Comments on Rare Variants Analyses Ryo Yamada Kyoto




















































- Slides: 103

Comments on Rare Variants Analyses Ryo Yamada Kyoto University 2012/08/27 IBC 2012@Kobe, Japan

Many difficulties to detect true signals in rare variant analyses Type 1 error, Power-control, Various Statistics with/without Weights, Data quality, Data load, Platform variations, Missing data, Permutations, Allele freq. , Rares with Commons, Replication, Neutral or not,

Many difficulties to detect true signals in rare variant analyses Type 1 error, Power-control, Various Statistics with/without Weights, Data quality, Data load, Platform variations, Missing data, Permutations, Allele freq. , Replication, Rares with Commons integrating all variations, So many. .

Many difficulties to detect true signals in rare variant analyses • My discussing points – Some problems in rare variant analyses are the unsolved problems in common variant analyses.

Many difficulties to detect true signals in rare variant analyses • My discussing points – Some problems in rare variant analyses are the unsolved problems in common variant analyses.

Many difficulties to detect true signals in rare variant analyses • My discussing points – Some problems in rare variant analyses are the unsolved problems in common variant analyses. – Changes in genetic studies along with rare variant analyses • Next-generation sequencing technologies-driven changes

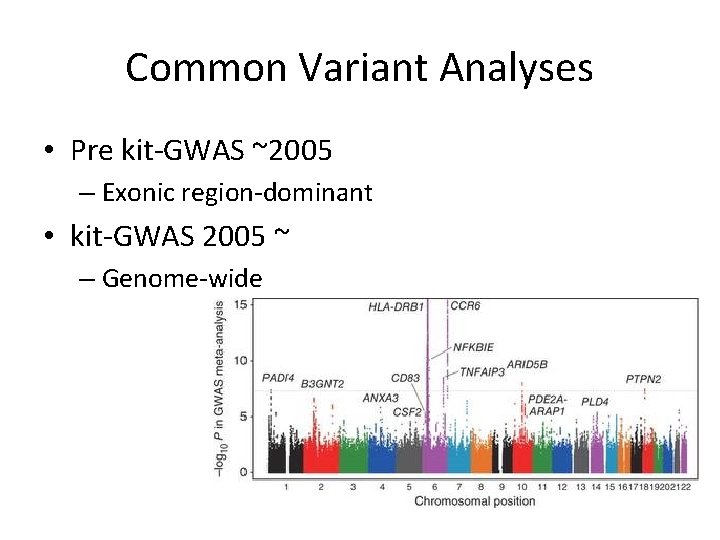
Common Variant Analyses • Pre kit-GWAS ~2005 – Exonic region-dominant • kit-GWAS 2005 ~ – Genome-wide

Common Variant Analyses • Pre kit-GWAS ~2005 – Exonic region-dominant • kit-GWAS 2005 ~ – Genome-wide Rare Variant Analyses • Exome • Whole-genome sequencing

Common Variant Analyses • Pre kit-GWAS ~2005 – Exonic region-dominant • kit-GWAS 2005 ~ – Genome-wide déjà vu Rare Variant Analyses • Exome • Whole-genome sequencing

What we can learn from SNP LD mapping • Hypothesis-free approach • All markers vs. 1 trait

What we can learn from SNP LD mapping • Hypothesis-free approach • All markers vs. 1 trait

What we can learn from SNP LD mapping • Hypothesis-free approach • All markers vs. 1 trait

Set of Hypotheses • Hypothesis-free approach • All markers vs. 1 trait • Almost all hypotheses are null. – They work as negative controls. – We obtain distribution under null hypothesis. • A few hypotheses are “positive”. • Pick up “outliers” from “null distribution” as positive signals

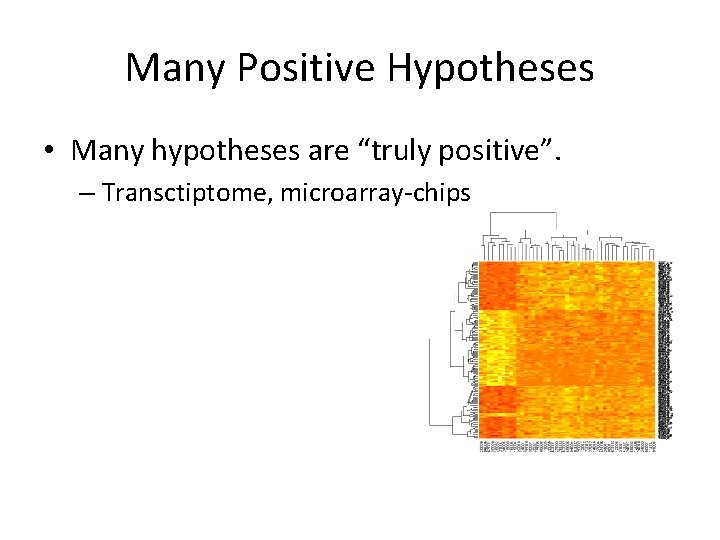
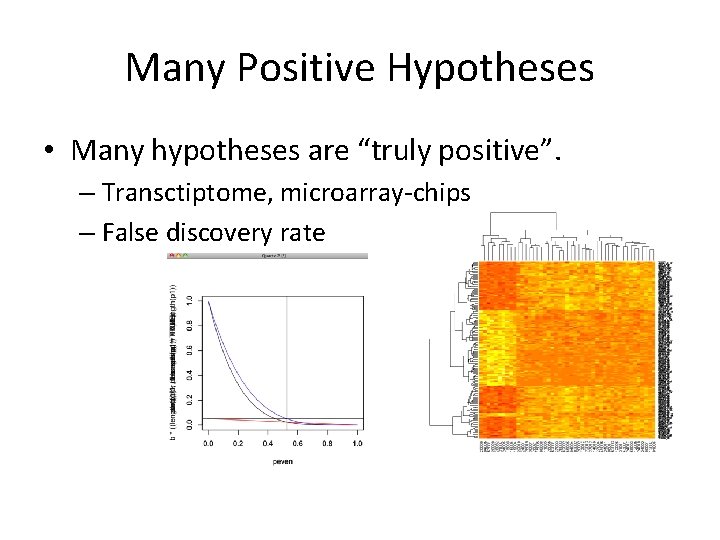
Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate

Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate

Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate

Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate • Different approach to multiple testings from GWAS

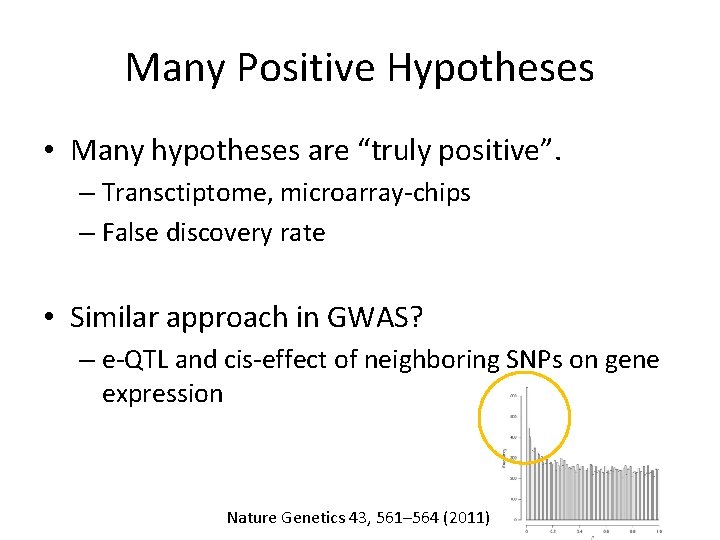
Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate • Similar approach to multiple testings in GWAS?

Many Positive Hypotheses • Many hypotheses are “truly positive”. – Transctiptome, microarray-chips – False discovery rate • Similar approach in GWAS? – e-QTL and cis-effect of neighboring SNPs on gene expression Nature Genetics 43, 561– 564 (2011)

Both are GWAS but different • Almost all hypotheses are null • Many positive hypotheses

Both are GWAS but different • Almost all hypotheses are null – All markers vs. 1 trait • Many positive hypotheses – All markers vs. genes (traits)

Both are GWAS but different • Almost all hypotheses are null – All markers vs. 1 trait • Many positive hypotheses – All markers vs. genes (traits) Many traits

Both are GWAS but different • Almost all hypotheses are null – All markers vs. 1 trait • Many positive hypotheses – All markers vs. genes (traits) Many traits Positives : Neighboring and cis effects Negatives : Remote or trans effects

Many Positive Hypotheses with Rare Variant Analyses

Many Positive Hypotheses with Rare Variant Analyses • Distribution of statistics is like what?

Many Positive Hypotheses with Rare Variant Analyses • Distribution of statistics is like what? • Strategy to detect signals should be cared.

Both are GWAS but different • Almost all hypotheses are null – All markers vs. 1 trait • Many Positive Hypotheses – All markers vs. genes (traits) Many traits Positives : Neighboring and cis effects Negatives : Remote or trans effects

Same GWAS but difference • Almost all hypotheses are null – All markers vs. 1 phenotype • Many Positive Hypotheses – All markers vs. genes (traits) Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Almost all hypotheses are null – All markers vs. 1 phenotype • Many Positive Hypotheses – All markers vs. cis-neighboring genes Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Biobank projects with multiple traits EHR (Electrical Health Record)-driven genetic • • Almost all hypotheses are null –studies All markers vs. 1 phenotype • – Very many phenotypes Many Positive Hypotheses Nature Reviews Genetics 12, 417 -428 (June 2011) – All markers vs. cis-neighboring genes Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Biobank projects with multiple traits EHR (Electrical Health Record)-driven genetic • • Almost all hypotheses are null –studies All markers vs. 1 phenotype • – Very many phenotypes Many Positive Hypotheses Nature Reviews Genetics 12, 417 -428 (June 2011) – All markers vs. cis-neighboring genes Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Physical / pathological conditions vary. Developmental stages vary. • • Almost all hypotheses are null • –Cell types/ tissue types/ organs vary. All markers vs. 1 phenotype • Many Positive Hypotheses – All markers vs. cis-neighboring genes Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Physical / pathological conditions vary. Developmental stages vary. • • Almost all hypotheses are null • –Cell types/ tissue types/ organs vary. All markers vs. 1 phenotype • Many Positive Hypotheses – All markers vs. cis-neighboring genes Many traits Positives : Neighboring Negatives : Trans / Remote genes

Where are many traits? Same GWAS but difference • Physical / pathological conditions vary. • Developmental stages vary. • Cell types/ tissue types/ organs vary. Intra-individual diversity Many traits Positives : Neighboring Negatives : Trans / Remote genes

Same GWAS but difference Many traits

Same GWAS but difference ts oin p w ie ge v Chan Many traits

Same GWAS but difference Many genotypes
Where are many genotypes? Same GWAS but difference Many genotypes

Where are many genotypes? Same GWAS but difference • Physical / pathological conditions vary. • Developmental stages vary. • Cell types/ tissue types/ organs vary. Many genotypes

Where are many genotypes? Same GWAS but difference • Physical / pathological conditions vary. • Developmental stages vary. • Cell types/ tissue types/ organs vary. in a g A Intra-individual diversity Many genotypes

Where are many genotypes? Same GWAS but difference • Physical / pathological conditions vary. • Developmental stages vary. • Cell types/ tissue types/ organs vary. Intra-individual diversity Many genotypes Next generation sequencing technology makes these possible.

Changes by Next Generation Sequencing Technologies • Individual cell-sequencing detects – Hereditary • Variants from parents to offsprings – Non-hereditary but genetic • de novo mutations in gamates • Somatic mutations in fetus • Somatic mutations after birth

Changes by Next Generation Sequencing Technologies • Individual cell-sequencing detects – Hereditary rth bi e r fo • Variants from parents to offsprings Be – Non-hereditary but genetic • de novo mutations in gamates • Somatic mutations in fetus • Somatic mutations after birth A bi r e ft rth

Changes by Next Generation Sequencing Technologies • Individual cell-sequencing detects – Hereditary rth bi e r fo • Variants from parents to offsprings Be – Non-hereditary but genetic • de novo mutations in gamates • Somatic mutations in fetus • Somatic mutations after birth A bi r e ft rth Hereditary ~ Genetic ~ Somatic Discriminations are becoming vague.


“Collapsing methods”

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure • Information on functionality of variants might be used when collapse to modify the “measure”.

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure • Is this problem NEW to rare variant analyses?

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure • Is this problem NEW to rare variant analyses?

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional measure • Is this problem NEW to rare variant analyses? – NO

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be tested easily; one-dimensional value • Is this problem NEW to rare variant analyses? – We do NOT have adequate solutions for patterns of common variants. Genetics September 2007 vol. 177 no. 1 535 -547

“Collapsing methods” • Patterns of possession of rare variants vary with phenotypes. • “Collapsing methods” make the patterns into a style that can be test easily; one-dimensional value • Is this problem NEW to rare variant analyses? – We do NOT have adequate solutions for patterns of common variants. How to handle patterns of variants, BOTH common and rare, should be solved.

Patterns of variants

Patterns of variants • Rephrase as “Genetic heterogeneity PER GENES”

Notes on Genetic Heterogeneity PER GENES • Genes vary in their size.

Notes on Genetic Heterogeneity PER GENES • Genes vary in their size.

Notes on Genetic Heterogeneity PER GENES • Genes vary in their size. • Number of variants vary among genes.

Notes on Genetic Heterogeneity PER GENES • Genes vary in their size. • Number of variants vary among genes. • Allele frequencies vary among genes.

Notes on Genetic Heterogeneity PER GENES • • Genes vary in their size. Number of variants vary among genes. Allele frequencies vary among genes. Haplotype frequencies vary among genes.

Notes on Genetic Heterogeneity PER GENES • • • Genes vary in their size. Number of variants vary among genes. Allele frequencies vary among genes. Haplotype frequencies vary among genes. Functional variations due to genetic variants vary among genes.

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS – Hypothesis-free approach, targetting genes evenly

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS – Hypothesis-free approach, targetting genes evenly – But. . .

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS – Hypothesis-free approach, targetting genes evenly – But. . . – All genes are studied UNEVENLY.

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS – Hypothesis-free approach, targetting genes evenly – But. . . – All genes are studied UNEVENLY. • Genetic heterogeneity PER GENES affects on sensitivity and specificity • HLA region and its extreme heterogeneity. . .

Unsolved Issues in Common Variant-GWAS on Genetic Heterogeneity PER GENES • GWAS – Hypothesis-free approach, targetting genes evenly – But. . . – All genes are studied UNEVENLY. • Genetic heterogeneity PER GENES affects on sensitivity and specificity • HLA region and its extreme heterogeneity. . . Am J Hum Genet. 2005 April; 76(4): 634– 646.

UNEVENNESS Genetic Importance • GWAS identifies genes of “Genetic Importance” that – are Important for a phenotype, – have functional variants.

UNEVENNESS Genetic Importance • GWAS identifies genes of “Genetic Importance” that – are Important for a phenotype, – have functional variants.

UNEVENNESS Genetic Importance • GWAS identifies genes of “Genetic Importance” that – are Important for a phenotype, – have functional variants.

UNEVENNESS Genetic Importance • GWAS identifies genes of “Genetic Importance” that – are Important for a phenotype, – have functional variants.

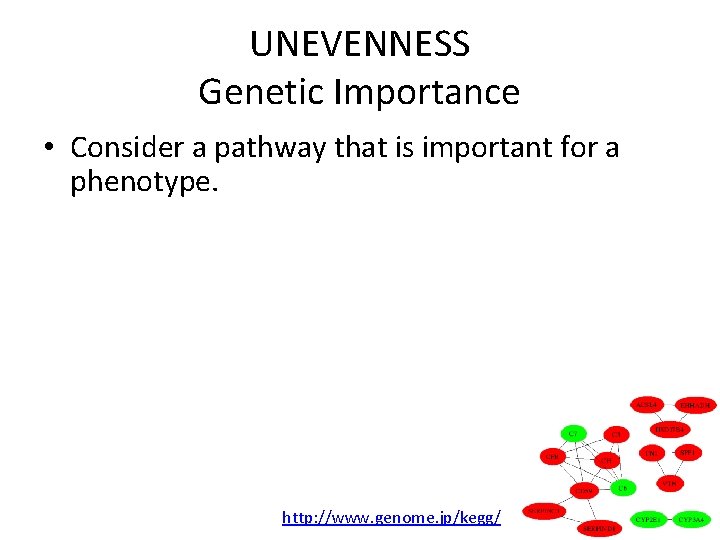
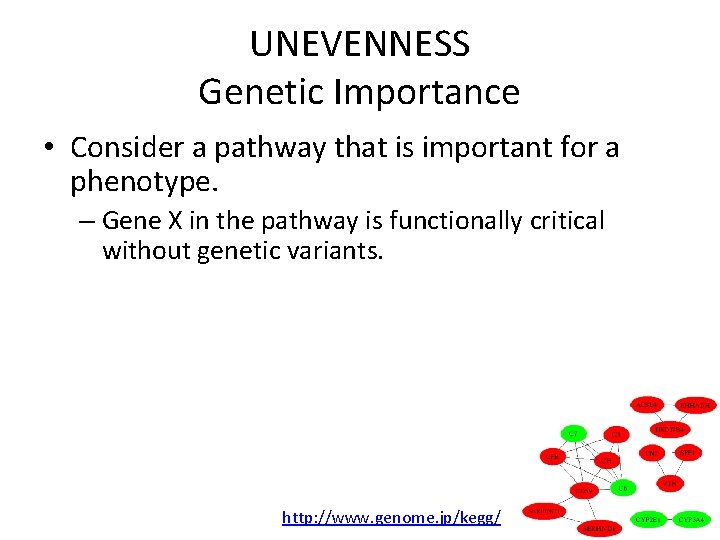
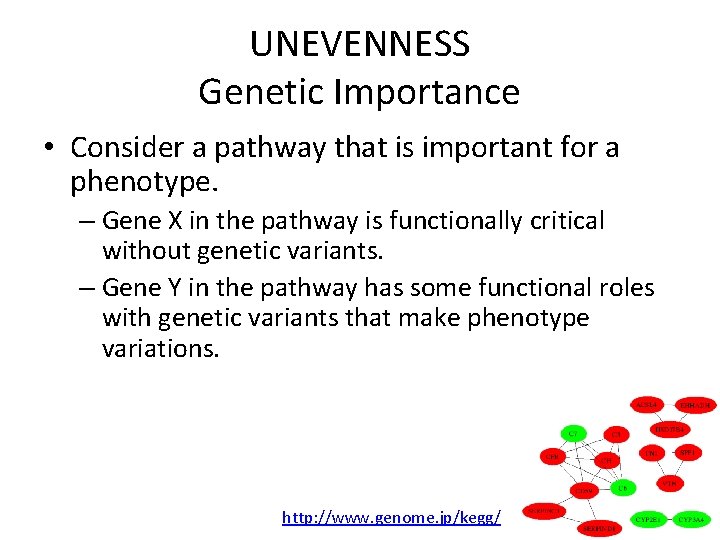
UNEVENNESS Genetic Importance • Consider a pathway that is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional roles with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important. http: //www. genome. jp/kegg/

UNEVENNESS Genetic Importance • Consider a pathway that is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional roles with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important. http: //www. genome. jp/kegg/

UNEVENNESS Genetic Importance • Consider a pathway that is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional roles with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important. http: //www. genome. jp/kegg/

UNEVENNESS Genetic Importance • Consider a pathway that is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional roles with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important.

UNEVENNESS Genetic Importance • Consider a pathway that is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional roles with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important.

UNEVENNESS Genetic Importance • Consider a pathway is important for a phenotype. – Gene X in the pathway is functionally critical without genetic variants. – Gene Y in the pathway has some functional role with genetic variants that make phenotype variations. • Gene X is “Functionally” important. • Gene Y is “Genetically” important. Genetic studies work only on “genetic” importance.

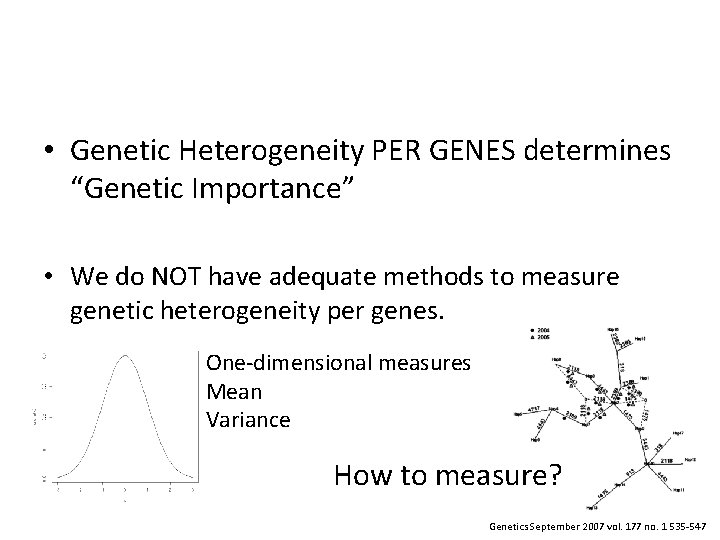
• Genetic Heterogeneity PER GENES determines “Genetic Importance” • We do NOT have adequate methods to measure genetic heterogeneity per genes.

• Genetic Heterogeneity PER GENES determines “Genetic Importance” • But, . . .

• Genetic Heterogeneity PER GENES determines “Genetic Importance” • We do NOT have adequate methods to measure genetic heterogeneity per genes.

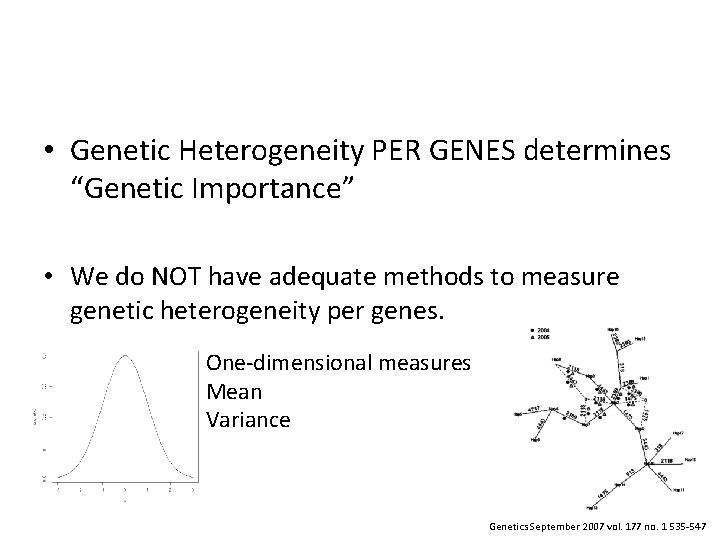
• Genetic Heterogeneity PER GENES determines “Genetic Importance” • We do NOT have adequate methods to measure genetic heterogeneity per genes. One-dimensional measures Mean Variance

• Genetic Heterogeneity PER GENES determines “Genetic Importance” • We do NOT have adequate methods to measure genetic heterogeneity per genes. One-dimensional measures Mean Variance Genetics September 2007 vol. 177 no. 1 535 -547

• Genetic Heterogeneity PER GENES determines “Genetic Importance” • We do NOT have adequate methods to measure genetic heterogeneity per genes. One-dimensional measures Mean Variance How to measure? Genetics September 2007 vol. 177 no. 1 535 -547
Genetic Heterogeneity PER GENES For Species Or For Individuals

Genetic Heterogeneity PER GENES For Populations Or For Individuals

Genetic Heterogeneity PER GENES For Populations Or For Individuals • Heterogeneity in an absolute scale

Genetic Heterogeneity PER GENES For Populations Or For Individuals • Heterogeneity in an absolute scale

Genetic Heterogeneity PER GENES For Populations Or For Individuals • Heterogeneity in an absolute scale • Heterogeneity in a relative scale inside of individuals

Genetic Heterogeneity PER GENES For Populations Or For Individuals • Heterogeneity in an absolute scale • Heterogeneity in a relative scale inside of individuals y t i e n e g o r e t e h c i t e n e g l u d i v i d n i a r t In

Intra-individul Genetic Heterogeneity • They are the source of phenotypic heterogeneity.

Intra-individul Genetic Heterogeneity • They are the source of phenotypic heterogeneity. • Individuals have to show phenotypic variations that are the basics of living creatures.

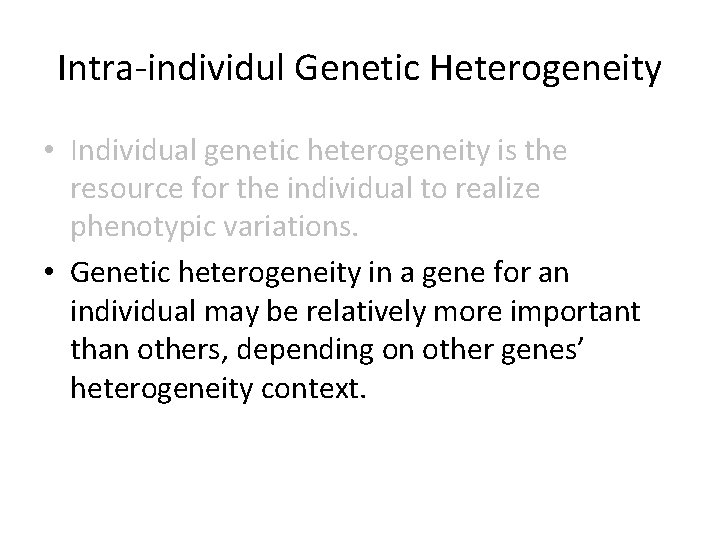
Intra-individul Genetic Heterogeneity • They are the source of phenotypic heterogeneity. • Individuals have to show phenotypic variations that are the basics of living creatures. • Individual genetic heterogeneity is the resource for the individual to realize phenotypic variations.

Intra-individul Genetic Heterogeneity • Individual genetic heterogeneity is the resource for the individual to realize phenotypic variations. • Genetic heterogeneity in a gene for an individual may be relatively more important than others, depending on other genes’ heterogeneity context.

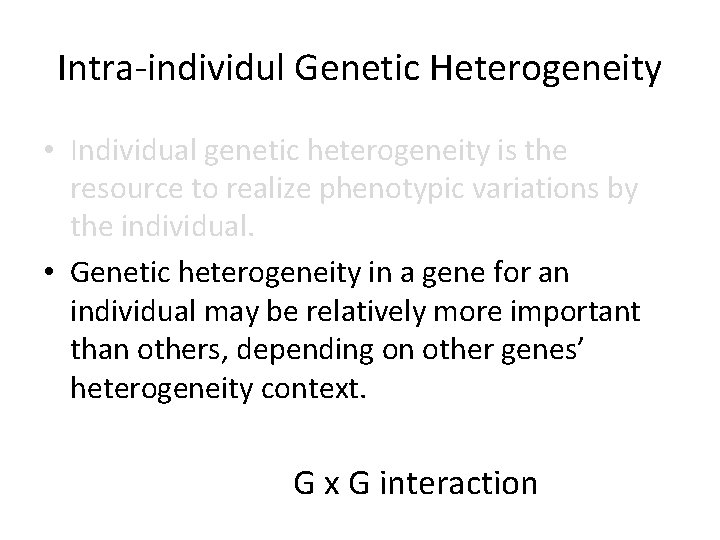
Intra-individul Genetic Heterogeneity • Individual genetic heterogeneity is the resource to realize phenotypic variations by the individual. • Genetic heterogeneity in a gene for an individual may be relatively more important than others, depending on other genes’ heterogeneity context. G x G interaction

G x G with rare variants • G x G approach in general – Gene set in a functional pathway – Problems with too many combinations • Same both for common variants and rare variants – Many and scarce variants might G x G approach less realistic. – Scarce but Many variants could open new ways to G x G approach ? ?

G x G with rare variants • G x G approach in general – Gene set in a functional pathway – Problems with too many combinations • Same both for common variants and rare variants – Many and scarce variants might G x G approach less realistic. – Scarce but Many variants could open new ways to G x G approach ? ?

G x G with rare variants • G x G approach in general – Gene set in a functional pathway – Problems with too many combinations • Same both for common variants and rare variants – Many and scarce variants might make G x G approach less realistic. – Scarce but Many variants could open new ways to G x G approach ? ?

G x G with rare variants • G x G approach in general – Gene set in a functional pathway – Problems with too many combinations • Same both for common variants and rare variants – Many and scarce variants might make G x G approach less realistic. – Scarce but Many variants could open new ways to G x G approach with new measures? ? ?

Summary • Next generation sequencing enabled rare variant analyses. • Use what next generation sequencing brought us. – Expansion of phenotypes and genotypes – Set of hypotheses and multiple testing – Measures of genetic heterogeneity – Intra-individual heterogeneities

Thank you

Thank you Announcement Symposium and Data Analysis Hands-on Seminar Jan. 16 – Jan. 19, 2013 @ Kyoto University, Kyoto, Japan Organized by Marc Lathrop @ CEPH Jurg Ott @ Rockefeller Univ. Fumihiko Matsuda @ Kyoto Univ. Details will be announced shortly. If you are interested in this seminar or need updated timely for the details or registration, e-mail to: ryamada@genome. med. kyoto-u. ac. jp (Ryo Yamada, Kyoto Univ. )