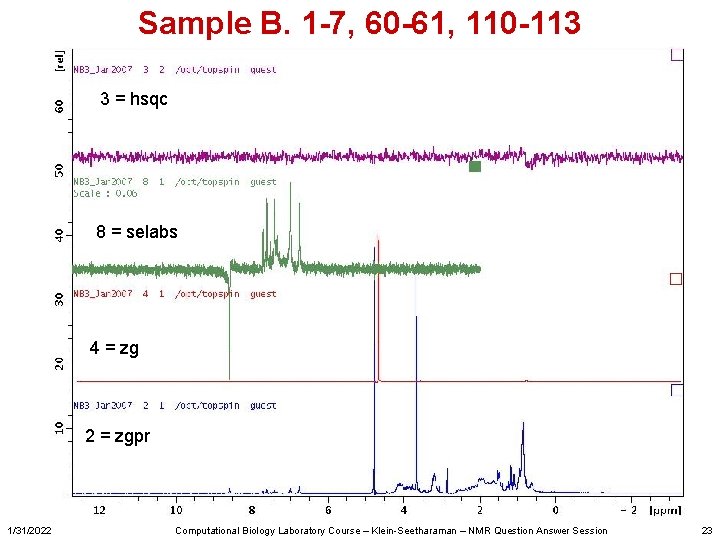
Sample B. 1 -7, 60 -61, 110 -113 3 = hsqc 8 = selabs 4 = zg 2 = zgpr 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 23

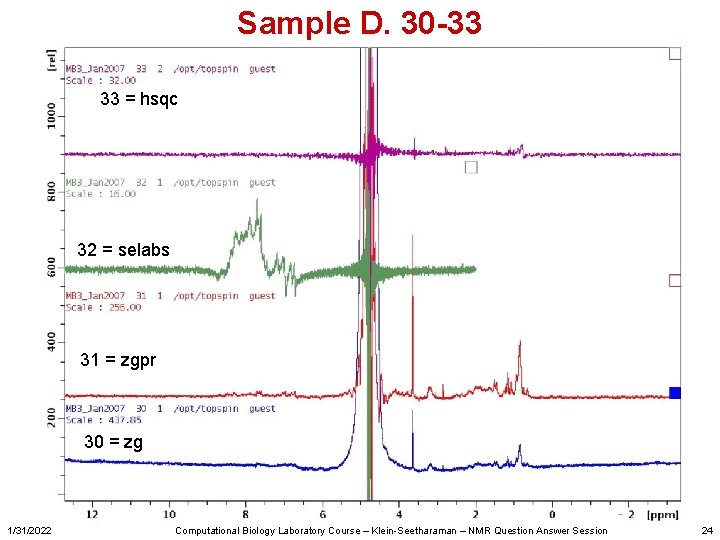
Sample D. 30 -33 33 = hsqc 32 = selabs 31 = zgpr 30 = zg 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 24

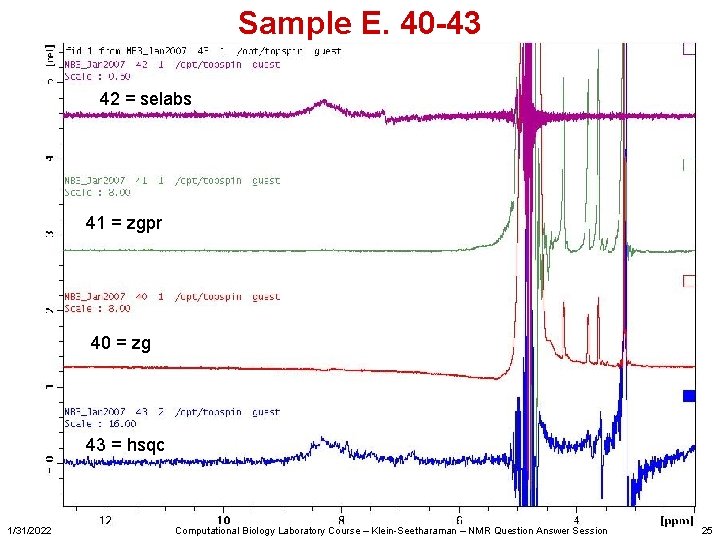
Sample E. 40 -43 42 = selabs 41 = zgpr 40 = zg 43 = hsqc 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 25

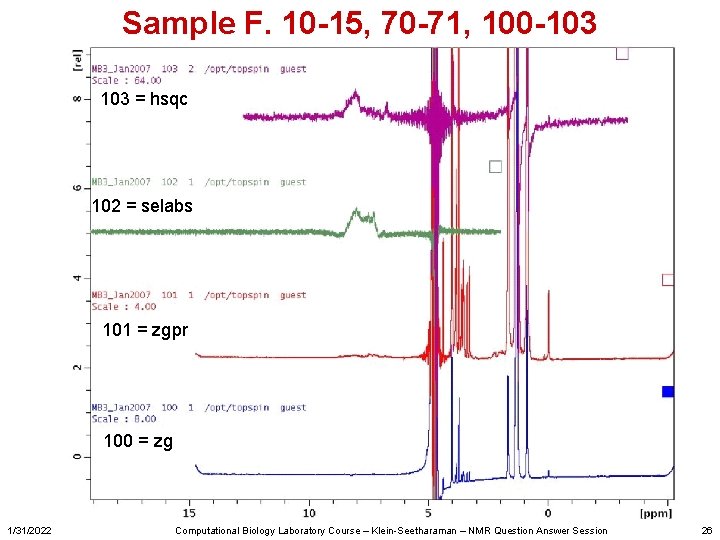
Sample F. 10 -15, 70 -71, 100 -103 = hsqc 102 = selabs 101 = zgpr 100 = zg 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 26

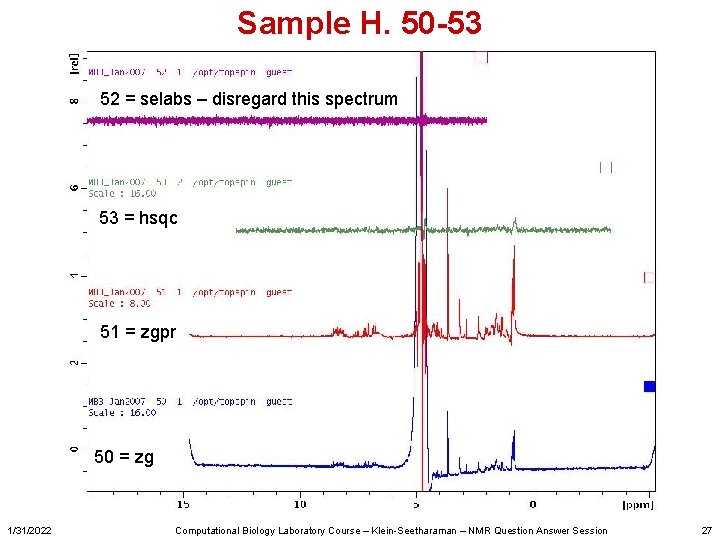
Sample H. 50 -53 52 = selabs – disregard this spectrum 53 = hsqc 51 = zgpr 50 = zg 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 27

Parameters to watch out for • ns • rg • will affect actual intensity measured. 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 28

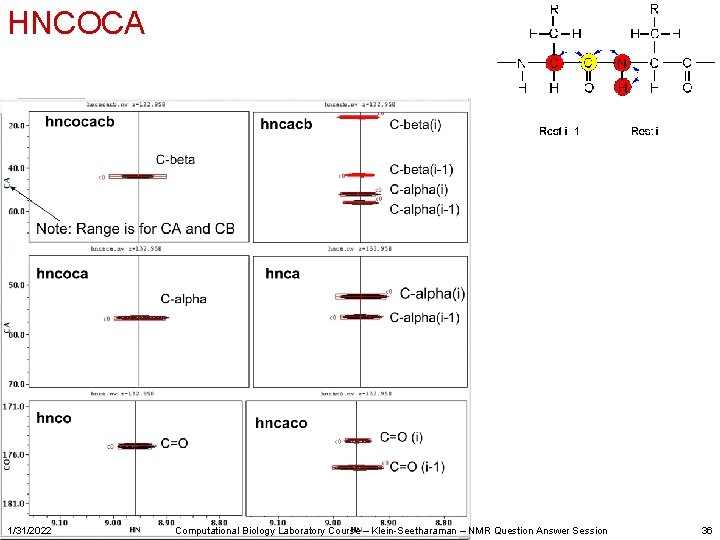
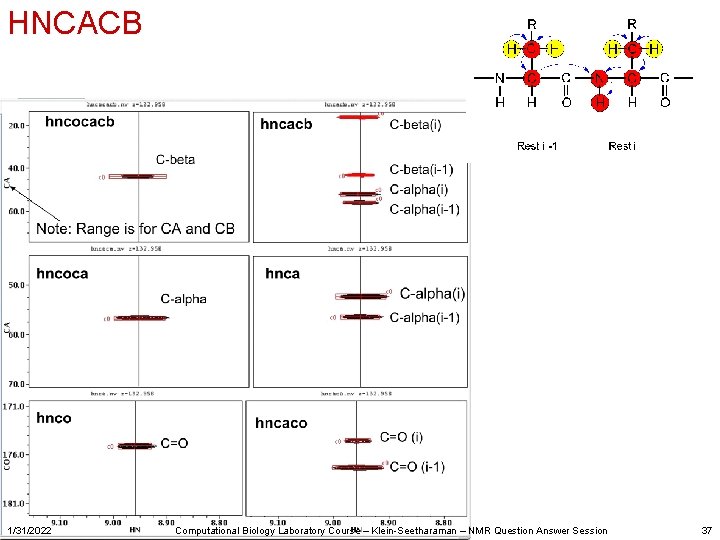
Several different assignment strategies exist Most easily automated: • • • 1/31/2022 HNCOCACB HNCOCA HNCACB HNCACO Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 29

Homework Part 2 • Converting raw spectral formats to other formats using NMRpipe – Example: HIV protease relaxation data • Assignment of signals with one strategy – – – HNCOCACBG HNCOCA HNCACB HNCACO • Example: Ubiquitin assignment with NMRview. J 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 30

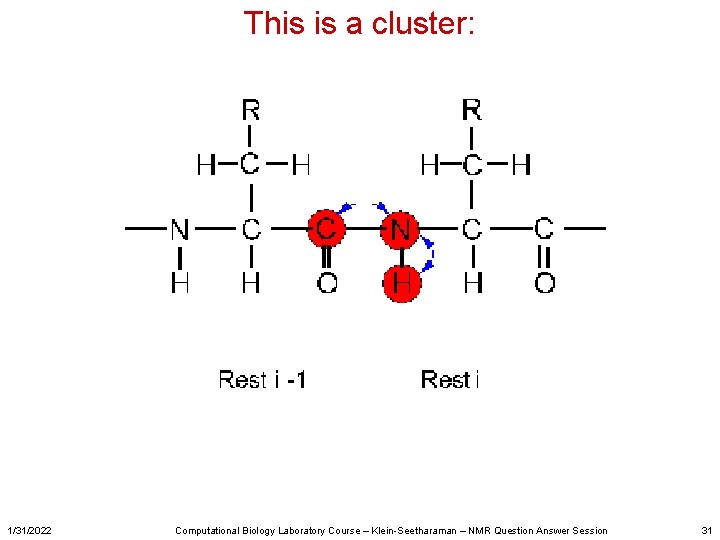
This is a cluster: 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 31

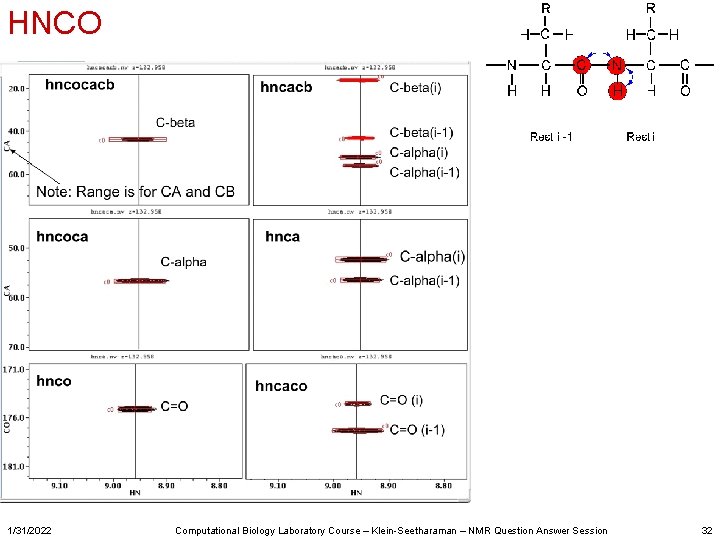
HNCO 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 32

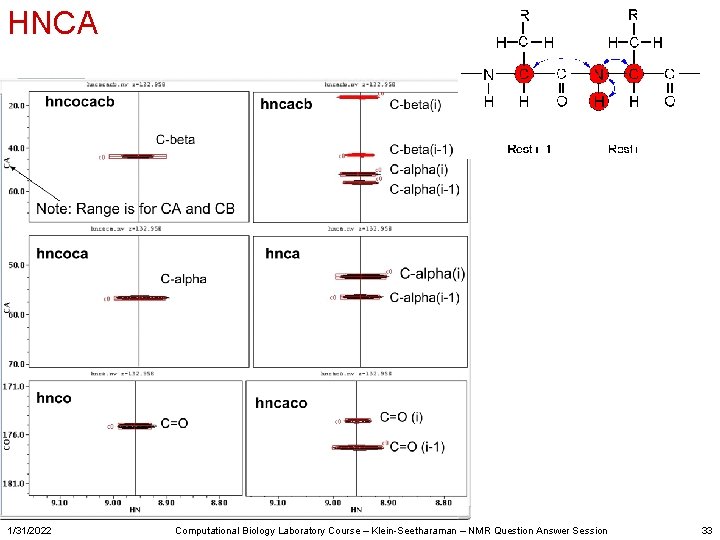
HNCA 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 33

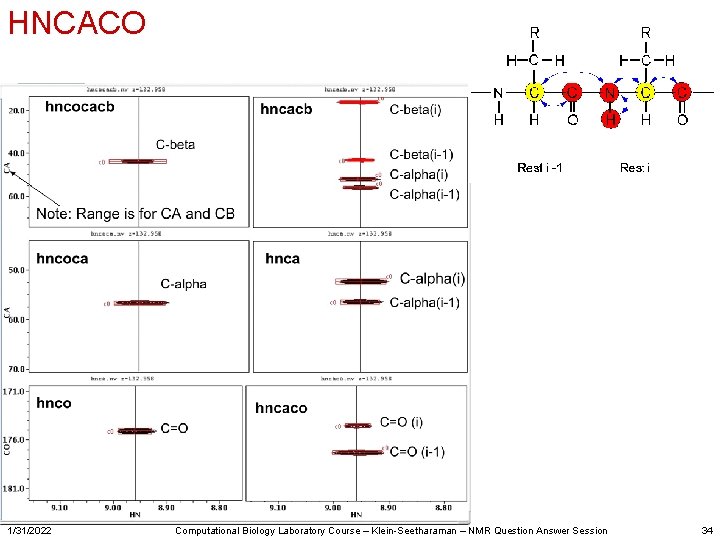
HNCACO 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 34

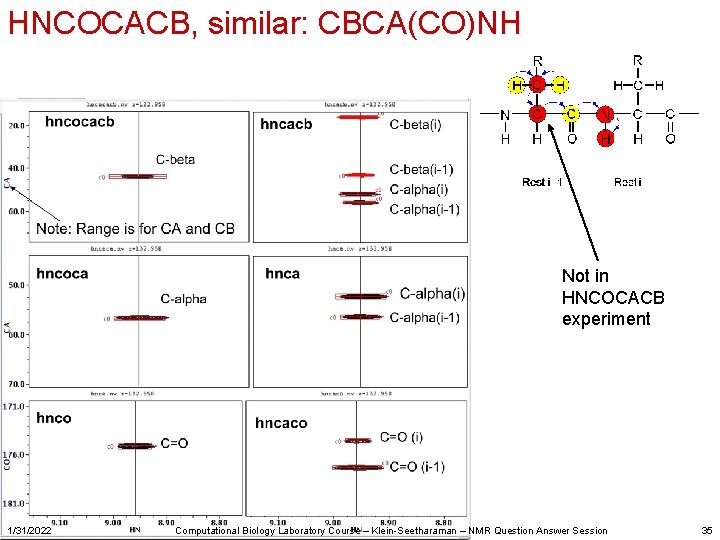
HNCOCACB, similar: CBCA(CO)NH Not in HNCOCACB experiment 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 35

HNCOCA 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 36

HNCACB 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 37

What are other things one can do with NMR? • For study of dynamics – Measure and analyze T 1, T 2, Het. NOE data – We went through T 1 analysis in computer lab (conversion of raw data with NMRpipe, analysis with NMRview. J) – T 2 was your homework • Other studies might include chemical shift perturbations • Temperature titrations • … 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 38

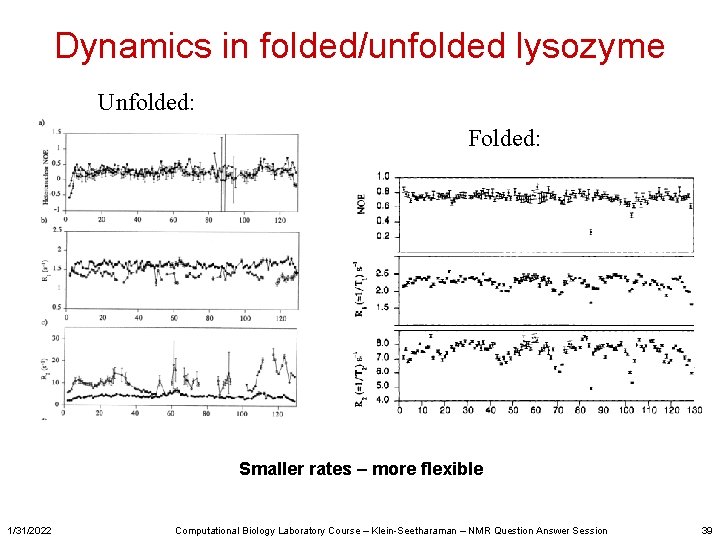
Dynamics in folded/unfolded lysozyme Unfolded: Folded: Smaller rates – more flexible 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 39

T 1 for HIV protease 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 40

T 2 for HIV protease 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 41

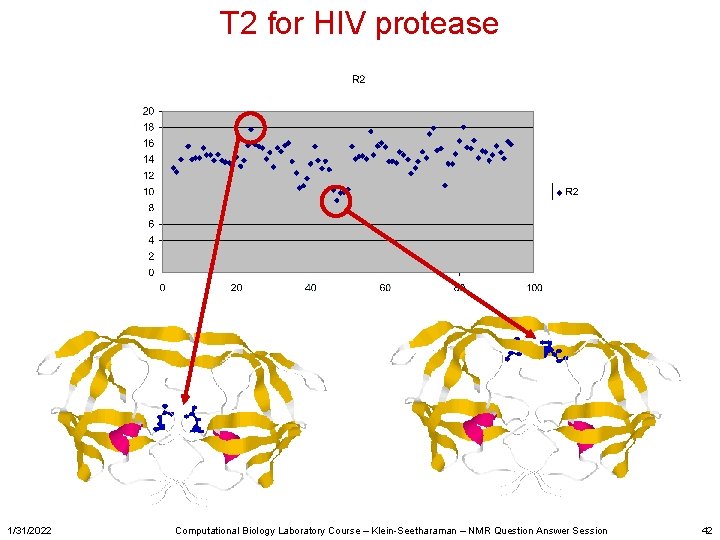
T 2 for HIV protease 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 42

• From last class in case there are questions 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 43

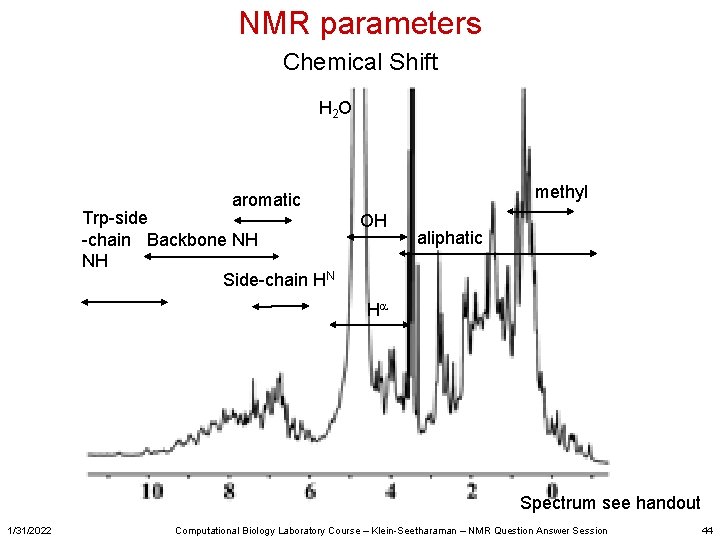
NMR parameters Chemical Shift H 2 O methyl aromatic Trp-side -chain Backbone NH NH Side-chain HN OH aliphatic Ha Spectrum see handout 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 44

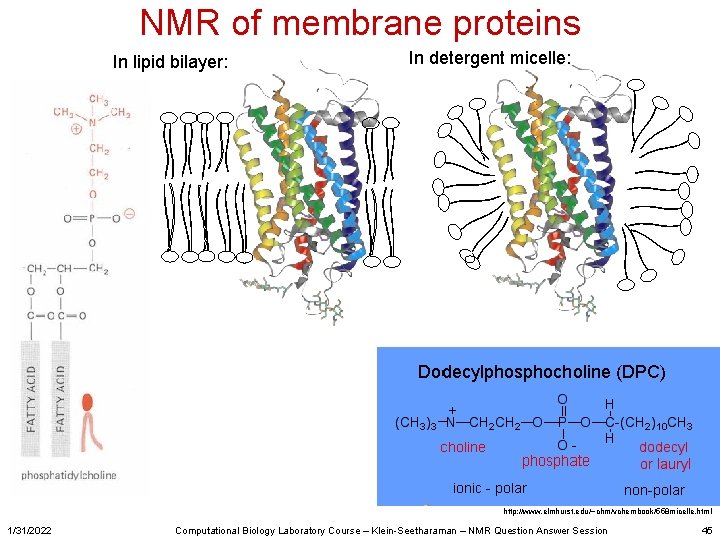
NMR of membrane proteins In lipid bilayer: In detergent micelle: http: //www. elmhurst. edu/~chm/vchembook/558 micelle. html 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 45

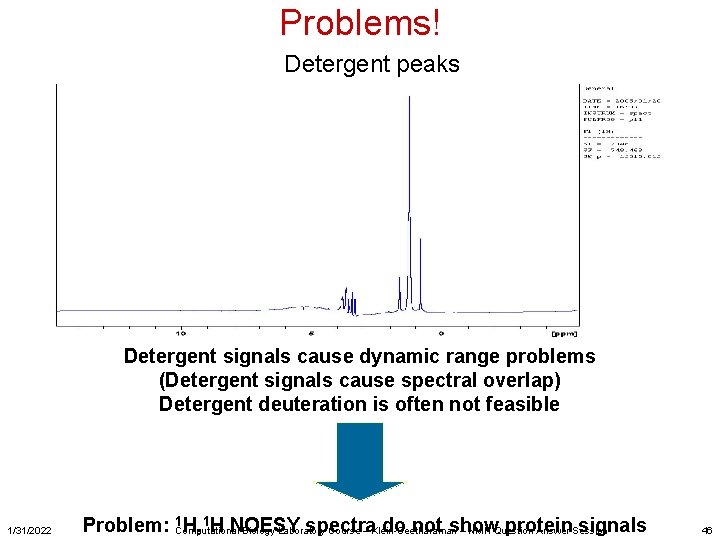
Problems! Detergent peaks Detergent signals cause dynamic range problems (Detergent signals cause spectral overlap) Detergent deuteration is often not feasible 1/31/2022 1 H, 1 H NOESY spectra do not show protein signals Problem: Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 46

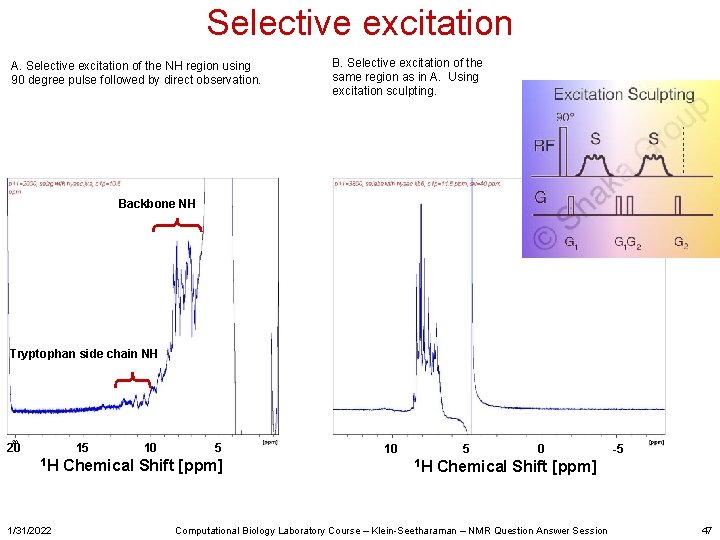
Selective excitation A. Selective excitation of the NH region using 90 degree pulse followed by direct observation. B. Selective excitation of the same region as in A. Using excitation sculpting. Backbone NH Tryptophan side chain NH 20 15 1 H 1/31/2022 10 5 Chemical Shift [ppm] 10 5 1 H 0 -5 Chemical Shift [ppm] Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 47

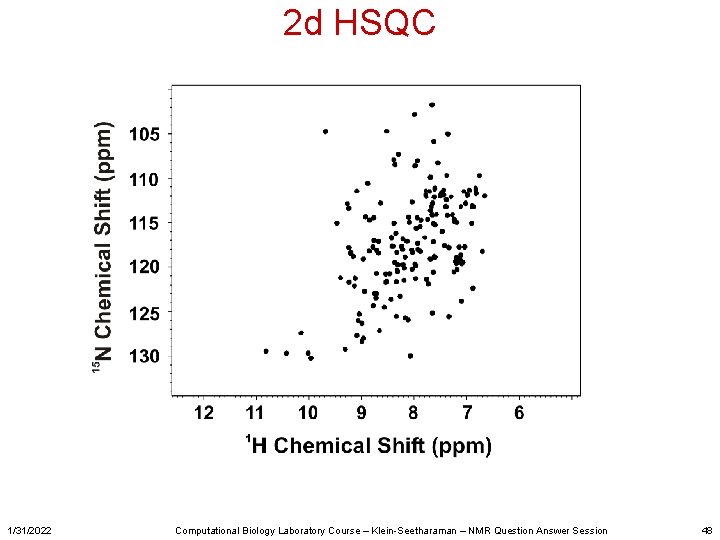
2 d HSQC 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 48

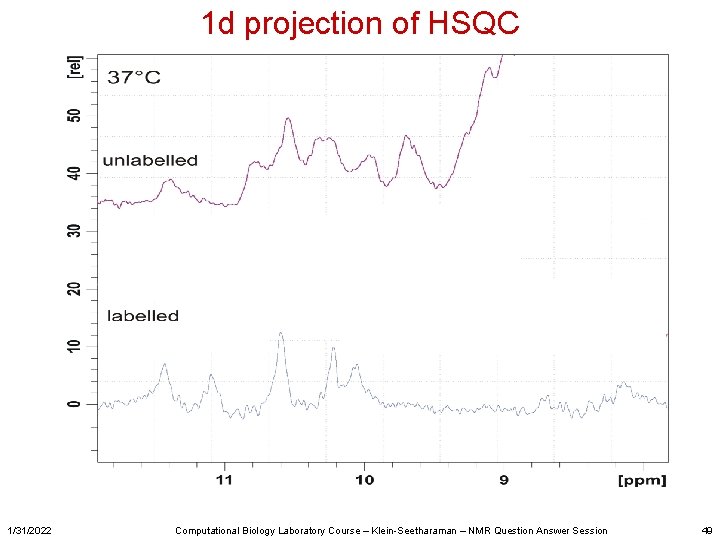
1 d projection of HSQC 1/31/2022 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Question Answer Session 49