Prioritizing somatic variants Approaches to identifying key variants
![[Ferreiro et al. , PNAS (’ 07)] 18 - Lectures. Gerstein. Lab. org What [Ferreiro et al. , PNAS (’ 07)] 18 - Lectures. Gerstein. Lab. org What](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-18.jpg)
![Lectures. Gerstein. Lab. org 19 - [Kumar et al. NAR (2016)] Workflow for evaluating Lectures. Gerstein. Lab. org 19 - [Kumar et al. NAR (2016)] Workflow for evaluating](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-19.jpg)
![[Lochovsky et al. NAR (’ 15)] 31 - Lectures. Gerstein. Lab. org Cancer Somatic [Lochovsky et al. NAR (’ 15)] 31 - Lectures. Gerstein. Lab. org Cancer Somatic](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-31.jpg)
![mr. TADFinder: [Yan et al. , PLOS Comp. Bio. (‘ 17)] 33 - Lectures. mr. TADFinder: [Yan et al. , PLOS Comp. Bio. (‘ 17)] 33 - Lectures.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-33.jpg)
![[Lochovsky et al. under review] 36 - Lectures. Gerstein. Lab. org MOAT-a: Annotation-based permutation [Lochovsky et al. under review] 36 - Lectures. Gerstein. Lab. org MOAT-a: Annotation-based permutation](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-36.jpg)
![MOAT-v: Variant-based Permutation [Lochovsky et al. under review] 37 - Lectures. Gerstein. Lab. org MOAT-v: Variant-based Permutation [Lochovsky et al. under review] 37 - Lectures. Gerstein. Lab. org](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-37.jpg)
![[Kumar & Gerstein, Nature ('17)] Lectures. Gerstein. Lab. org Better annotation or large number [Kumar & Gerstein, Nature ('17)] Lectures. Gerstein. Lab. org Better annotation or large number](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-45.jpg)
![[Li et al. PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org supported by expr. [Li et al. PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org supported by expr.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-48.jpg)
![[S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. [S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-51.jpg)
![[S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. [S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-52.jpg)
![53 - Lectures. Gerstein. Lab. org [Li et al. , PLOS Genetics (‘ 17)] 53 - Lectures. Gerstein. Lab. org [Li et al. , PLOS Genetics (‘ 17)]](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-53.jpg)

- Slides: 57

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence Mark Gerstein Yale & “tweetable” (via @markgerstein). See last slide for more info. 1 - downloadable from Lectures. Gerstein. Lab. org Slides freely

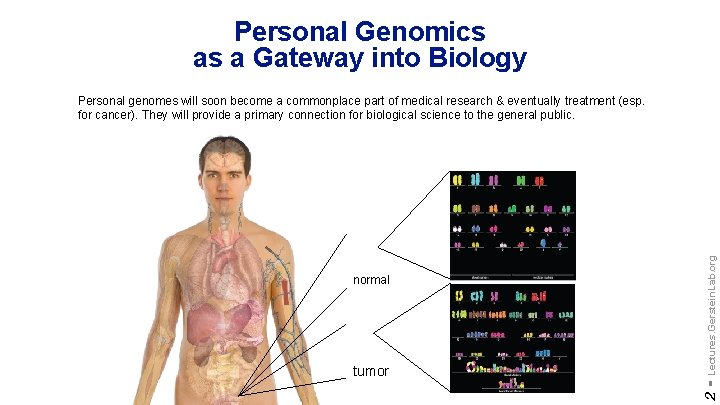
Personal Genomics as a Gateway into Biology tumor 2 - normal Lectures. Gerstein. Lab. org Personal genomes will soon become a commonplace part of medical research & eventually treatment (esp. for cancer). They will provide a primary connection for biological science to the general public.

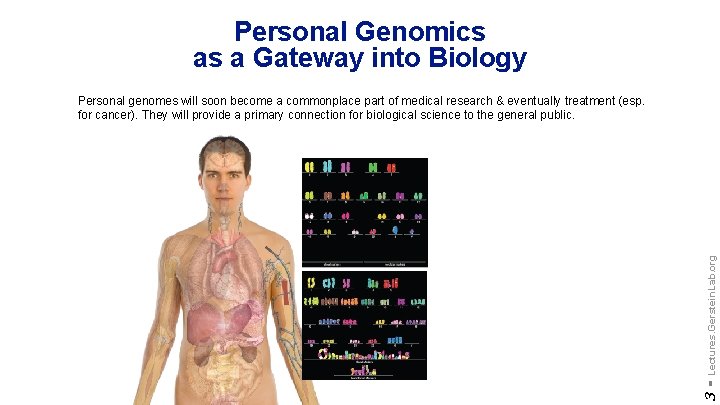
Personal Genomics as a Gateway into Biology 3 - Lectures. Gerstein. Lab. org Personal genomes will soon become a commonplace part of medical research & eventually treatment (esp. for cancer). They will provide a primary connection for biological science to the general public.

4 - Lectures. Gerstein. Lab. org

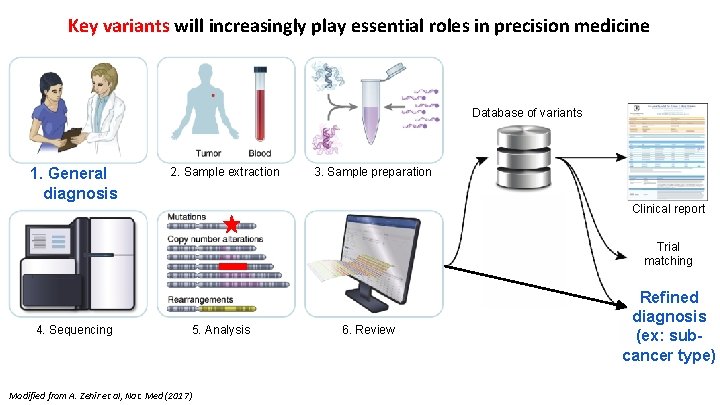
Key variants will increasingly play essential roles in precision medicine Database of variants 1. General diagnosis 2. Sample extraction 3. Sample preparation Trial matching Modified from A. Zehir et al, Nat. Med (2017) 5. Analysis 6. Review Refined diagnosis (ex: subcancer type) 5 - 4. Sequencing Lectures. Gerstein. Lab. org Clinical report

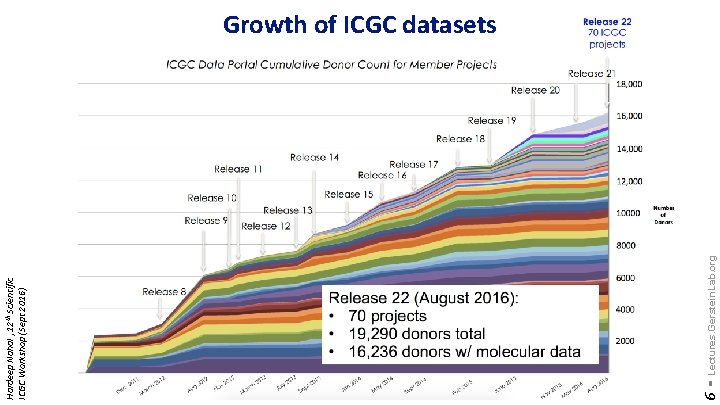
6 - Lectures. Gerstein. Lab. org Hardeep Nahal , 12 th Scientific ICGC Workshop (Sept 2016) Growth of ICGC datasets

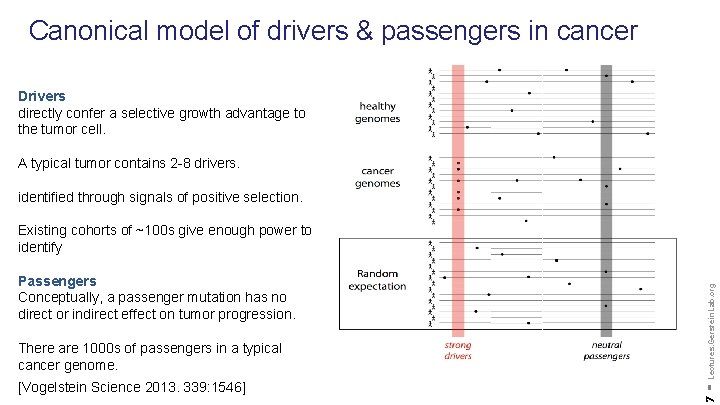
Canonical model of drivers & passengers in cancer Drivers directly confer a selective growth advantage to the tumor cell. A typical tumor contains 2 -8 drivers. identified through signals of positive selection. There are 1000 s of passengers in a typical cancer genome. [Vogelstein Science 2013. 339: 1546] 7 Passengers Conceptually, a passenger mutation has no direct or indirect effect on tumor progression. - Lectures. Gerstein. Lab. org Existing cohorts of ~100 s give enough power to identify

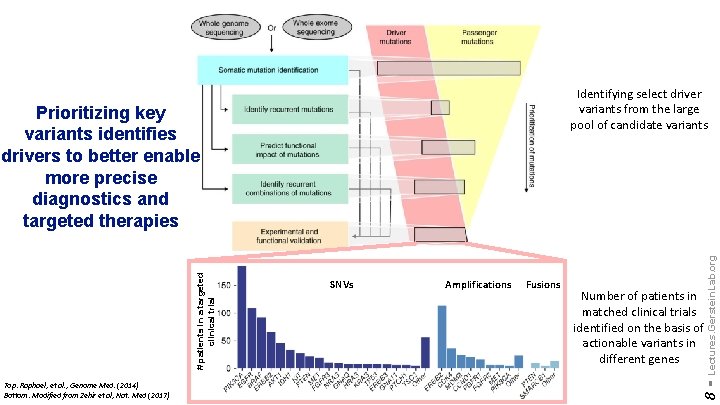
Top: Raphael, et al. , Genome Med. (2014) Bottom: Modified from Zehir et al, Nat. Med (2017) SNVs Amplifications Fusions Number of patients in matched clinical trials identified on the basis of actionable variants in different genes 8 - # patients in a targeted clinical trial Prioritizing key variants identifies drivers to better enable more precise diagnostics and targeted therapies Lectures. Gerstein. Lab. org Identifying select driver variants from the large pool of candidate variants

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 9 - • Introduction

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 10 - • Introduction

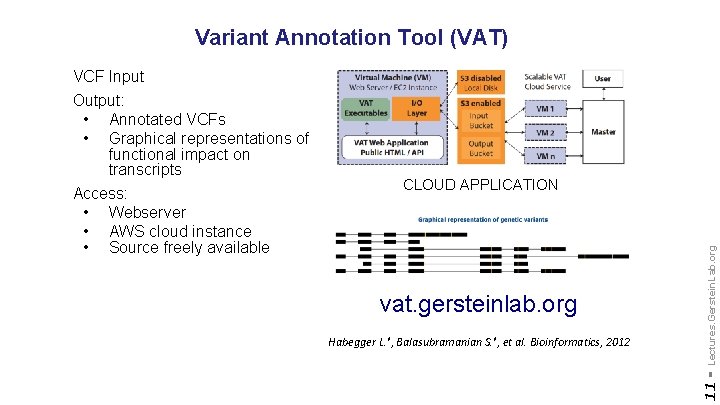
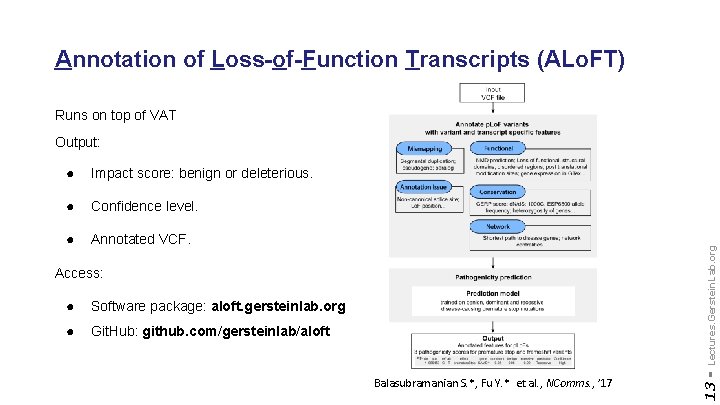
Variant Annotation Tool (VAT) vat. gersteinlab. org Habegger L. *, Balasubramanian S. *, et al. Bioinformatics, 2012 Lectures. Gerstein. Lab. org CLOUD APPLICATION 11 - VCF Input Output: • Annotated VCFs • Graphical representations of functional impact on transcripts Access: • Webserver • AWS cloud instance • Source freely available

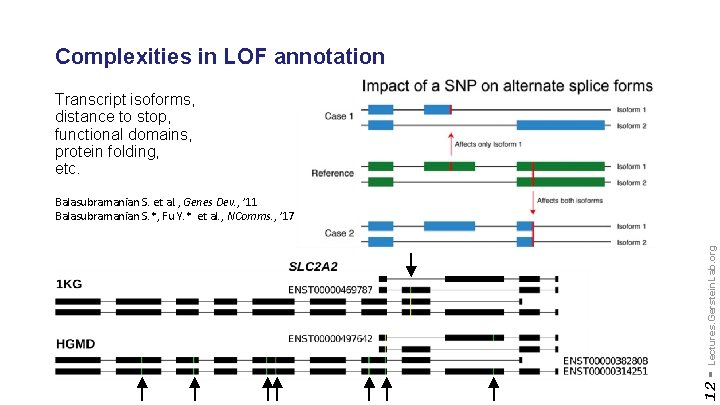
Complexities in LOF annotation Transcript isoforms, distance to stop, functional domains, protein folding, etc. 12 - Lectures. Gerstein. Lab. org Balasubramanian S. et al. , Genes Dev. , ’ 11 Balasubramanian S. *, Fu Y. * et al. , NComms. , ’ 17

Annotation of Loss-of-Function Transcripts (ALo. FT) Runs on top of VAT Impact score: benign or deleterious. ● Confidence level. ● Annotated VCF. Access: ● Software package: aloft. gersteinlab. org ● Git. Hub: github. com/gersteinlab/aloft Balasubramanian S. *, Fu Y. * et al. , NComms. , ’ 17 13 - ● Lectures. Gerstein. Lab. org Output:

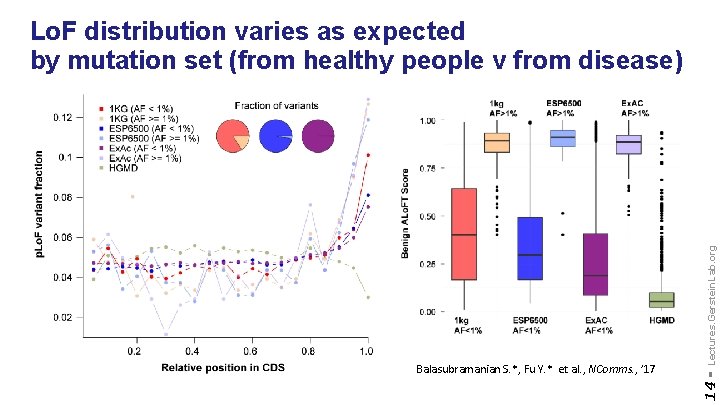
14 - Balasubramanian S. *, Fu Y. * et al. , NComms. , ’ 17 Lectures. Gerstein. Lab. org Lo. F distribution varies as expected by mutation set (from healthy people v from disease)

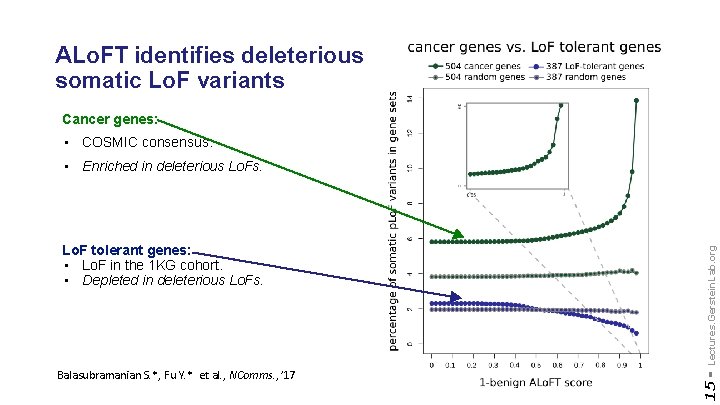
ALo. FT identifies deleterious somatic Lo. F variants Cancer genes: • COSMIC consensus. Balasubramanian S. *, Fu Y. * et al. , NComms. , ’ 17 15 - Lo. F tolerant genes: • Lo. F in the 1 KG cohort. • Depleted in deleterious Lo. Fs. Lectures. Gerstein. Lab. org • Enriched in deleterious Lo. Fs.

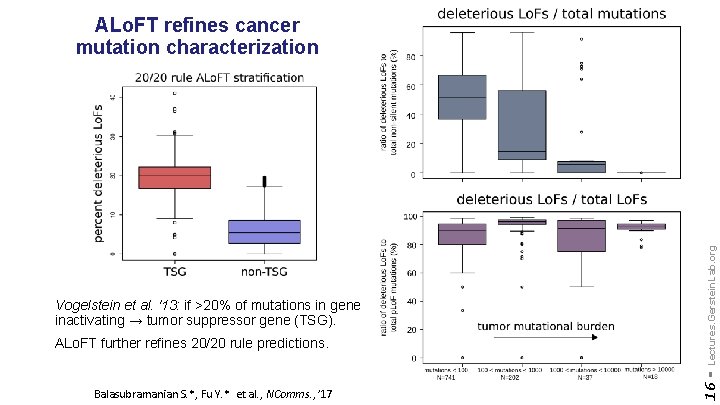
ALo. FT further refines 20/20 rule predictions. Balasubramanian S. *, Fu Y. * et al. , NComms. , ’ 17 16 - Vogelstein et al. '13: if >20% of mutations in gene inactivating → tumor suppressor gene (TSG). Lectures. Gerstein. Lab. org ALo. FT refines cancer mutation characterization

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 17 - • Introduction
![Ferreiro et al PNAS 07 18 Lectures Gerstein Lab org What [Ferreiro et al. , PNAS (’ 07)] 18 - Lectures. Gerstein. Lab. org What](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-18.jpg)
[Ferreiro et al. , PNAS (’ 07)] 18 - Lectures. Gerstein. Lab. org What is localized frustration?
![Lectures Gerstein Lab org 19 Kumar et al NAR 2016 Workflow for evaluating Lectures. Gerstein. Lab. org 19 - [Kumar et al. NAR (2016)] Workflow for evaluating](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-19.jpg)
Lectures. Gerstein. Lab. org 19 - [Kumar et al. NAR (2016)] Workflow for evaluating localized frustration changes (∆F)

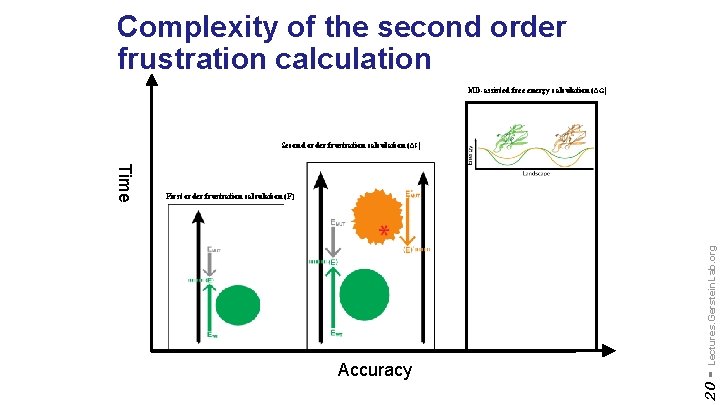
Complexity of the second order frustration calculation MD-assisted free energy calculation (∆G) Second order frustration calculation (∆F) Lectures. Gerstein. Lab. org Accuracy 20 - Time First order frustration calculation (F)

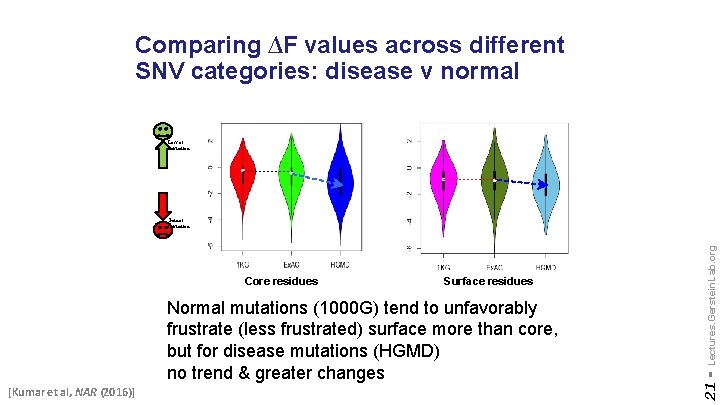
Comparing ∆F values across different SNV categories: disease v normal Loss of frustration Surface residues Normal mutations (1000 G) tend to unfavorably frustrate (less frustrated) surface more than core, but for disease mutations (HGMD) no trend & greater changes [Kumar et al, NAR (2016)] 21 - Core residues Lectures. Gerstein. Lab. org Gain of frustration

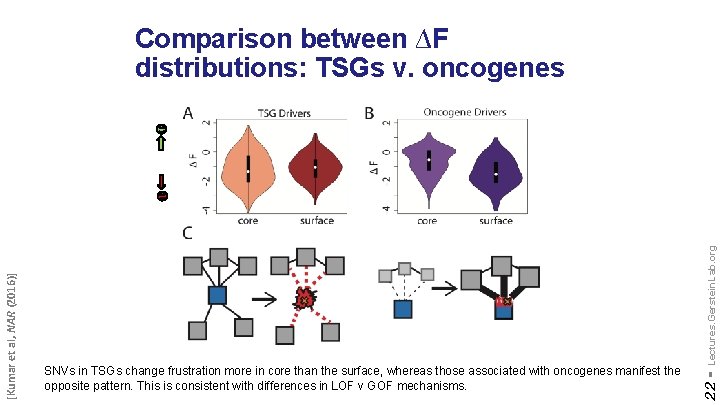
Lectures. Gerstein. Lab. org SNVs in TSGs change frustration more in core than the surface, whereas those associated with oncogenes manifest the opposite pattern. This is consistent with differences in LOF v GOF mechanisms. 22 - [Kumar et al, NAR (2016)] Comparison between ∆F distributions: TSGs v. oncogenes

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 23 - • Introduction

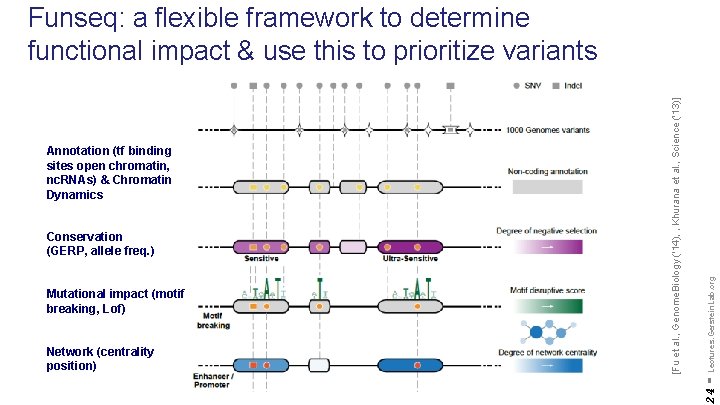
Mutational impact (motif breaking, Lof) Network (centrality position) - Lectures. Gerstein. Lab. org Conservation (GERP, allele freq. ) 24 Annotation (tf binding sites open chromatin, nc. RNAs) & Chromatin Dynamics [Fu et al. , Genome. Biology ('14), , Khurana et al. , Science ('13)] Funseq: a flexible framework to determine functional impact & use this to prioritize variants

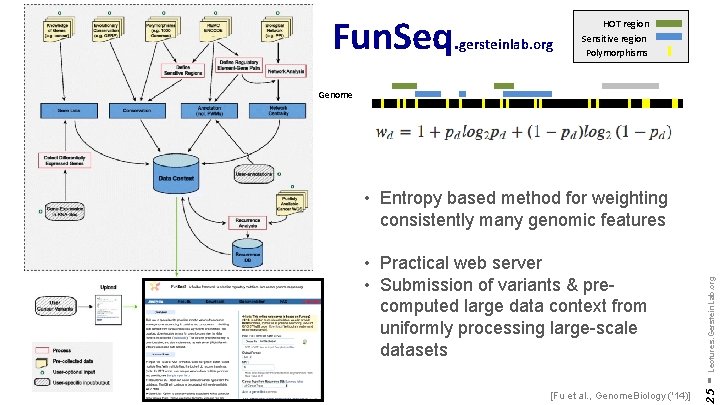
Fun. Seq. gersteinlab. org HOT region Sensitive region Polymorphisms Genome [Fu et al. , Genome. Biology ('14)] 25 • Practical web server • Submission of variants & precomputed large data context from uniformly processing large-scale datasets - Lectures. Gerstein. Lab. org • Entropy based method for weighting consistently many genomic features

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 26 - • Introduction

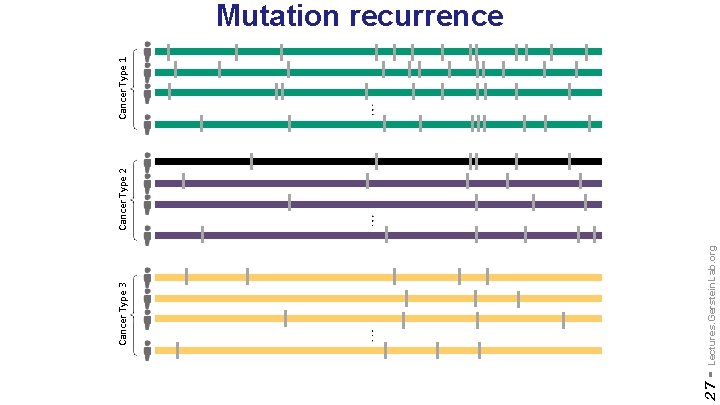
27 - Lectures. Gerstein. Lab. org Cancer Type 3 Cancer Type 2 Cancer Type 1 Mutation recurrence

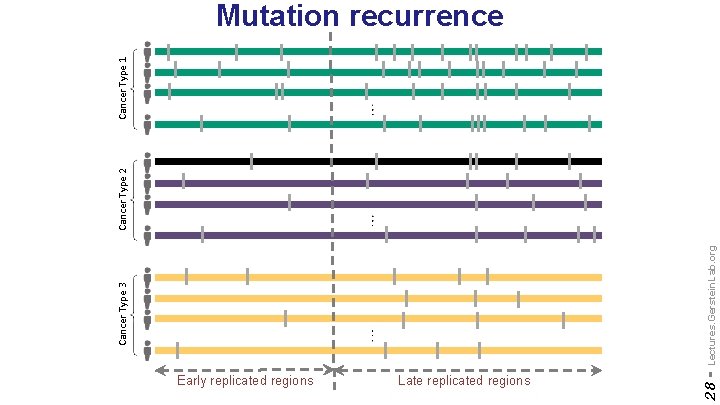
Early replicated regions Late replicated regions 28 - Lectures. Gerstein. Lab. org Cancer Type 3 Cancer Type 2 Cancer Type 1 Mutation recurrence

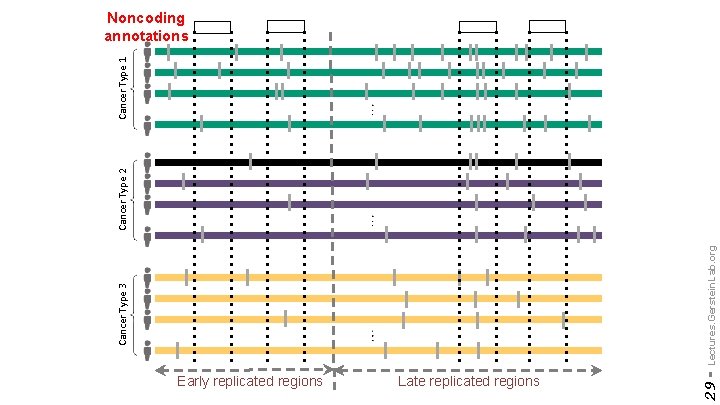
Early replicated regions Late replicated regions 29 - Cancer Type 3 Lectures. Gerstein. Lab. org Cancer Type 2 Cancer Type 1 Noncoding annotations

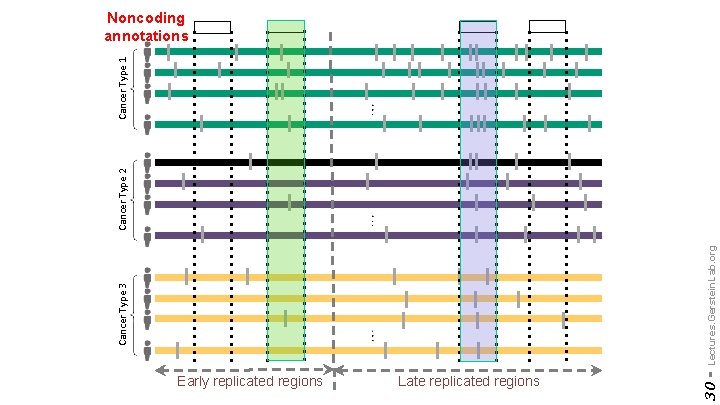
Early replicated regions Late replicated regions 30 - Cancer Type 3 Lectures. Gerstein. Lab. org Cancer Type 2 Cancer Type 1 Noncoding annotations
![Lochovsky et al NAR 15 31 Lectures Gerstein Lab org Cancer Somatic [Lochovsky et al. NAR (’ 15)] 31 - Lectures. Gerstein. Lab. org Cancer Somatic](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-31.jpg)
[Lochovsky et al. NAR (’ 15)] 31 - Lectures. Gerstein. Lab. org Cancer Somatic Mutational Heterogeneity, across cancer types, samples & regions

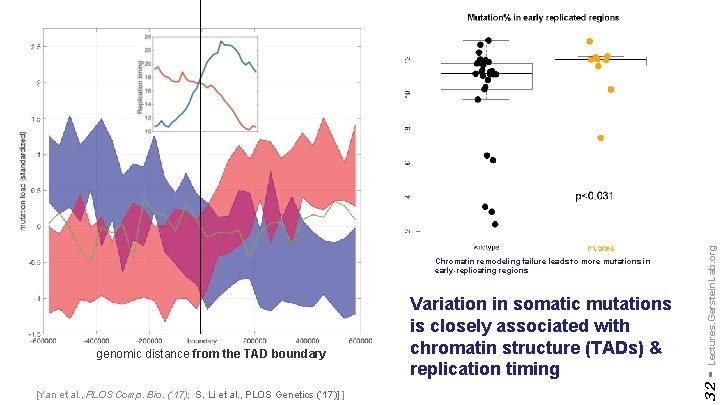
[Yan et al. , PLOS Comp. Bio. (‘ 17); S. Li et al. , PLOS Genetics (‘ 17)] ] Lectures. Gerstein. Lab. org genomic distance from the TAD boundary Variation in somatic mutations is closely associated with chromatin structure (TADs) & replication timing 32 - Chromatin remodeling failure leads to more mutations in early-replicating regions
![mr TADFinder Yan et al PLOS Comp Bio 17 33 Lectures mr. TADFinder: [Yan et al. , PLOS Comp. Bio. (‘ 17)] 33 - Lectures.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-33.jpg)
mr. TADFinder: [Yan et al. , PLOS Comp. Bio. (‘ 17)] 33 - Lectures. Gerstein. Lab. org Identifying TADs at multiple resolutions by maximizing modularity vs appropriate null

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 34 - • Introduction

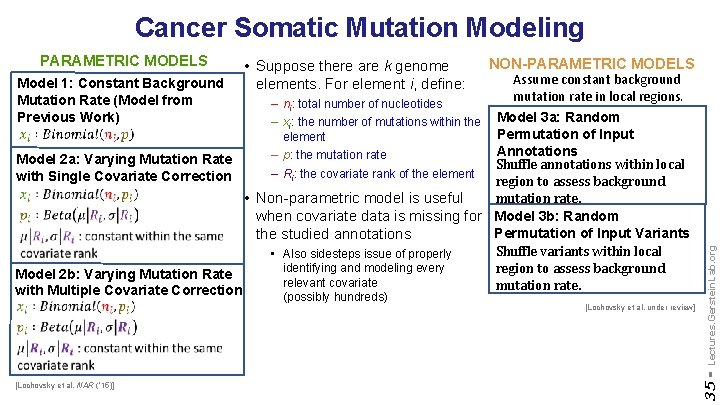
Cancer Somatic Mutation Modeling – ni: total number of nucleotides – xi: the number of mutations within the element – p: the mutation rate – Ri: the covariate rank of the element NON-PARAMETRIC MODELS Assume constant background mutation rate in local regions. Model 3 a: Random Permutation of Input Annotations Model 2 a: Varying Mutation Rate Shuffle annotations within local with Single Covariate Correction region to assess background mutation rate. • Non-parametric model is useful when covariate data is missing for Model 3 b: Random Permutation of Input Variants the studied annotations Shuffle variants within local • Also sidesteps issue of properly identifying and modeling every region to assess background Model 2 b: Varying Mutation Rate relevant covariate mutation rate. with Multiple Covariate Correction (possibly hundreds) [Lochovsky et al. under review] [Lochovsky et al. NAR (’ 15)] Lectures. Gerstein. Lab. org Model 1: Constant Background Mutation Rate (Model from Previous Work) • Suppose there are k genome elements. For element i, define: 35 - PARAMETRIC MODELS
![Lochovsky et al under review 36 Lectures Gerstein Lab org MOATa Annotationbased permutation [Lochovsky et al. under review] 36 - Lectures. Gerstein. Lab. org MOAT-a: Annotation-based permutation](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-36.jpg)
[Lochovsky et al. under review] 36 - Lectures. Gerstein. Lab. org MOAT-a: Annotation-based permutation
![MOATv Variantbased Permutation Lochovsky et al under review 37 Lectures Gerstein Lab org MOAT-v: Variant-based Permutation [Lochovsky et al. under review] 37 - Lectures. Gerstein. Lab. org](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-37.jpg)
MOAT-v: Variant-based Permutation [Lochovsky et al. under review] 37 - Lectures. Gerstein. Lab. org Can preserve tri-nt context in shuffle

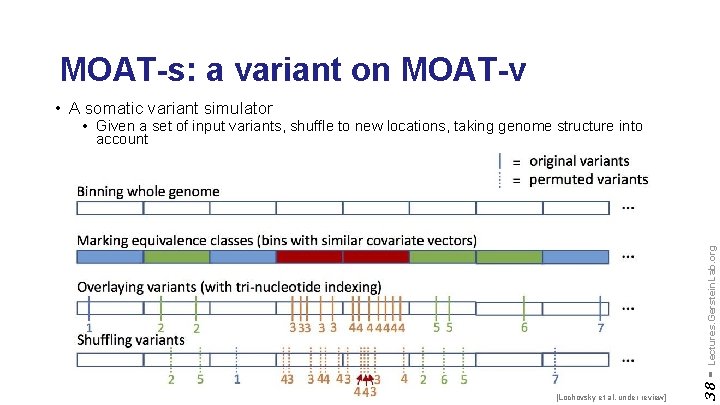
MOAT-s: a variant on MOAT-v • A somatic variant simulator [Lochovsky et al. under review] 38 - Lectures. Gerstein. Lab. org • Given a set of input variants, shuffle to new locations, taking genome structure into account

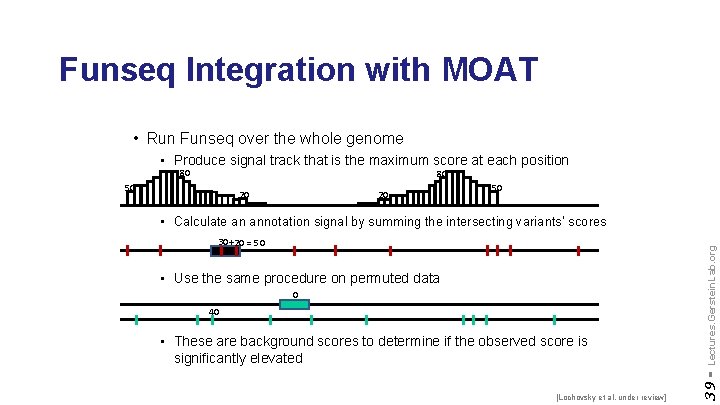
Funseq Integration with MOAT • Run Funseq over the whole genome • Produce signal track that is the maximum score at each position 80 80 50 20 20 50 • Use the same procedure on permuted data 0 40 • These are background scores to determine if the observed score is significantly elevated [Lochovsky et al. under review] 39 - 30 + 20 = 50 Lectures. Gerstein. Lab. org • Calculate an annotation signal by summing the intersecting variants’ scores

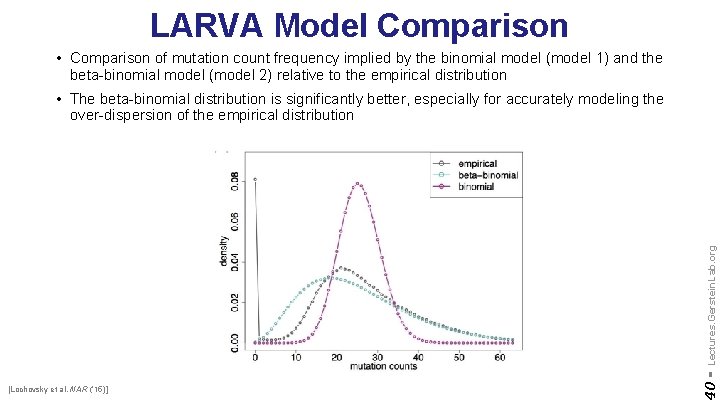
LARVA Model Comparison • Comparison of mutation count frequency implied by the binomial model (model 1) and the beta-binomial model (model 2) relative to the empirical distribution [Lochovsky et al. NAR (’ 15)] 40 - Lectures. Gerstein. Lab. org • The beta-binomial distribution is significantly better, especially for accurately modeling the over-dispersion of the empirical distribution

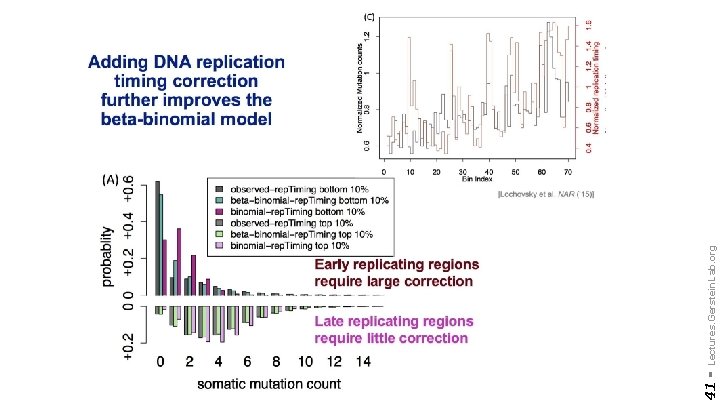
41 - Lectures. Gerstein. Lab. org

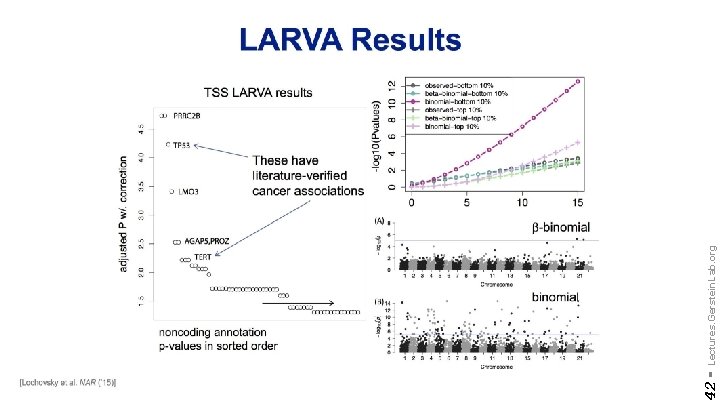
42 - Lectures. Gerstein. Lab. org

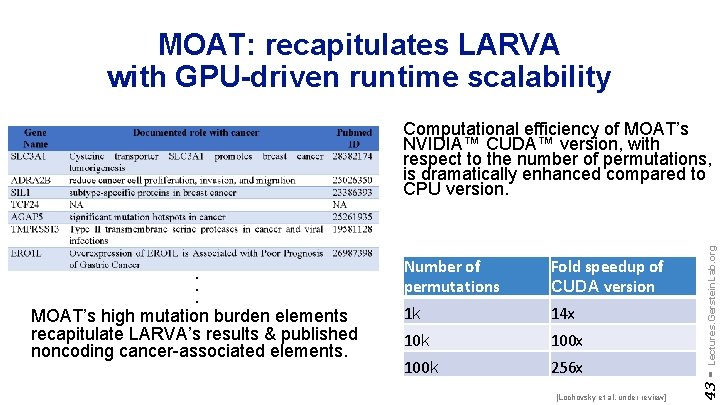
MOAT: recapitulates LARVA with GPU-driven runtime scalability Number of permutations Fold speedup of CUDA version 1 k 14 x 10 k 100 x 100 k 256 x [Lochovsky et al. under review] 43 - . . . MOAT’s high mutation burden elements recapitulate LARVA’s results & published noncoding cancer-associated elements. Lectures. Gerstein. Lab. org Computational efficiency of MOAT’s NVIDIA™ CUDA™ version, with respect to the number of permutations, is dramatically enhanced compared to CPU version.

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 44 - • Introduction
![Kumar Gerstein Nature 17 Lectures Gerstein Lab org Better annotation or large number [Kumar & Gerstein, Nature ('17)] Lectures. Gerstein. Lab. org Better annotation or large number](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-45.jpg)
[Kumar & Gerstein, Nature ('17)] Lectures. Gerstein. Lab. org Better annotation or large number of samples could help. 45 - Power, as an issue in driver discovery

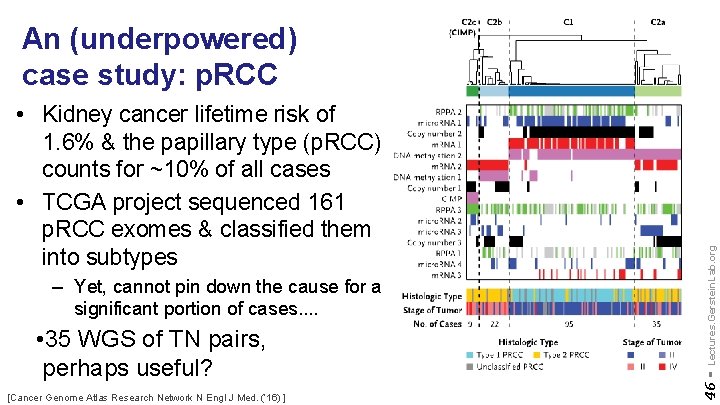
– Yet, cannot pin down the cause for a significant portion of cases. . • 35 WGS of TN pairs, perhaps useful? [Cancer Genome Atlas Research Network N Engl J Med. (‘ 16) ] 46 - • Kidney cancer lifetime risk of 1. 6% & the papillary type (p. RCC) counts for ~10% of all cases • TCGA project sequenced 161 p. RCC exomes & classified them into subtypes Lectures. Gerstein. Lab. org An (underpowered) case study: p. RCC

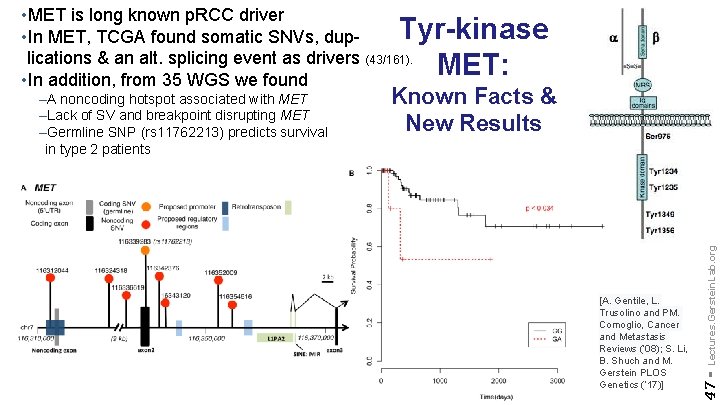
• MET is long known p. RCC driver • In MET, TCGA found somatic SNVs, duplications & an alt. splicing event as drivers (43/161). • In addition, from 35 WGS we found Tyr-kinase MET: [A. Gentile, L. Trusolino and PM. Comoglio, Cancer and Metastasis Reviews (‘ 08); S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org Known Facts & New Results 47 - –A noncoding hotspot associated with MET –Lack of SV and breakpoint disrupting MET –Germline SNP (rs 11762213) predicts survival in type 2 patients
![Li et al PLOS Genetics 17 Lectures Gerstein Lab org supported by expr [Li et al. PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org supported by expr.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-48.jpg)
[Li et al. PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org supported by expr. changes & survival analysis 48 - Beyond MET: 2 noncoding hotspots in NEAT & ERRFl 1,

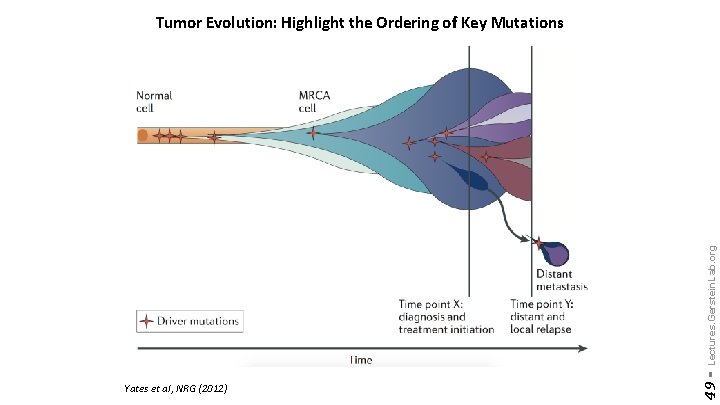
Yates et al, NRG (2012) 49 - Lectures. Gerstein. Lab. org Tumor Evolution: Highlight the Ordering of Key Mutations

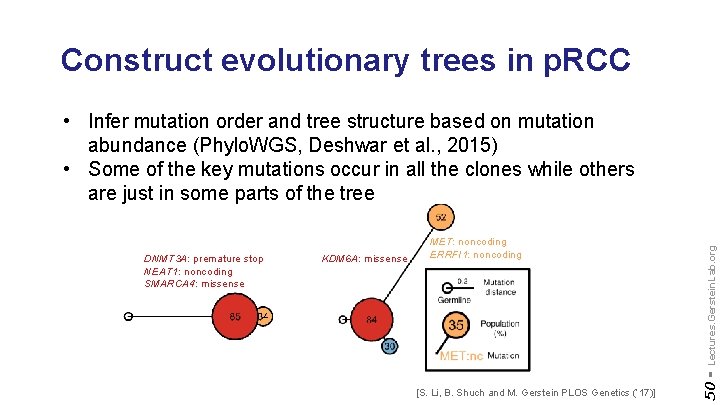
Construct evolutionary trees in p. RCC KDM 6 A: missense [S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] 50 - DNMT 3 A: premature stop NEAT 1: noncoding SMARCA 4: missense MET: noncoding ERRFI 1: noncoding Lectures. Gerstein. Lab. org • Infer mutation order and tree structure based on mutation abundance (Phylo. WGS, Deshwar et al. , 2015) • Some of the key mutations occur in all the clones while others are just in some parts of the tree
![S Li B Shuch and M Gerstein PLOS Genetics 17 Lectures Gerstein Lab [S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-51.jpg)
[S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org Germline 51 - Mutation Populations distance (%) 0. 5
![S Li B Shuch and M Gerstein PLOS Genetics 17 Lectures Gerstein Lab [S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab.](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-52.jpg)
[S. Li, B. Shuch and M. Gerstein PLOS Genetics (‘ 17)] Lectures. Gerstein. Lab. org Germline 52 - Mutation Populations distance (%) 0. 5
![53 Lectures Gerstein Lab org Li et al PLOS Genetics 17 53 - Lectures. Gerstein. Lab. org [Li et al. , PLOS Genetics (‘ 17)]](https://slidetodoc.com/presentation_image_h2/44d73cc3961fe5f2a3e1fe8b89a8db1b/image-53.jpg)
53 - Lectures. Gerstein. Lab. org [Li et al. , PLOS Genetics (‘ 17)] Tree topology correlates with molecular subtypes

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 54 - • Introduction

Prioritizing somatic variants: Approaches to identifying key variants through functional impact & recurrence • Functional impact #1: Coding • ALo. FT: Annotation of Loss-of-Function Transcripts. LOF annotation as a complex problem. • Finding deleterious Lo. F SNVs • Frustration as a localized metric of SNV impact. Differential profiles for oncogenes vs. TSGs • Functional impact #2: Non-coding • Fun. Seq integrates evidence, with an entropy based weighting scheme • Background mutation rate significantly varies & is correlated with replication timing & TADs • Developed a variety of parametric & non-parametric methods taking this into account • LARVA uses parametric beta-binomial model, explicitly modeling covariates • MOAT does a variety of non-parm. shuffles (annotation, variants, &c). Useful when explicit covariates not available. Slower than but speeded up w/ GPUs • Recurrence #2: (Low-power) application to p. RCC • WGS finds additional facts on the canonical driver, MET. Other suggestive non-coding hotspots. • Tumor evolution analysis of the timing of key mutations helps with classification Lectures. Gerstein. Lab. org • Large growth in cancer genome data • Mining the data to prioritize variants for key drivers • Recurrence #1: Statistics for driver identification 55 - • Introduction

Acknowledgments github. com/gersteinlab/Frustration S Kumar, D Clarke Hiring Postdocs. See Jobs. gersteinlab. org github. com/gersteinlab/Mr. TADfinder KK Yan, S Lou ALo. FT. gersteinlab. org S Balasubramanian, Y Fu, M Pawashe, P Mc. Gillivray, M Jin, J Liu, KJ Karczewski, DG Mac. Arthur Fun. Seq. gersteinlab. org Y Fu, E Khurana, Z Liu, S Lou, J Bedford, XJ Mu, KY Yip, LARVA. gersteinlab. org L Lochovsky, J Zhang, Y Fu, E Khurana MOAT. gersteinlab. org L Lochovsky, J Zhang Lectures. Gerstein. Lab. org DZ Chen, E Khurana, A Sboner, A Harmanci, J Rozowsky, D Clarke, M Snyder p. RCC S Li, B Shuch 56 - VAT. gersteinlab. org L Habegger, S Balasubramanian,

Info about this talk General PERMISSIONS • This Presentation is copyright Mark Gerstein, Yale University, 2016. • Please read permissions statement at gersteinlab. org/misc/permissions. html. For thoughts on the source and permissions of many of the photos and clipped images in this presentation see streams. gerstein. info. In particular, many of the images have particular EXIF tags, such as kwpotppt , that can be easily queried from flickr, viz: flickr. com/photos/mbgmbg/tags/kwpotppt 57 - PHOTOS & IMAGES Lectures. Gerstein. Lab. org • Feel free to use slides & images in the talk with PROPER acknowledgement (via citation to relevant papers or link to gersteinlab. org). Paper references in the talk were mostly from Papers. Gerstein. Lab. org.