NMR Spectroscopy Part 2 Judith KleinSeetharaman Department of





- Slides: 27

NMR Spectroscopy – Part 2 Judith Klein-Seetharaman Department of Structural Biology jks 33@pitt. edu Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture

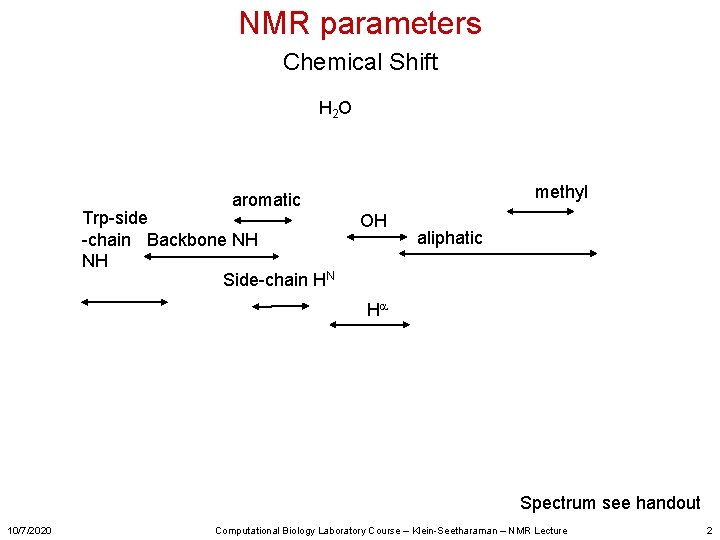
NMR parameters Chemical Shift H 2 O methyl aromatic Trp-side -chain Backbone NH NH Side-chain HN OH aliphatic Ha Spectrum see handout 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 2

1 d 1 H NMR spectra 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 3

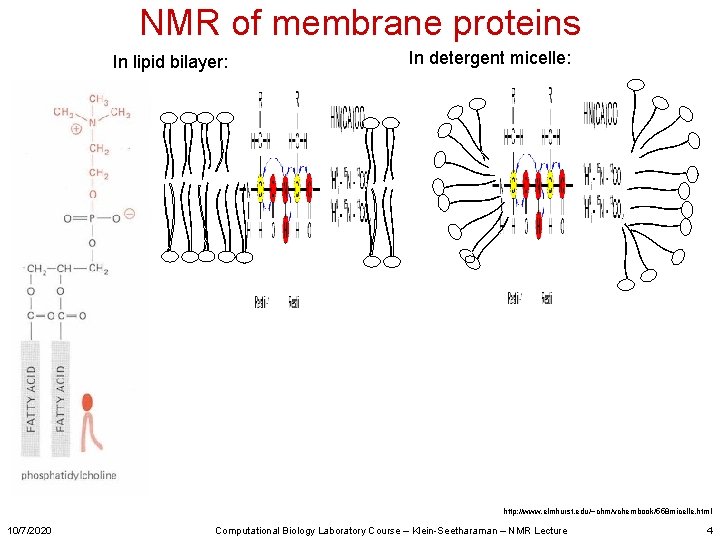
NMR of membrane proteins In lipid bilayer: In detergent micelle: http: //www. elmhurst. edu/~chm/vchembook/558 micelle. html 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 4

Problems! Detergent peaks Detergent signals cause dynamic range problems (Detergent signals cause spectral overlap) Detergent deuteration is often not feasible 10/7/2020 Problem: 1 H, 1 HComputational NOESY spectra do– Klein-Seetharaman not show–protein Biology Laboratory Course NMR Lecture signals 5

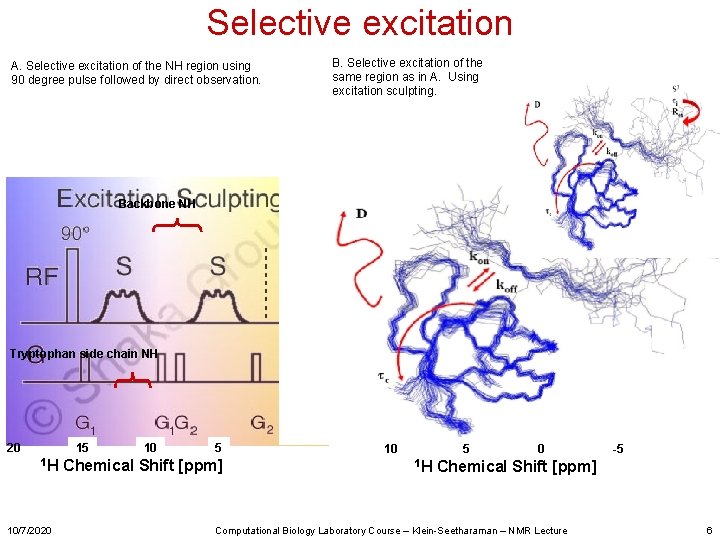
Selective excitation A. Selective excitation of the NH region using 90 degree pulse followed by direct observation. B. Selective excitation of the same region as in A. Using excitation sculpting. Backbone NH Tryptophan side chain NH 20 15 1 H 10/7/2020 10 5 Chemical Shift [ppm] 10 5 1 H 0 -5 Chemical Shift [ppm] Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 6

2 d HSQC 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 7

1 d projection of HSQC 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 8

HSQC spectra 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 9

Chemical shift perturbation Figure 2 in “Cap-free structure of e. IF 4 E suggests a basis for conformational regulation by its ligands Laurent Volpon, Michael J Osborne, Ivan Topisirovic, Nadeem Siddiqui and Katherine LB Borden The EMBO Journal (2006) 25, 5138– 5149 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 10

Assignment is needed! 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 11

Several different assignment strategies exist Most easily automated: • • • 10/7/2020 HNCOCACB HNCOCA HNCACB HNCACO Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 12

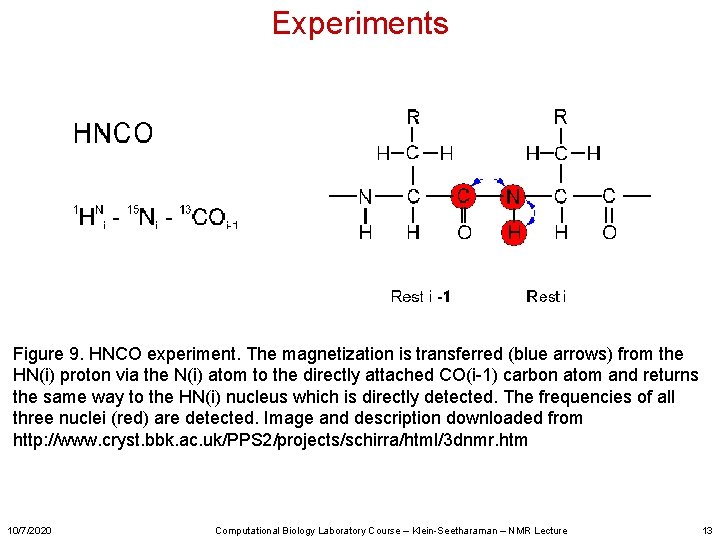
Experiments Figure 9. HNCO experiment. The magnetization is transferred (blue arrows) from the HN(i) proton via the N(i) atom to the directly attached CO(i-1) carbon atom and returns the same way to the HN(i) nucleus which is directly detected. The frequencies of all three nuclei (red) are detected. Image and description downloaded from http: //www. cryst. bbk. ac. uk/PPS 2/projects/schirra/html/3 dnmr. htm 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 13

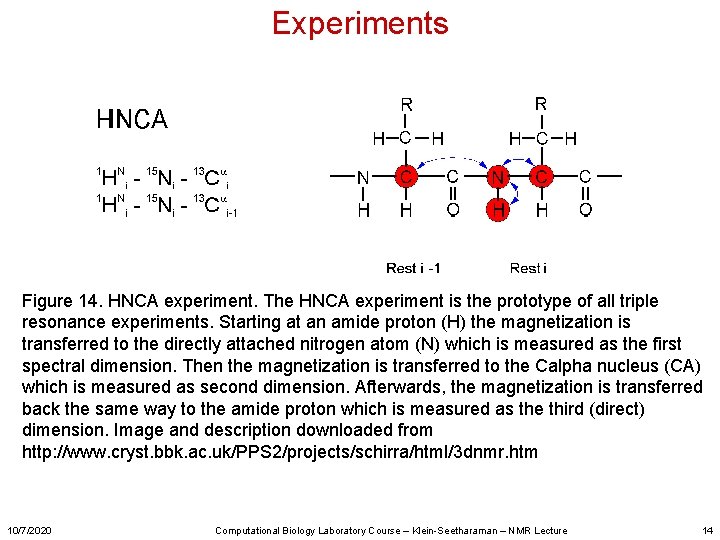
Experiments Figure 14. HNCA experiment. The HNCA experiment is the prototype of all triple resonance experiments. Starting at an amide proton (H) the magnetization is transferred to the directly attached nitrogen atom (N) which is measured as the first spectral dimension. Then the magnetization is transferred to the Calpha nucleus (CA) which is measured as second dimension. Afterwards, the magnetization is transferred back the same way to the amide proton which is measured as the third (direct) dimension. Image and description downloaded from http: //www. cryst. bbk. ac. uk/PPS 2/projects/schirra/html/3 dnmr. htm 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 14

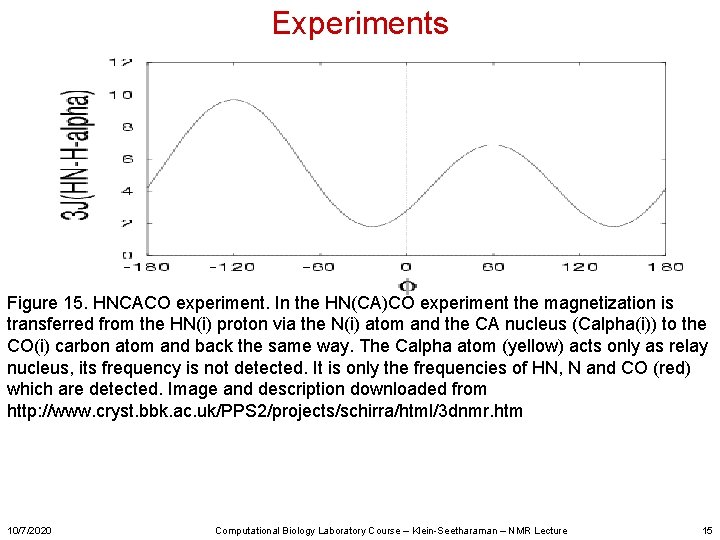
Experiments Figure 15. HNCACO experiment. In the HN(CA)CO experiment the magnetization is transferred from the HN(i) proton via the N(i) atom and the CA nucleus (Calpha(i)) to the CO(i) carbon atom and back the same way. The Calpha atom (yellow) acts only as relay nucleus, its frequency is not detected. It is only the frequencies of HN, N and CO (red) which are detected. Image and description downloaded from http: //www. cryst. bbk. ac. uk/PPS 2/projects/schirra/html/3 dnmr. htm 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 15

Assignments 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 16

Structure Prediction by NMR 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 17

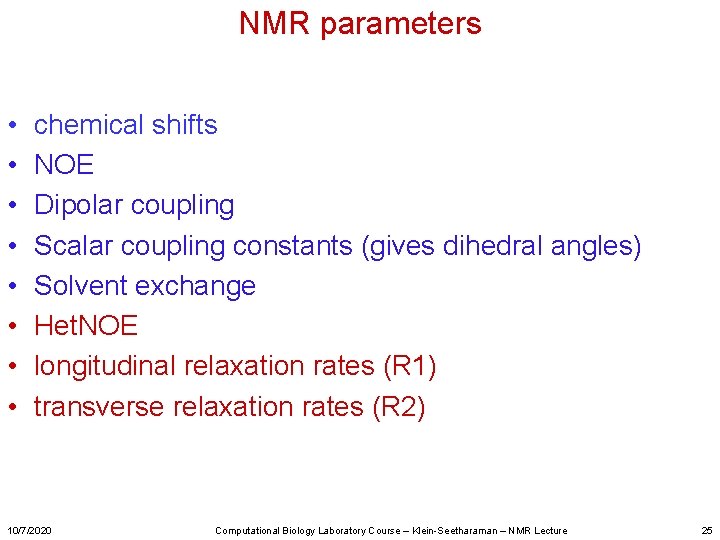
NMR parameters • • chemical shifts NOE Dipolar coupling Scalar coupling constants (gives dihedral angles) Solvent exchange Het. NOE longitudinal relaxation rates (R 1) transverse relaxation rates (R 2) 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 18

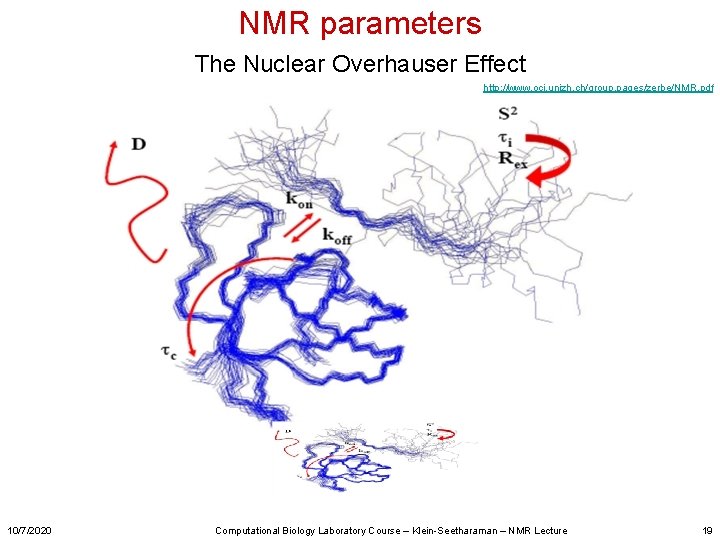
NMR parameters The Nuclear Overhauser Effect http: //www. oci. unizh. ch/group. pages/zerbe/NMR. pdf 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 19

HSQC TOCSY http: //www. oci. unizh. ch/group. pages/zerbe/NMR. pdf 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 20

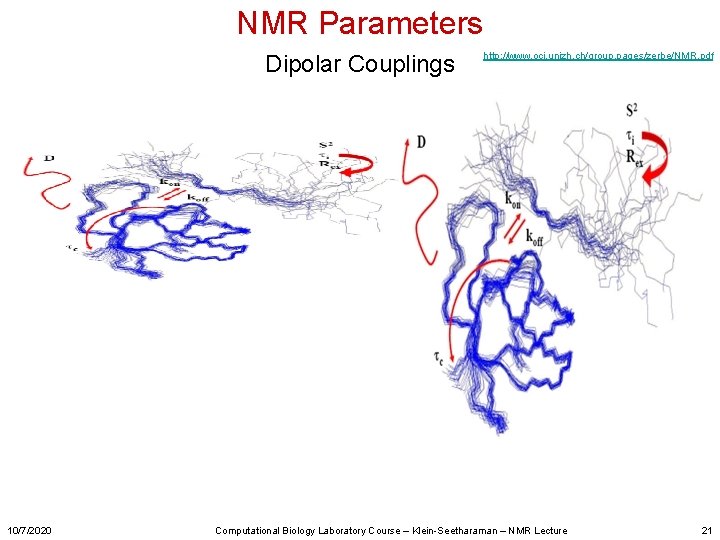
NMR Parameters Dipolar Couplings 10/7/2020 http: //www. oci. unizh. ch/group. pages/zerbe/NMR. pdf Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 21

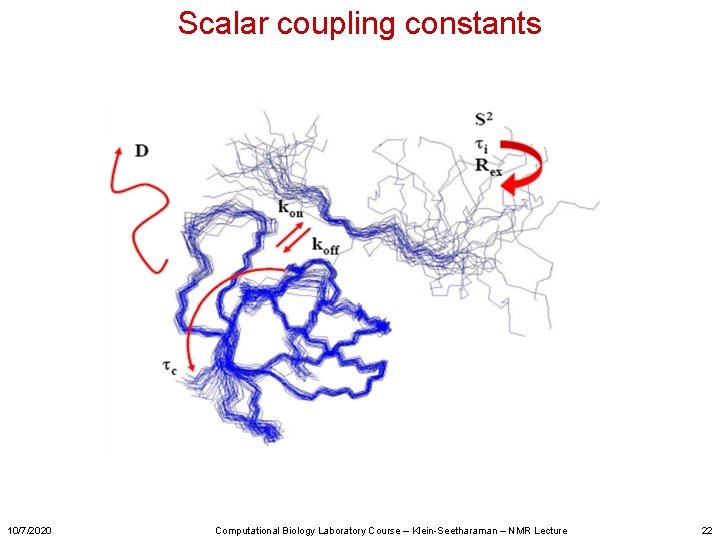
Scalar coupling constants 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 22

Structure Calculations • Distance geometry – Determines ensembles of structures consistent with an incomplete set of distance restraints • Metric matrix algorithm • Variable target function approach • Restrained molecular dynamics – Cartesian or torsion-angle coordinate systems – Molecular dynamics force fields are supplemented by pseudo energy terms based on the NMR-derived restraints 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 23

Structure Prediction From NMR Parameters Most widely used software suites • CNS – http: //cns. csb. yale. edu/v 1. 1/ • XPLOR – http: //xplor. csb. yale. edu/xplor/ 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 24

NMR parameters • • chemical shifts NOE Dipolar coupling Scalar coupling constants (gives dihedral angles) Solvent exchange Het. NOE longitudinal relaxation rates (R 1) transverse relaxation rates (R 2) 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 25

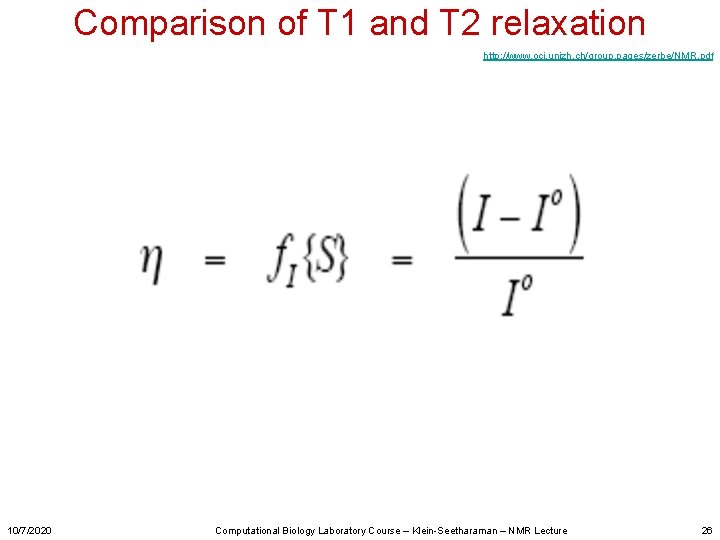
Comparison of T 1 and T 2 relaxation http: //www. oci. unizh. ch/group. pages/zerbe/NMR. pdf 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 26

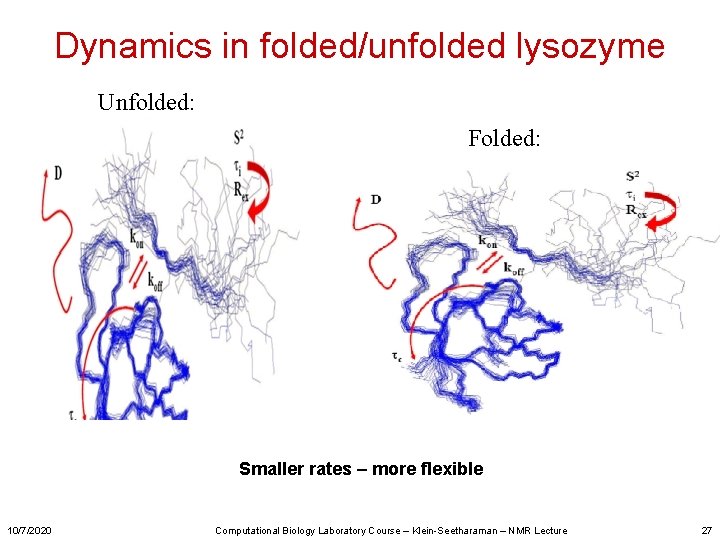
Dynamics in folded/unfolded lysozyme Unfolded: Folded: Smaller rates – more flexible 10/7/2020 Computational Biology Laboratory Course – Klein-Seetharaman – NMR Lecture 27
 Nmr instrumentation
Nmr instrumentation Factors influencing chemical shift
Factors influencing chemical shift Dept nmr spectroscopy
Dept nmr spectroscopy Advantages of nmr spectroscopy
Advantages of nmr spectroscopy Chch3cl
Chch3cl Nmr lipoprofile
Nmr lipoprofile Pople notation examples
Pople notation examples Borrow heavily
Borrow heavily Acorn nmr
Acorn nmr Hydrohalogenation mechanism
Hydrohalogenation mechanism 1 bromopropane nmr
1 bromopropane nmr Evans method
Evans method Ortho meta para h nmr
Ortho meta para h nmr Spektra nmr
Spektra nmr Nmr lipoprofile
Nmr lipoprofile Multipletowość nmr
Multipletowość nmr Mezomer effektus
Mezomer effektus Nmr sample requirements
Nmr sample requirements Efekt dachowy nmr
Efekt dachowy nmr Singlet doublet triplet quartet quintet
Singlet doublet triplet quartet quintet Tabel pergeseran kimia h nmr
Tabel pergeseran kimia h nmr Pola splitting nmr
Pola splitting nmr Acetylferrocene ir spectrum labeled
Acetylferrocene ir spectrum labeled Nmr nedir
Nmr nedir Nmr polymer
Nmr polymer Function of nmr
Function of nmr Hexane 1h nmr
Hexane 1h nmr Factors affecting chemical shift
Factors affecting chemical shift