Common variants vs Rare variants The 1000 genome





- Slides: 30

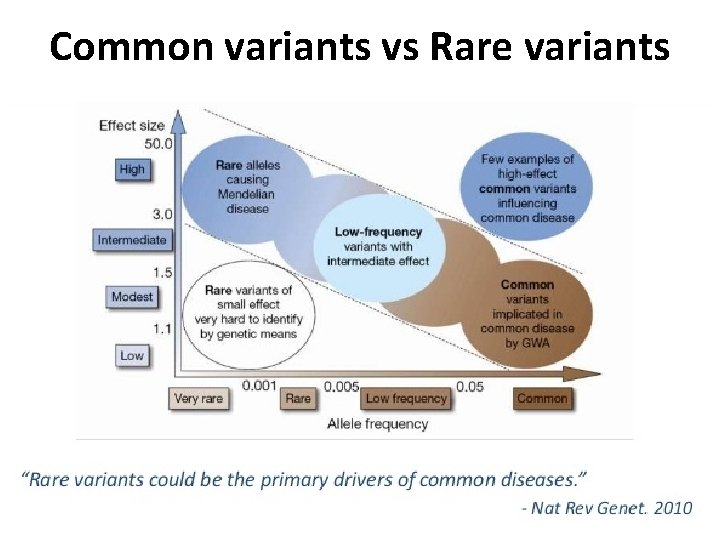
Common variants vs Rare variants

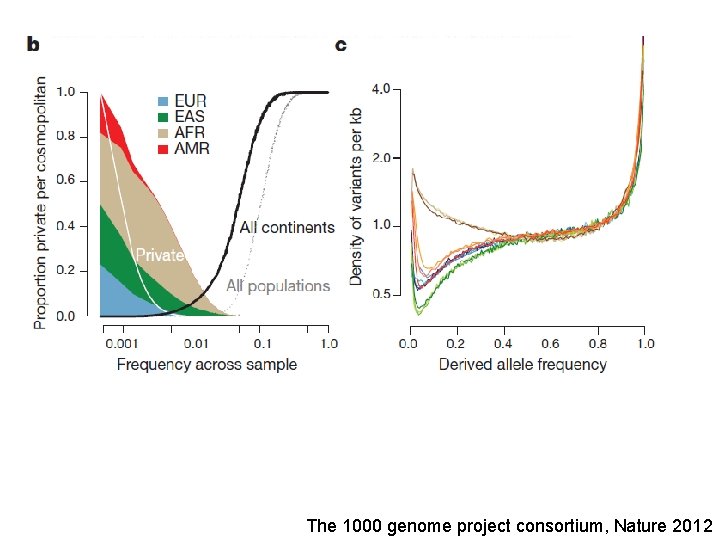
The 1000 genome project consortium, Nature 2012

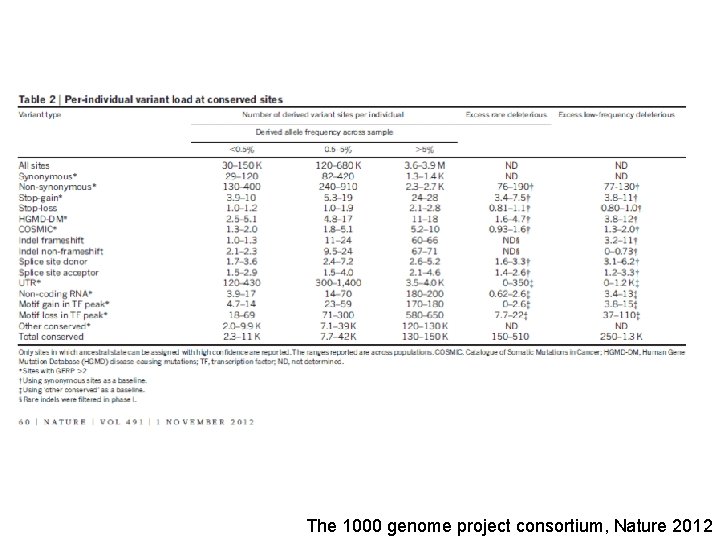
The 1000 genome project consortium, Nature 2012

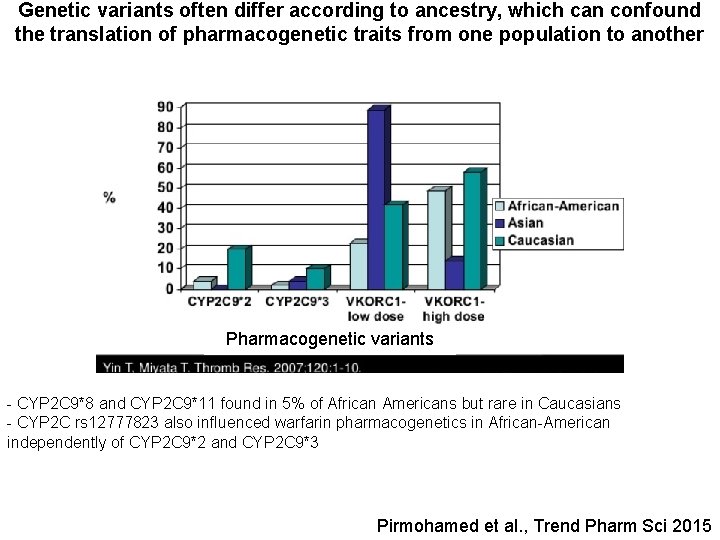
Genetic variants often differ according to ancestry, which can confound the translation of pharmacogenetic traits from one population to another Pharmacogenetic variants - CYP 2 C 9*8 and CYP 2 C 9*11 found in 5% of African Americans but rare in Caucasians - CYP 2 C rs 12777823 also influenced warfarin pharmacogenetics in African-American independently of CYP 2 C 9*2 and CYP 2 C 9*3 Pirmohamed et al. , Trend Pharm Sci 2015

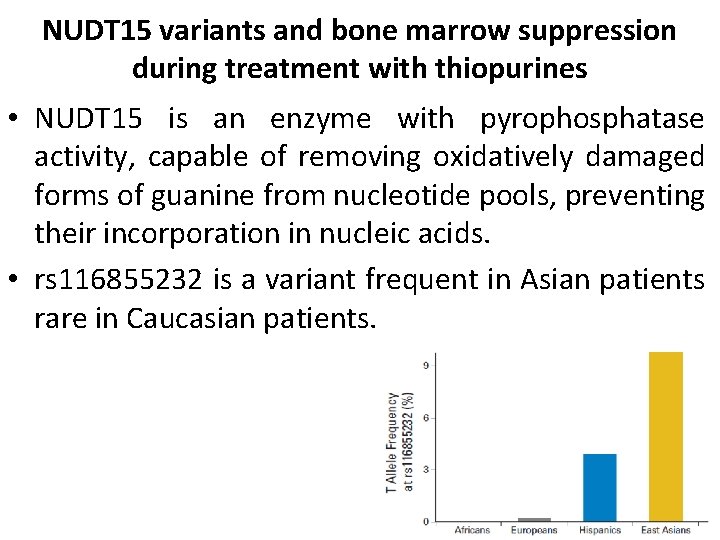
NUDT 15 variants and bone marrow suppression during treatment with thiopurines • NUDT 15 is an enzyme with pyrophosphatase activity, capable of removing oxidatively damaged forms of guanine from nucleotide pools, preventing their incorporation in nucleic acids. • rs 116855232 is a variant frequent in Asian patients rare in Caucasian patients.

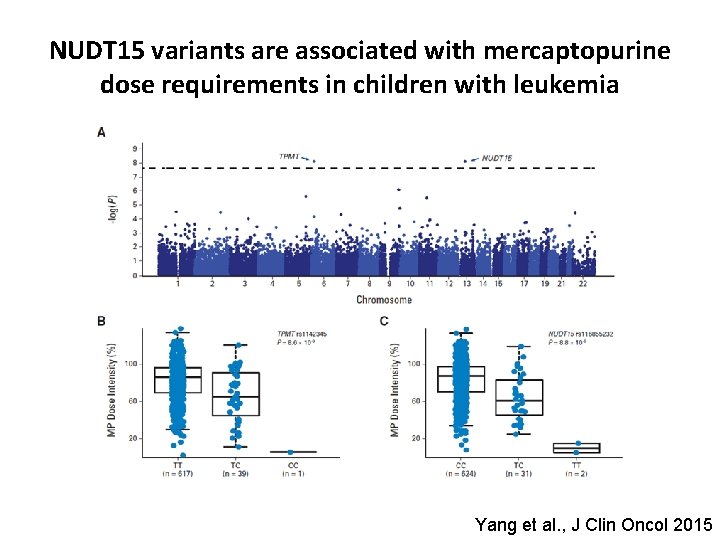
NUDT 15 variants are associated with mercaptopurine dose requirements in children with leukemia Yang et al. , J Clin Oncol 2015

Effetti delle varianti genetiche sui fenotipi (anche farmacogenomici) Dipendono da: - tipo di variante: SNP? indel? CNA? - posizione nel genoma: esonico? intronico? regione regolatrice? intergenico? - effetto sulla sequenza: sinonimo/non sinonimo? - frequenza della variante: rara? frequente? varianti più frequenti in genere hanno effetti più deboli sul fenotipo rispetto alle varianti più rare - modello di ereditarietà: omozigosi/eterozigosi? dominante/recessivo/co-dominante? - linkage: diretto? da linkage disequilibrium? effetto

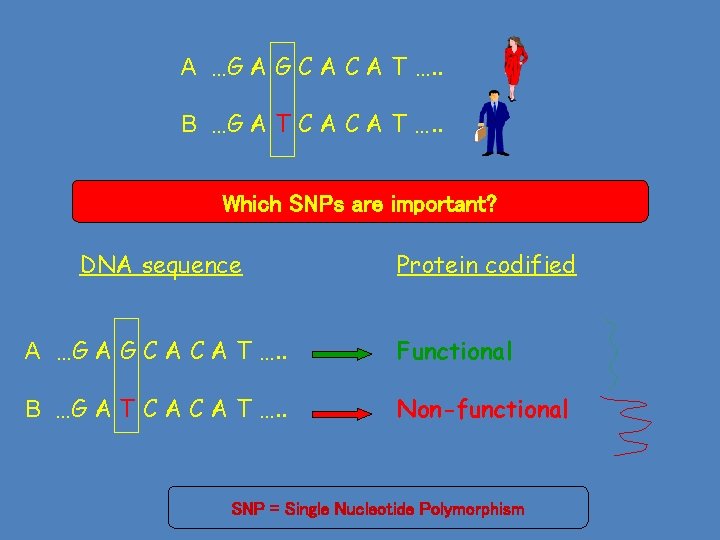
A …G A G C A T …. . B …G A T C A T …. . Which SNPs are important? DNA sequence Protein codified A …G A G C A T …. . Functional B …G A T C A T …. . Non-functional SNP = Single Nucleotide Polymorphism

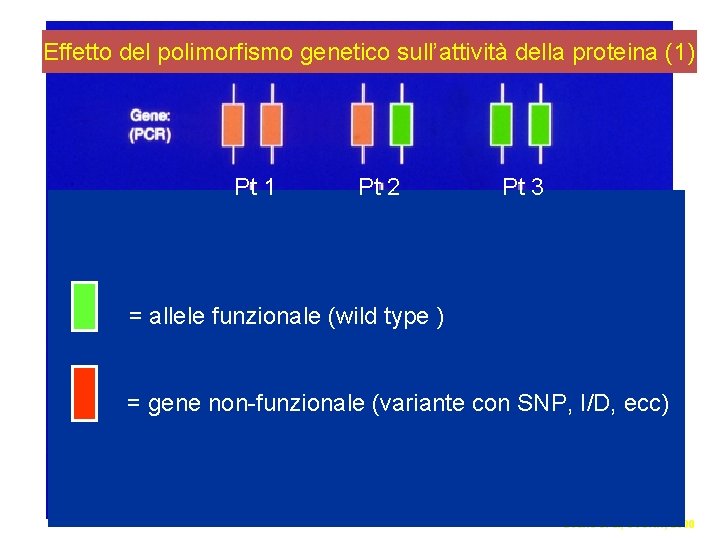
Effetto del polimorfismo genetico sull’attività della proteina (1) Pt 1 Pt 2 Pt 3 = allele funzionale (wild type ) = gene non-funzionale (variante con SNP, I/D, ecc) Evans et al, SJCRH, 2000

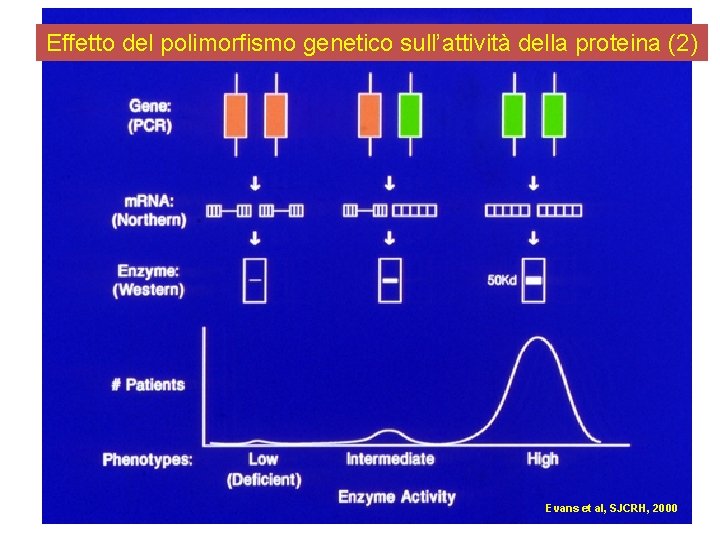
Effetto del polimorfismo genetico sull’attività della proteina (2) Evans et al, SJCRH, 2000

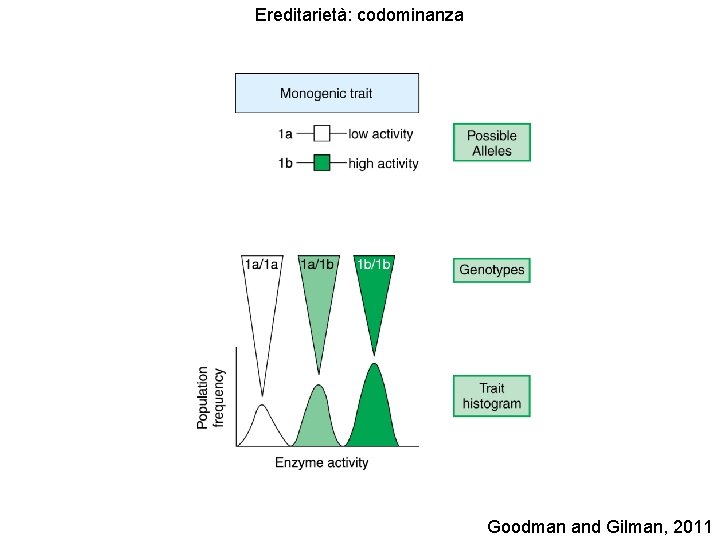
Ereditarietà: codominanza Goodman and Gilman, 2011

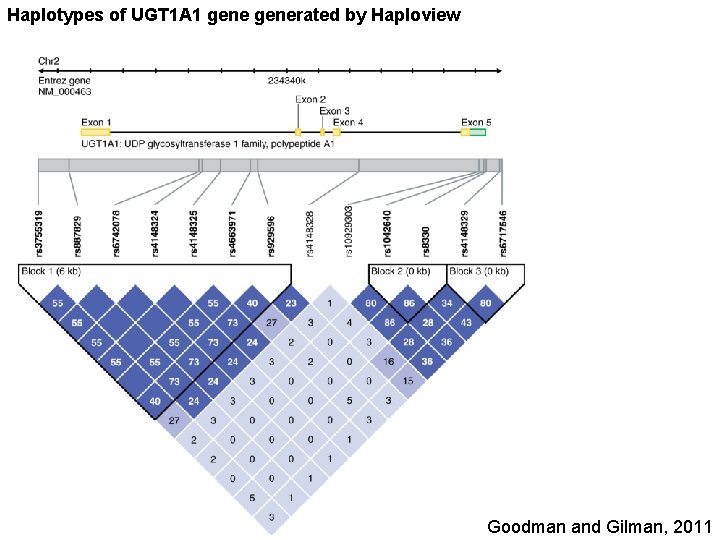
Haplotypes of UGT 1 A 1 generated by Haploview Goodman and Gilman, 2011

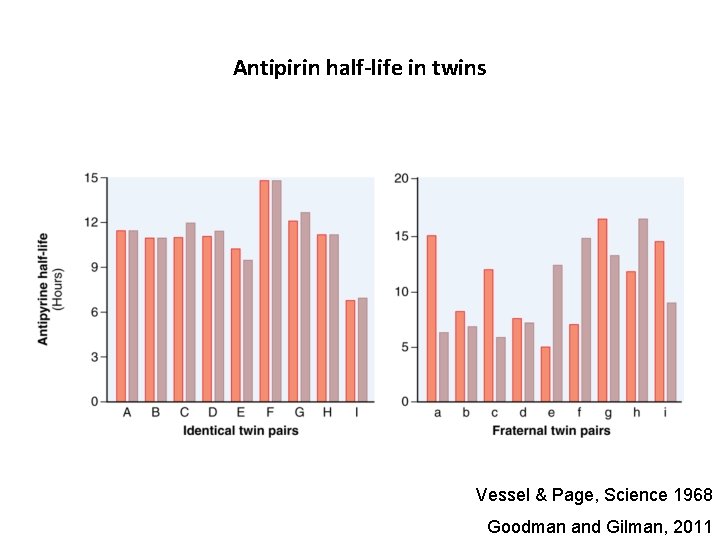
Antipirin half-life in twins Vessel & Page, Science 1968 Goodman and Gilman, 2011

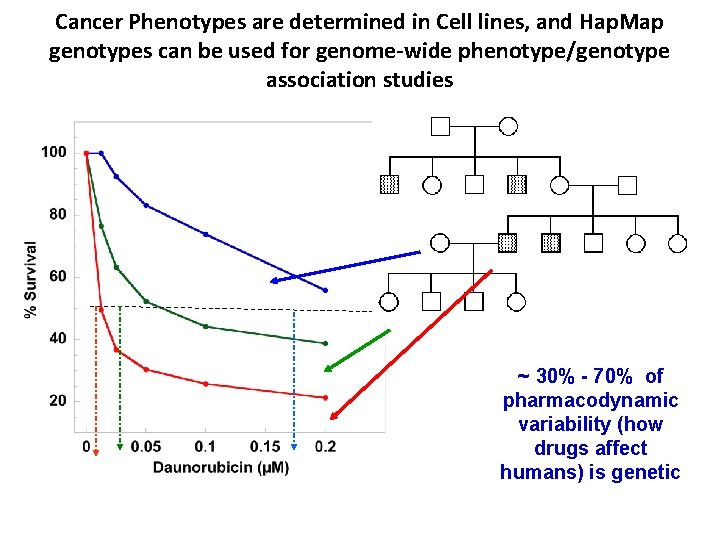
Cancer Phenotypes are determined in Cell lines, and Hap. Map genotypes can be used for genome-wide phenotype/genotype association studies ~ 30% - 70% of pharmacodynamic variability (how drugs affect humans) is genetic (Dolan et al Cancer Res 2004; Watters et al PNAS 2004)

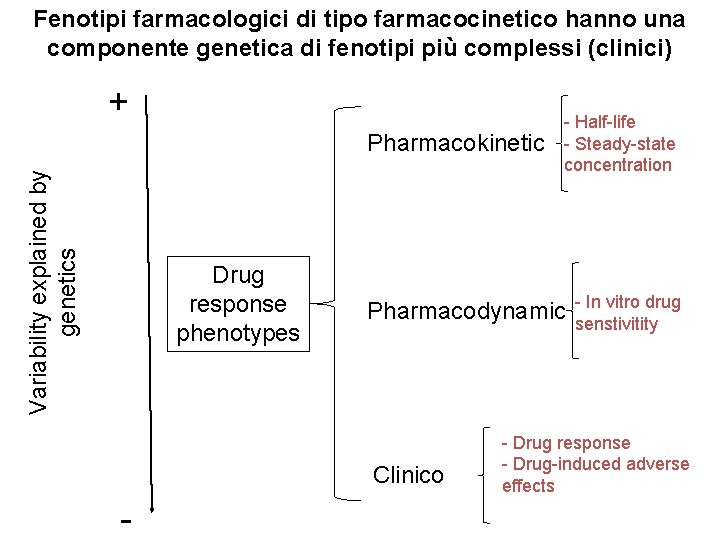
Fenotipi farmacologici di tipo farmacocinetico hanno una componente genetica di fenotipi più complessi (clinici) + Variability explained by genetics Pharmacokinetic Drug response phenotypes Pharmacodynamic Clinico - - Half-life - Steady-state concentration - In vitro drug senstivitity - Drug response - Drug-induced adverse effects

Personalized medicine for pediatric inflammatory bowel disease The aim of personalized medicine is to provide the most appropriate cure to the right patient, at the right dose and at the right time Personalized medicine should help clinicians in: - Indentifying patients who will not respond to therapies - Finding patients at risk of adverse effects - Choosing ideal drug, dose, and employing less expensive agents.

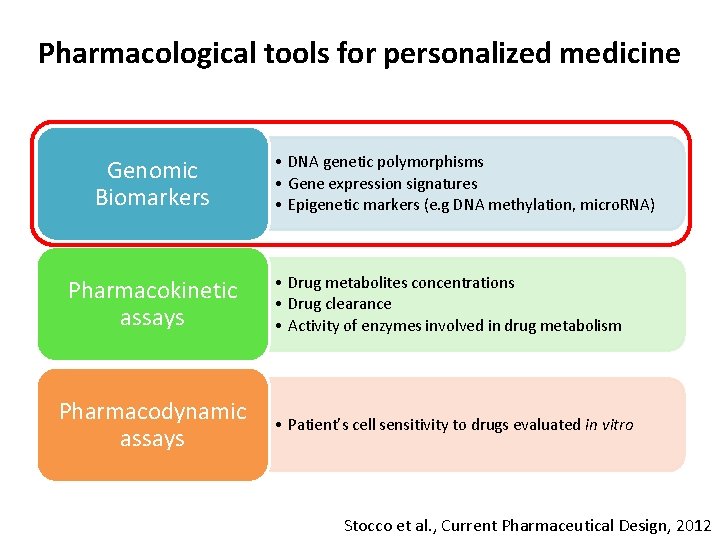
Pharmacological tools for personalized medicine Genomic Biomarkers Pharmacokinetic assays Pharmacodynamic assays • DNA genetic polymorphisms • Gene expression signatures • Epigenetic markers (e. g DNA methylation, micro. RNA) • Drug metabolites concentrations • Drug clearance • Activity of enzymes involved in drug metabolism • Patient’s cell sensitivity to drugs evaluated in vitro Stocco et al. , Current Pharmaceutical Design, 2012

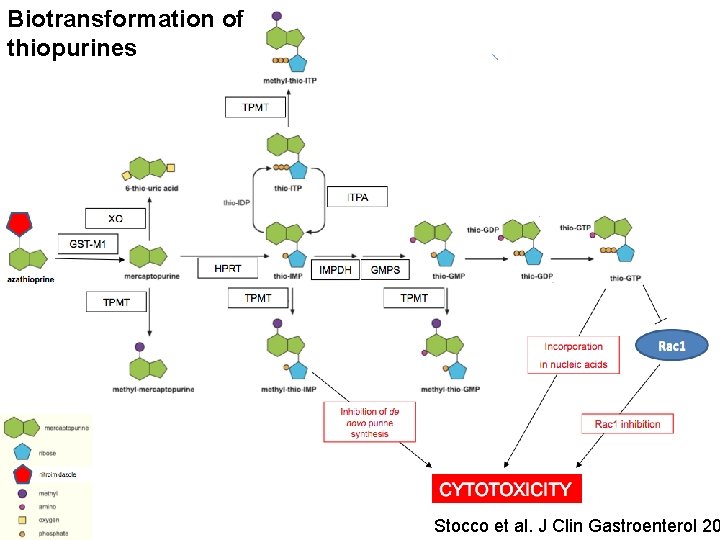
Biotransformation of thiopurines Stocco et al. J Clin Gastroenterol 20

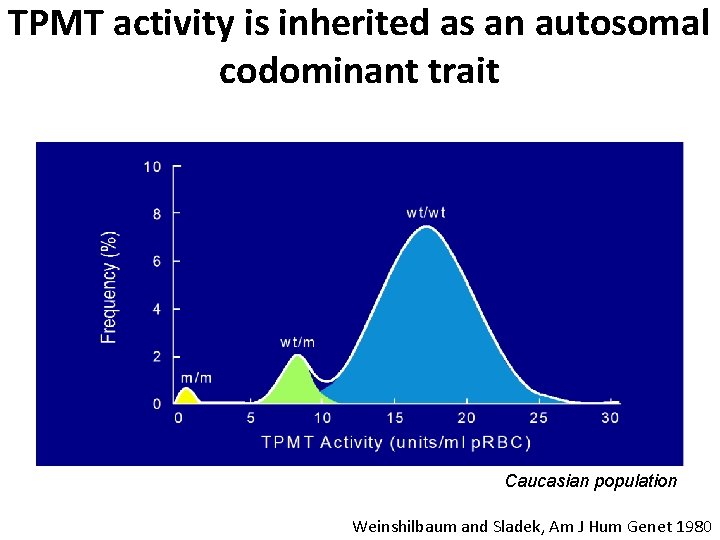
TPMT activity is inherited as an autosomal codominant trait Caucasian population Weinshilbaum and Sladek, Am J Hum Genet 1980

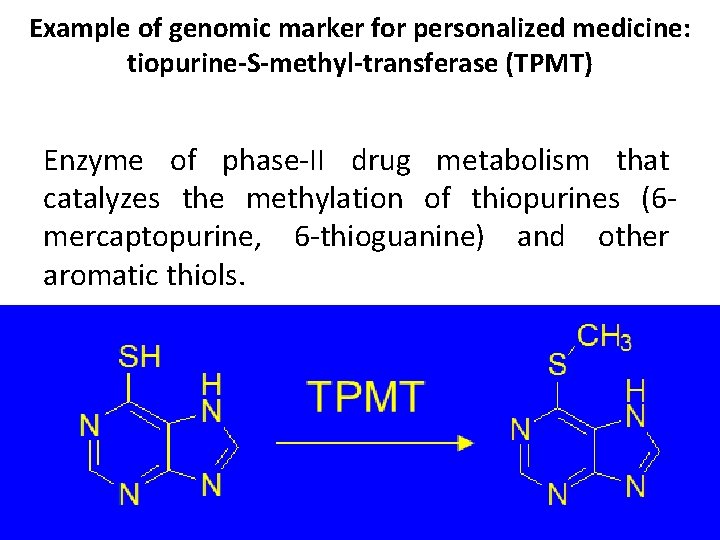
Example of genomic marker for personalized medicine: tiopurine-S-methyl-transferase (TPMT) Enzyme of phase-II drug metabolism that catalyzes the methylation of thiopurines (6 mercaptopurine, 6 -thioguanine) and other aromatic thiols.

Tiopurine-S-methyl-transferase (TPMT) Physiologically, TPMT is involved in the biosynthesis of a cofactor that confers molibdenum binding propreties to proteins. MDO meeting, Stuttgart 2014

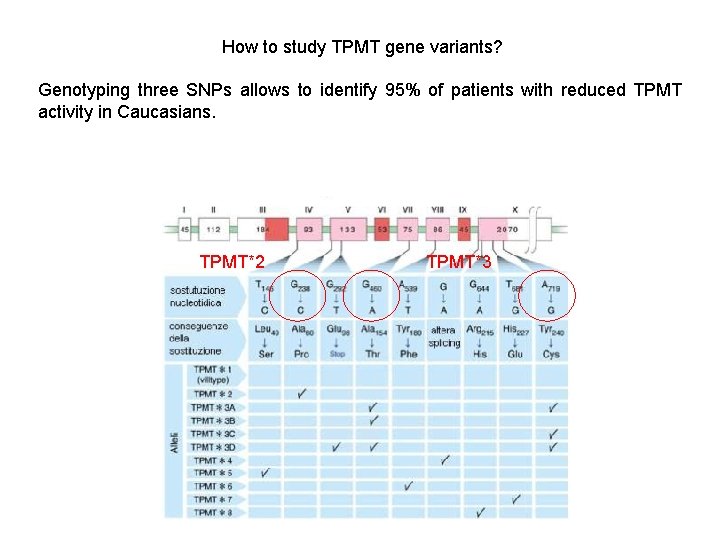
How to study TPMT gene variants? Genotyping three SNPs allows to identify 95% of patients with reduced TPMT activity in Caucasians. TPMT*2 TPMT*3

Official database of TPMT variant alleles http: //www. imh. liu. se/tpmtalleles? l=en

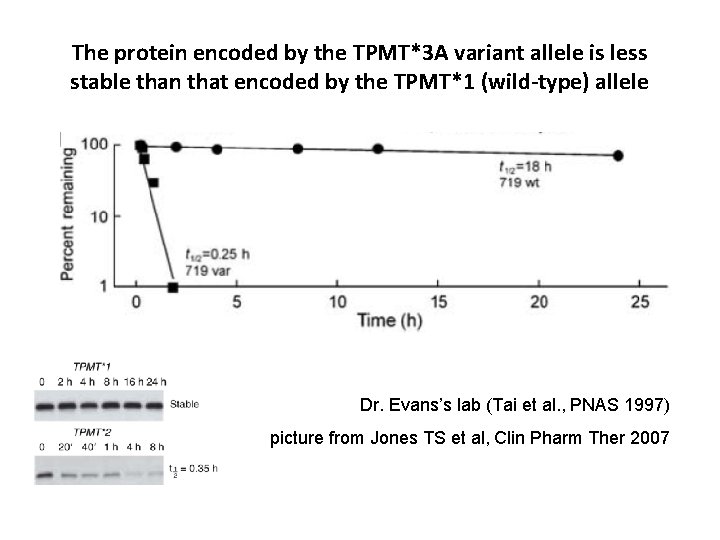
The protein encoded by the TPMT*3 A variant allele is less stable than that encoded by the TPMT*1 (wild-type) allele Dr. Evans’s lab (Tai et al. , PNAS 1997) picture from Jones TS et al, Clin Pharm Ther 2007

Cheok & Evans, 2006

Cheok & Evans, 2006


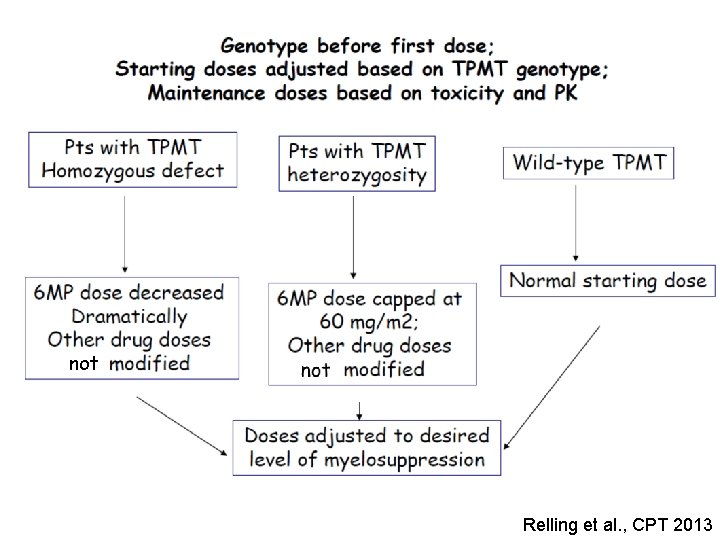
not Relling et al. , CPT 2013

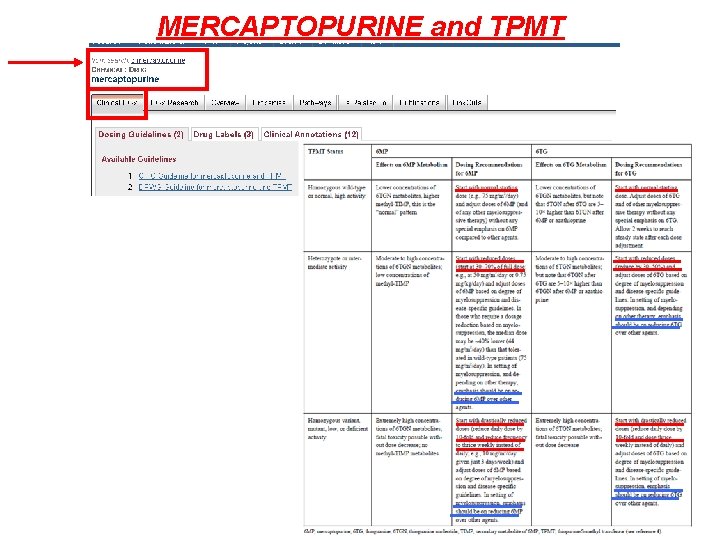
MERCAPTOPURINE and TPMT GUIDELINES are designed to help clinicians by • recommending the best pharmacogenetic test to perform • Interpreting the pharmacogentic test results to optimize drug therapy

MERCAPTOPURINE and TPMT