Computational Pharmacology Judith KleinSeetharaman Assistant Professor Department of























- Slides: 47

Computational Pharmacology Judith Klein-Seetharaman Assistant Professor, Department of Pharmacology

High Points of the Case Study: The Development of Cox-2 Inhibitors • High points regarding the success of the drugs… • High points regarding drug discovery principles…

High Points of the Case Study: The Development of Cox-2 Inhibitors • High points regarding the success of the drugs: – Isoforms with differential properties – Differential expression • High points regarding drug discovery principles: – Anomalous drug effects (don’t ignore compounds that have therapeutic profiles that do not “fit”) – Molecular biology in isozyme discovery and characterization – Transgenic disease models were not useful here – Importance of differential expression for drug action and sideeffects – Significance of structural data (importance of sequence to structure mapping)

Case study COX A Wonder Drug: What is the most commonly-taken drug today? It is an effective painkiller. It reduces fever and inflammation when the body gets overzealous in its defenses against infection and damage. It slows blood clotting, reducing the chance of stroke and heart attack in susceptible individuals. It may be an effective addition to the fight against cancer. Aspirin has been used professionally for a century, and traditionally since ancient times. A similar compound found in willow bark, salicylic acid, has a long history of use in herbal treatment. But only in the last few decades have we understood how aspirin works, and how it might be improved http: //www. rcsb. org/pdb/molecules/pdb 17_1. html

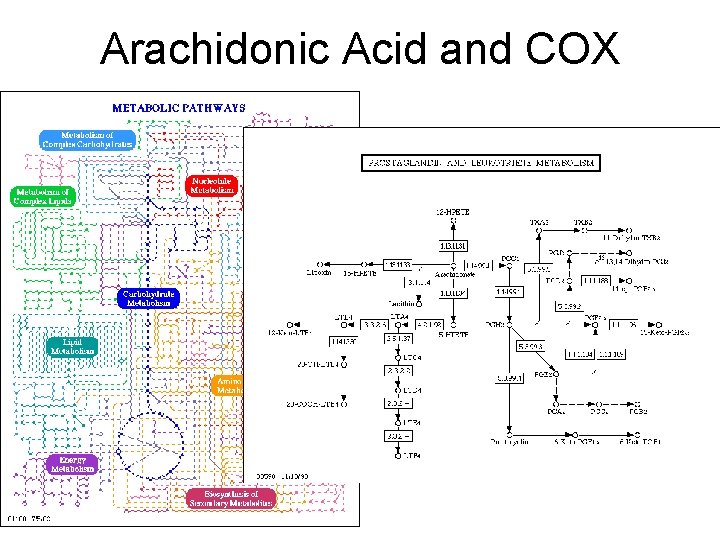
Prostaglandins As you might expect from a drug with such diverse actions, aspirin blocks a central process in the body: Aspirin blocks the production of prostaglandins, key hormones that are used to carry local messages. Unlike most hormones, which are produced in specialized glands and then delivered throughout the body by the blood, prostaglandins are created by cells and then act only in the surrounding area before they are broken down. Prostaglandins control many of these neighborhood processes, including the constriction of muscle cells around blood vessels, aggregation of platelets during blood clotting, and constriction of the uterus during labor. Prostaglandins also deliver and strengthen pain signals and induce inflammation. These many different processes are all controlled by different prostaglandins, but all created from a common precursor molecule. http: //www. rcsb. org/pdb/molecules/pdb 17_1. html

Arachidonic Acid and COX

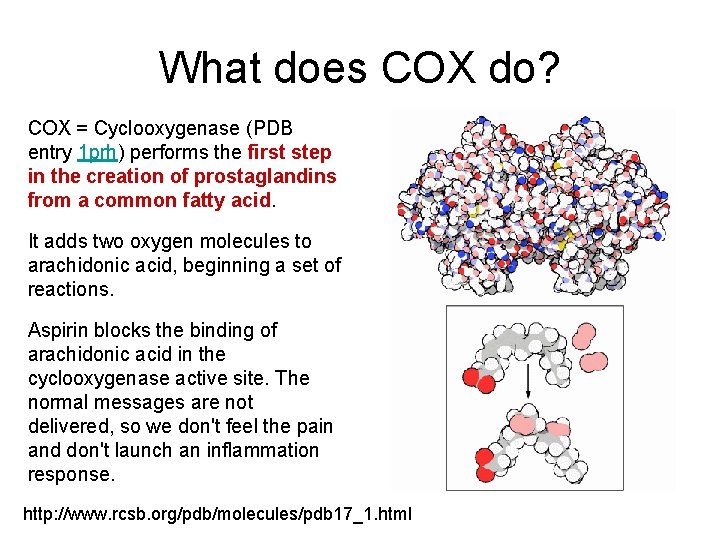
What does COX do? COX = Cyclooxygenase (PDB entry 1 prh) performs the first step in the creation of prostaglandins from a common fatty acid. It adds two oxygen molecules to arachidonic acid, beginning a set of reactions. Aspirin blocks the binding of arachidonic acid in the cyclooxygenase active site. The normal messages are not delivered, so we don't feel the pain and don't launch an inflammation response. http: //www. rcsb. org/pdb/molecules/pdb 17_1. html

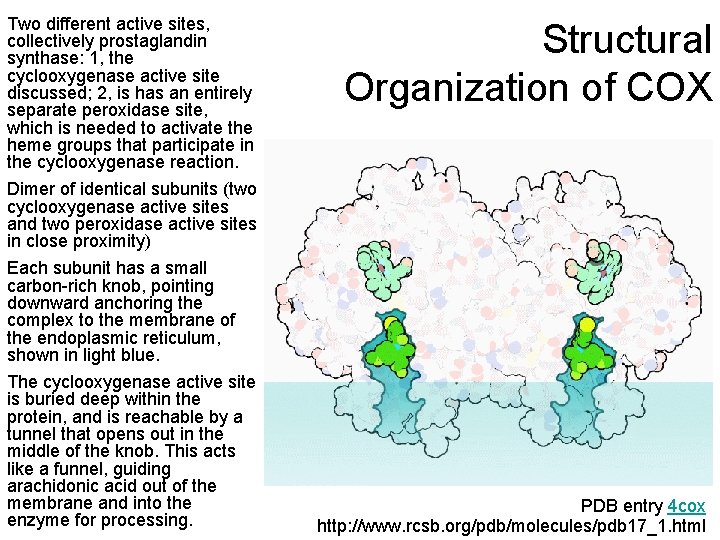
Two different active sites, collectively prostaglandin synthase: 1, the cyclooxygenase active site discussed; 2, is has an entirely separate peroxidase site, which is needed to activate the heme groups that participate in the cyclooxygenase reaction. Structural Organization of COX Dimer of identical subunits (two cyclooxygenase active sites and two peroxidase active sites in close proximity) Each subunit has a small carbon-rich knob, pointing downward anchoring the complex to the membrane of the endoplasmic reticulum, shown in light blue. The cyclooxygenase active site is buried deep within the protein, and is reachable by a tunnel that opens out in the middle of the knob. This acts like a funnel, guiding arachidonic acid out of the membrane and into the enzyme for processing. PDB entry 4 cox http: //www. rcsb. org/pdb/molecules/pdb 17_1. html

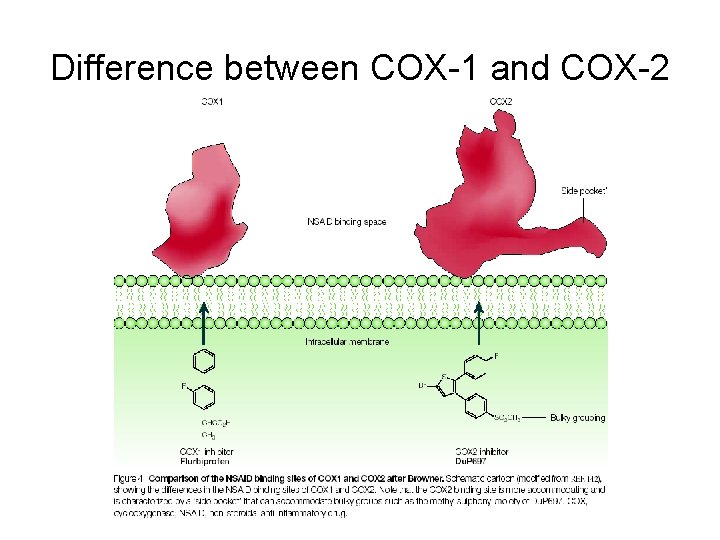
Why is there a COX-1 and COX-2? COX-1 and COX-2 are made for different purposes. COX-1 is built in many different cells to create prostaglandins used for basic housekeeping messages throughout the body. COX-2 is built only in special cells and is used for signaling pain and inflammation. Aspirin attacks both. Since COX-1 is targeted, aspirin can lead to unpleasant complications, such as stomach bleeding. Needed: specific compounds that block just COX-2, leaving COX-1 to perform its essential jobs. These drugs are selective pain-killers and fever reducers, without the unpleasant side-effects.

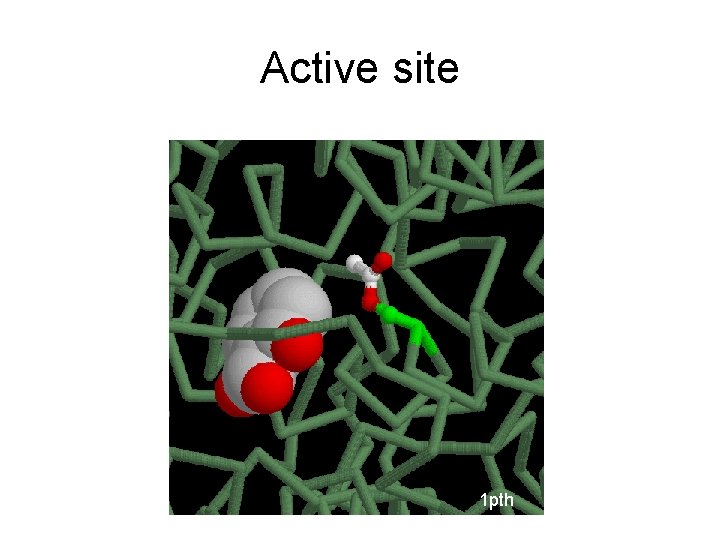
Active site 1 pth

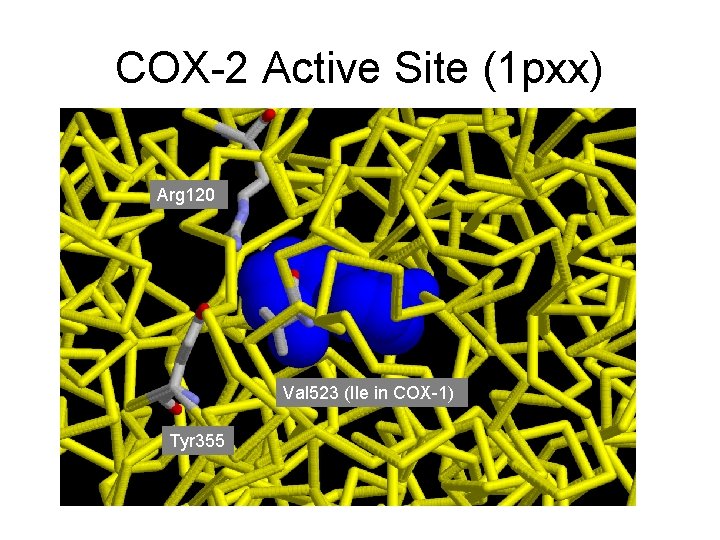
COX-2 Active Site (1 pxx) Arg 120 Val 523 (Ile in COX-1) Tyr 355

Difference between COX-1 and COX-2

Meaning of this case study for future drug discovery • If one could predict the structure of proteins from sequence, one could discover new drugs at a fast pace • If one could predict the relationship between isozyme and tissue expression, one could design drugs specific to certain tissues • If one could predict the interactions of proteins in different protein networks, one could interpret complex data such as animal models • If one could…

What is computational pharmacology? • Use of bioinformatics and computational biology with relevance to pharmacology, including understanding of – drug action – drug side effects – identification of drug targets – drug design

Background • View of living organisms as molecular circuitry: – intended modes of operation = healthy state – aberrant modes of operation = disease state • Diagnosis: – identify the molecular basis of disease • Therapy: – guide biochemical circuitry back to healthy state • Molecular circuitry: – biochemical processes, that form and recycle molecules in a coordinated and balanced fashion

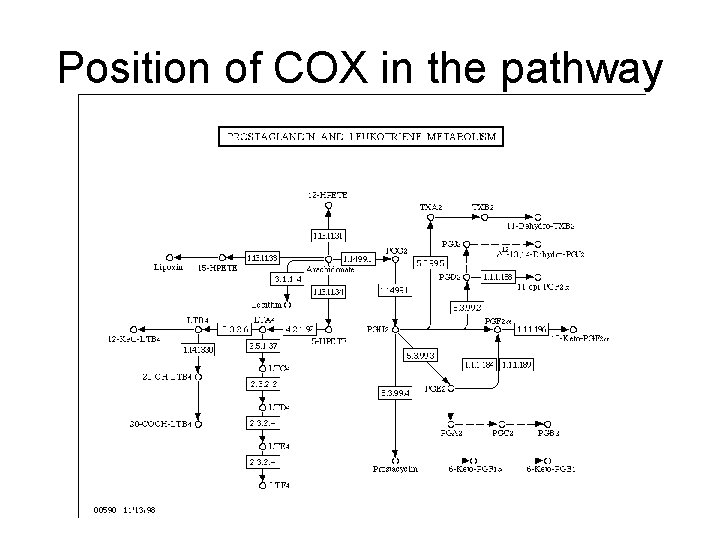
Where is COX?

Position of COX in the pathway

Information Sources • New technology generates massive amounts of data (often stored in publicly accessible databases): Genomics and Proteomics – Protein and DNA sequences / Whole genome sequences – Protein structure data – Protein pathways and networks – Protein interaction data – Expression data • How is this data useful for drug discovery? – Data needs to be organized, mined and visualized to allow scientific discovery

Basis for a role of computational approaches in drug discovery • help with the information in the databases and infer information that is not provided directly by genomics and proteomics data: higher level information => piece together all available information 1. To get detailed picture of a molecular process (or disease) 2. From 1, to identify new protein targets 3. From 2, to develop drugs • based on chemical similarity of known drugs • rational (structure-based) drug design interactively on computer screen • molecular docking (automatic, systematic computer-based prediction of structure and binding affinity of complex) • high-throughput screening and combinatorial chemistry

Areas of application: 7 hierarchical layers Layer 5. Predicting functional • Layer 1. Sequencing support structures (DNA - RNA – (physical mapping, fragment assembly proteins - lipids outcome: raw genome sequence) carbohydrates) • Layer 2. DNA sequence analysis 1. Homology modeling 1. Gene finding 2. ab initio 2. non-coding sequences 3. templates 3. regulatory sequences finding 4. partial information 4. orthologous and paralogous sequences 1. overall architecture 5. Evolution 2. binding pocket • Layer 3. Protein sequence analysis 3. protein backbone 1. homology detection Layer 6. Molecular 2. alignment interactions 3. functional annotation (Protein-ligand, -protein, -DNA, 4. cellular localization RNA, -lipid, -carbohydrate) • Layer 4. From linear sequence to threedimensional shapes Layer 7. Gene expression, – conformational space metabolic and regulatory – models for protein (mis)folding networks – discriminating structures – conformational ambiguity

General challenges Linking variety of databases Linking the different layers Interpretation Understanding drug action Drug discovery

Meaning of this case study for future drug discovery • If one could predict the structure of proteins from sequence, one could discover new drugs at a fast pace • If one could predict the relationship between isozyme and tissue expression, one could design drugs specific to certain tissues • if one could predict the interactions of proteins in different protein networks, one could interpret complex data such as animal models

Computational Approaches to Sequence-Structure Mapping in Drug Discovery • How would you go about comparing the sequences of COX-1 and COX-2? – Pairwise sequence alignment methods • Are there other isoforms? – Multiple sequence alignment – Protein family profiles and databases

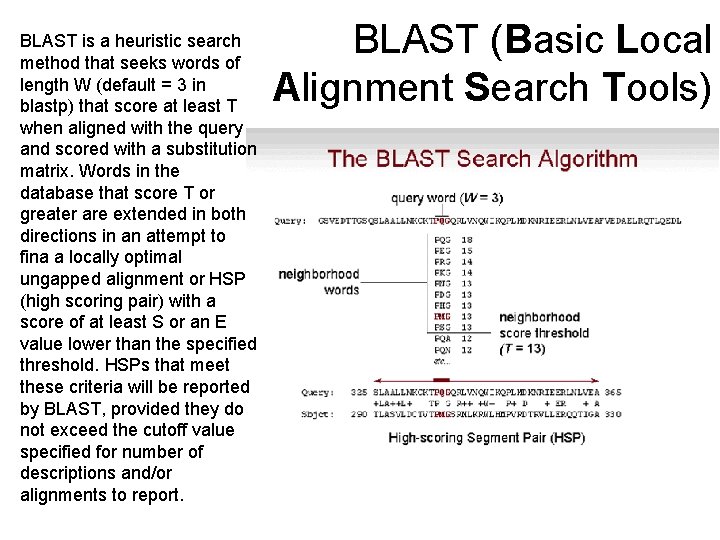
BLAST is a heuristic search method that seeks words of length W (default = 3 in blastp) that score at least T when aligned with the query and scored with a substitution matrix. Words in the database that score T or greater are extended in both directions in an attempt to fina a locally optimal ungapped alignment or HSP (high scoring pair) with a score of at least S or an E value lower than the specified threshold. HSPs that meet these criteria will be reported by BLAST, provided they do not exceed the cutoff value specified for number of descriptions and/or alignments to report. BLAST (Basic Local Alignment Search Tools)

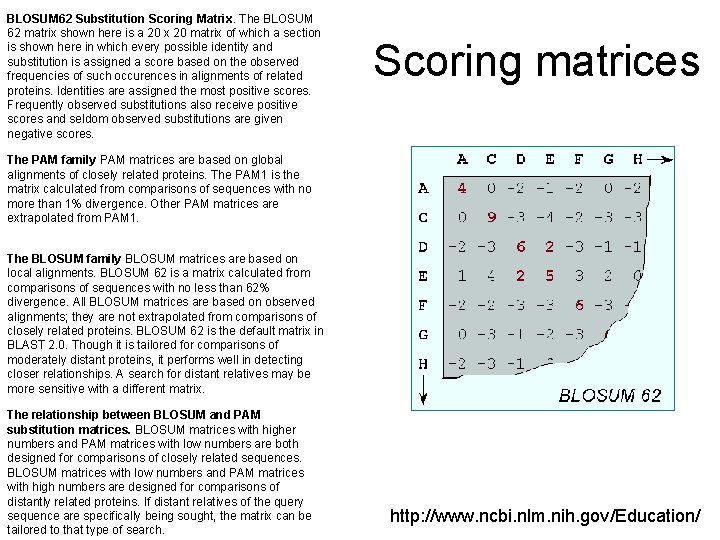
BLOSUM 62 Substitution Scoring Matrix. The BLOSUM 62 matrix shown here is a 20 x 20 matrix of which a section is shown here in which every possible identity and substitution is assigned a score based on the observed frequencies of such occurences in alignments of related proteins. Identities are assigned the most positive scores. Frequently observed substitutions also receive positive scores and seldom observed substitutions are given negative scores. Scoring matrices The PAM family PAM matrices are based on global alignments of closely related proteins. The PAM 1 is the matrix calculated from comparisons of sequences with no more than 1% divergence. Other PAM matrices are extrapolated from PAM 1. The BLOSUM family BLOSUM matrices are based on local alignments. BLOSUM 62 is a matrix calculated from comparisons of sequences with no less than 62% divergence. All BLOSUM matrices are based on observed alignments; they are not extrapolated from comparisons of closely related proteins. BLOSUM 62 is the default matrix in BLAST 2. 0. Though it is tailored for comparisons of moderately distant proteins, it performs well in detecting closer relationships. A search for distant relatives may be more sensitive with a different matrix. The relationship between BLOSUM and PAM substitution matrices. BLOSUM matrices with higher numbers and PAM matrices with low numbers are both designed for comparisons of closely related sequences. BLOSUM matrices with low numbers and PAM matrices with high numbers are designed for comparisons of distantly related proteins. If distant relatives of the query sequence are specifically being sought, the matrix can be tailored to that type of search. http: //www. ncbi. nlm. nih. gov/Education/

• Show blast result

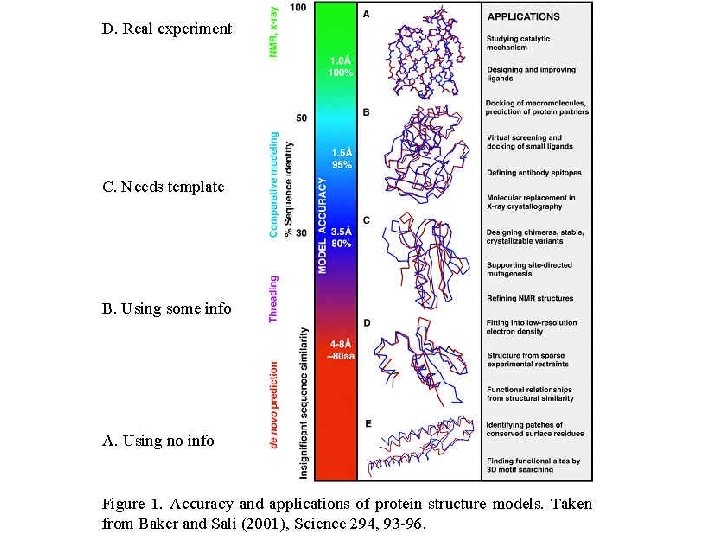
Structure Analysis • How does one determine structures? – Experimentally (X-ray, NMR) – Using computational methods (ab initio, Rosetta, threading, homology models) • How does one access structures? – pdb – SCOP/CATH • How does one analyze structural features? – Visualization programs (chime, rasmol, molmol, etc. ) – Demo with COX


Modeling Methods A. When no information but sequence and physical principles are used = ab initio structure prediction (Blue Gene IBM ) B. When other information is used (Survey of "ab initio" methods that use pdb information and their relation to protein folding) "fold recognition": requires a method for evaluating the compatibility of a given sequence with a given folding pattern B 0. 3 D profiles B 1. Rosetta: conformations from short segments in pdb B 2. Including experimental structural constraints B 3. Threading (=sequence-structure alignment), B 4. Inverse threading and folding experiments Reference Ivet B 4 a. using short-range information B 4 b. using short- and long-range information B 4. Predicting structural class only Reference Ivet B 5. Predicting active site only? B 6. Predicting protein-protein interaction sites? B 7. Predicting surface shape? C. When a template with known structure must be available homology modeling D. Modeling structures based on experimental data Both NMR and X-ray underdetermine the protein structure. To solve a structure one must minimize a combination of the deviation from the experimental data and the conformational energy: D 1. NMR (set of constraints on distances and angles) D 2. X-ray crystallography (Fourier transform of the electron density)

Evaluating structure prediction • Use rmsd to known structures - defines structural similarity • Critical Assessment of Structure Predictions (CASP) competitions • EVA, EVA submits sequences automatically to different prediction servers shortly before structures are published in pdb

Structures of High Sequence Similarity • Homology modeling

Basic Steps in Homology Modeling • Database searching for homologous proteins ( Blast the query sequence towards the pdb database ) • Alignment (Pairwise/ Multiple Alignments) – needs minimum 30% sequence identity, but to be useful usually need 40 -50% – note that ~30% of genomes have sequence identity of 20% • Model Building – Modeller , Composer etc • Model Refinement and Evaluation – Joy – Procheck etc

Model Building • Modeller (freeware, http: //www. salilab. org/modeller. html) • Spdbviewer Swissmodel–module (freeware, http: //us. expasy. org/spdbv/) • Composer (module of Insight. II, commercial version of Modeller)

Model Building Principles • Sequentially go from amino acid position to next position – if same amino acid, copy the coordinates – If different amino acid, if the new amino acid has atoms in common with the template, those atoms will be copied, and the rest are computed • At every step, check for steric clashes with previous amino acids – Minimization allowing the position of new amino acid to change – Only at the final stage, bond energy is minimized

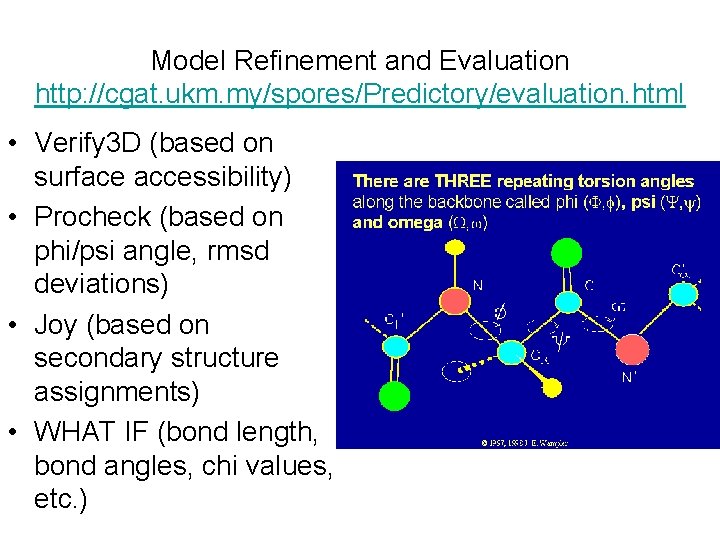
Model Refinement and Evaluation http: //cgat. ukm. my/spores/Predictory/evaluation. html • Verify 3 D (based on surface accessibility) • Procheck (based on phi/psi angle, rmsd deviations) • Joy (based on secondary structure assignments) • WHAT IF (bond length, bond angles, chi values, etc. )

WHAT IF Checklist A WHAT IF check report: what does it mean? ·General points ·Administrative checks ·Nomenclature ·Chain name ·Weights (occupancy) ·Missing atoms and C-terminal oxygens ·Symmetry ·Consistency ·Cell conventions ·Matthews' Coefficient ·Higher symmetry ·Non crystallographic symmetry ·Geometry ·Chirality ·Bond lengths ·Bond angles ·Torsion Angles: "Evaluation"; "Ramachandran"; "omega"; "Chi 1/2" ·Rings and planarity: "Planarity"; "Proline Puckering" ·Structure ·Inside/outside profile ·Bumps ·Packing quality ·Backbone: "number of hits"; "backbone normality"; "peptide flips" ·Sidechain rotamers ·Water molecules: "floating clusters"; "symmetry relations" ·B-factors: "average"; "low B-factors"; "B-factor distribution" ·Hydrogen bonds: "Flip check"; "HIS assignments"; "Unsatisfied"

Collection of homology models • MODBASE – uses PSI-BLAST plus MODELLER to model and stores coordinates in this database • SWISS-MODEL – automatic structure prediction

Similarity Measures • Sequence similarity – Alignment scores, identity, similarity, substitution matrices • Structural similarity – secondary structure – rmsd and alternative methods

Differences in Degrees of Similarity • High sequence similarity but structural differences – Impact on drug design : specificity of drugs for different isozymes (Example COX 1/2) • Low sequence similarity but structurally related – Impact on drug design: design based on existing drugs (Example GPCR)

GPCR Family and Their Ligands Class A: Rhodopsin-like Family Opsins, Odorants, Monoamines, Lipid messengers, Purines, Neuropeptides, Peptide hormones (e. g. platelet activating factor, gonadotropin -releasing hormone, th yrotropin releasing hormone & melatonin), Glycoprotein hormones, Chemokines, Proteases, Cannabis, Viral Class B: Secretin-like Family Glucagon, Calcitonin, parathyroid hormone, secretin Class C: Metabotropic glutamate and Chemosensor Family m. Glu. R 1 -7, Calcium sensors, GABA-B Class D: Fungal pheromone Family Class E: c-AMP receptor (Dictyostelium) Family Class F: Frizzled/Smoothened family Putative families: Ocular albinism proteins, Drosophila odorant receptors, Plant Mlo receptors, Nematode chemoreceptors, Vomeronasal receptors Putative/ unclassified orphans

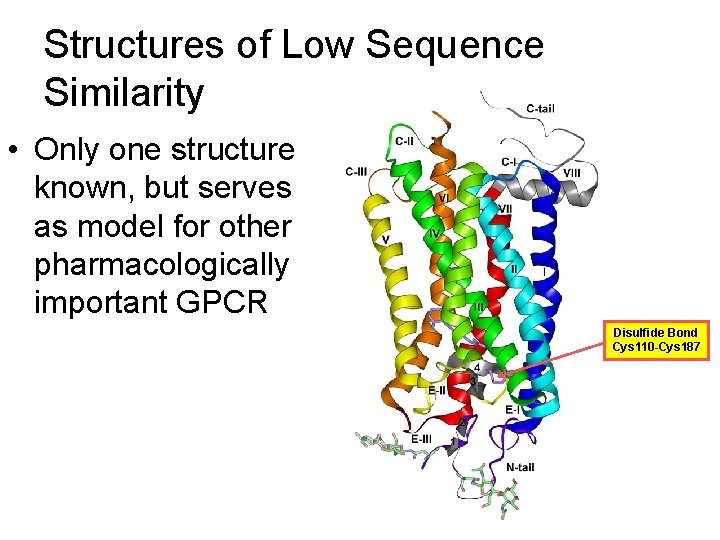
Structures of Low Sequence Similarity • Only one structure known, but serves as model for other pharmacologically important GPCR Disulfide Bond Cys 110 -Cys 187

• Demo if time permits

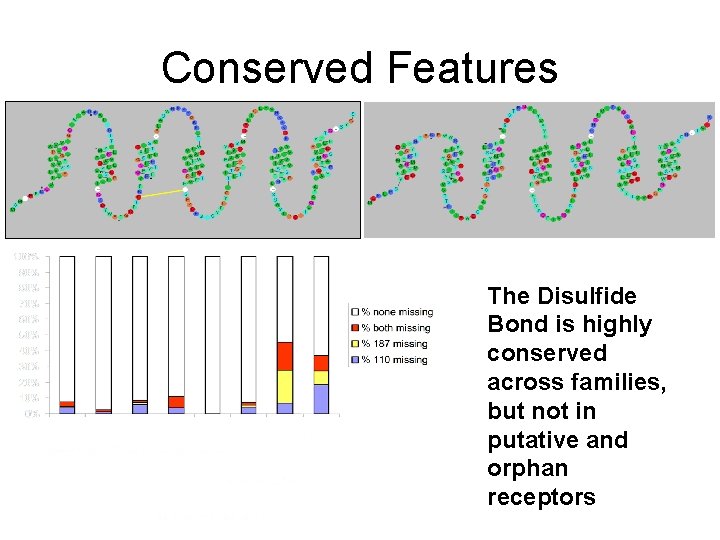
Conserved Features The Disulfide Bond is highly conserved across families, but not in putative and orphan receptors

Sequence Alignment when homology is low • Secondary structure prediction methods • Membrane protein prediction • Novel alignment method developed by us using phi/psi angles • Sequence conservation based on property conservation

Take home messages • Structural and functional effects of small changes in sequences • Conservation of structure despite large differences in sequences • Prediction of structural and functional effects using computational pharmacology to understand disease mechanisms and drug action with the goal of identifying targets and designing drugs against them – Example: Specific Structure of COX and of GPCR – Current hot topics: Complex interactions of proteins within their environment, differences between individuals

Play with homology models • www. cs. cmu. edu/~blmt/Seminar. Materials/COX 2 Modelling : Template structure : 1 PTH. pdb (cox 1 in ovis aries) query seq: sequence of 1 PXX. pdb (cox 2 in musculus) model generated using modeller: 2 cox. pdb COX 1 Modelling: Template structure : 1 PXX. pdb (cox 2 in musculus) query seq: sequence of 1 PTH. pdb (cox 1 in ovis aries) model generated using modeller: 1 cox. pdb • Rasmol is also in this directory, just click on the raswin icon to start program

A protein conformation has to satisfy three conditions: 1. Only stereochemically allowed conformations of all residues are acceptable (=avoid steric clashes). Model system: dialanine peptide Rotation of the polypeptide chain is permitted around the N-Calpha (angle Phi) and Calpha-C (angle Psi) bonds (except Proline) and the peptide bond (angle omega), which is either trans in most cases (omega=180 o) or cis (omega=0 o) in rare cases, i. e. at Proline residues. These angles define the backbone conformation, and specific conformations are allowed, as described by the Ramachandran plot. 2. The folded state must be energetically favorable The native state of globular protein is only 20 -60 k. J mol-1 (5 -15 kcal/mol) more stable than the denatured state. This is the equivalent of one or two water-water hydrogen bonds. It is unclear why this is the case, because the stability of proteins can be increased by adding stabilizing contacts. The main problem in achieving the native state is the loss of conformational freedom (entropy reduction), when going from many unfolded to a single folded conformation. This process is therefore thermodynamically unfavorable. Why does it still occur? Because the loss in entropy arising from conformational restriction is compensated by an increase in entropy arising from the hydrophobic effect. The fact that native protein structures are more stable than unfolded protein by 1 -2 H-bonds, means that 1 -2 unsatisfied H-bonds in a protein can make the native state unstable. 3. The folded state must be tightly packed. How tightly packed is the interior of a protein? In theory, relatively loose packing would ensure exclusion of water, since a <1. 4Å radius (=size of water molecule) hole is acceptable. However, attraction between atoms (van der Waals forces) cause closer packing than theoretically required by the hydrophobic effect alone. Thus, a protein is like a jigsaw puzzle, except that the pieces in a jigsaw puzzle are rigid, while the side chains in proteins are dynamic and can adopt many conformations. More details on requirements 2 and 3: The folded state must be energetically favorable and the folded state must be tightly packed. Terms used in the evaluation of the energy of a conformation (see page 253 in chapter 5, Ref_Lesk for equations): 1. Bond stretching 2. Bond angle bend 3. deviations from planarity and enforcement of correct chirality 4. Torsion angle 5. van der Waals interactions 6. Hydrogen bonds 7. Electrostatics 8. solvent => set of conformational energy potentials that fine tune these parameter sets "Potential functions" The potential functions satisfy necessary but not sufficient conditions for successful structure prediction. Multiple local minima cannot be distinguished reliably from the correct one on the basis of calculated conformational energies. (What does this mean in practice? How many possible structures are there as opposed to the real structures? How different are the structures? )
 Computational pharmacology
Computational pharmacology Cuhk assistant professor salary
Cuhk assistant professor salary Promotion from associate professor to professor
Promotion from associate professor to professor What is bioavailability
What is bioavailability Annual review of pharmacology and toxicology
Annual review of pharmacology and toxicology Factors affecting absorption of drug
Factors affecting absorption of drug Clinical pharmacology
Clinical pharmacology Pharmacology module
Pharmacology module Rate of elimination of drug
Rate of elimination of drug Slidetodoc
Slidetodoc Maintenance dose formula
Maintenance dose formula Bioavailability
Bioavailability Fundamentals of pharmacology for veterinary technicians
Fundamentals of pharmacology for veterinary technicians Ion trapping
Ion trapping Rationale meaning in pharmacology
Rationale meaning in pharmacology Pharmacology pay
Pharmacology pay Spremicides
Spremicides Tactcardia
Tactcardia Chapter 15 diagnostic procedures and pharmacology
Chapter 15 diagnostic procedures and pharmacology Fish pharmacology
Fish pharmacology Reverse pharmacology
Reverse pharmacology Dopamine blockers
Dopamine blockers Receptors in pharmacology
Receptors in pharmacology Basic & clinical pharmacology
Basic & clinical pharmacology First pass metabolism definition pharmacology
First pass metabolism definition pharmacology What is ion trapping in pharmacology
What is ion trapping in pharmacology Competitive antagonist
Competitive antagonist Mdi pharmacology
Mdi pharmacology Tachyphylaxis
Tachyphylaxis Analglesia
Analglesia What is pharmacology
What is pharmacology Pharmacology chapter 1
Pharmacology chapter 1 Pharmacology definition
Pharmacology definition Pharmacology for nurses: a pathophysiological approach
Pharmacology for nurses: a pathophysiological approach Pharmacology and venipuncture
Pharmacology and venipuncture Alpha 1 vs alpha 2 receptors
Alpha 1 vs alpha 2 receptors Pharmacology definition
Pharmacology definition Filtration pharmacology
Filtration pharmacology Define pharmacology
Define pharmacology First pass effect in pharmacology
First pass effect in pharmacology Basic & clinical pharmacology
Basic & clinical pharmacology Clinical pharmacology powered by clinicalkey
Clinical pharmacology powered by clinicalkey Metabolism of drug definition
Metabolism of drug definition Focus on pharmacology essentials for health professionals
Focus on pharmacology essentials for health professionals What is ion trapping in pharmacology
What is ion trapping in pharmacology Clinical pharmacology seminar
Clinical pharmacology seminar Therapeutic index
Therapeutic index Clinical pharmacology residency
Clinical pharmacology residency