Whole Genome Sequencing Lecture for CS 498 CXZ









- Slides: 32

Whole Genome Sequencing (Lecture for CS 498 -CXZ Algorithms in Bioinformatics) Sept. 13, 2005 Cheng. Xiang Zhai Department of Computer Science University of Illinois, Urbana-Champaign Most slides are taken/adapted from Serafim Batzoglou’s lectures

Outline • Practical challenges in genome sequencing • Whole genome sequencing strategies • Sequencing coverage (Lander-Waterman model) • Overlap-Layout-Consensus approach

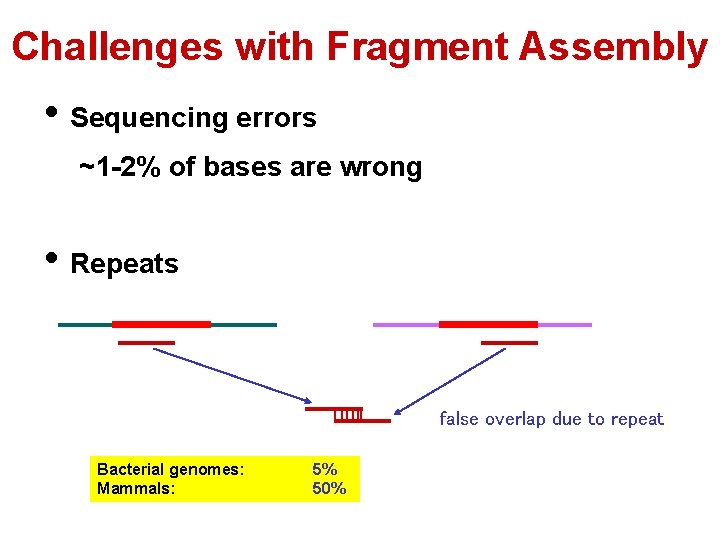
Challenges with Fragment Assembly • Sequencing errors ~1 -2% of bases are wrong • Repeats false overlap due to repeat Bacterial genomes: Mammals: 5% 50%

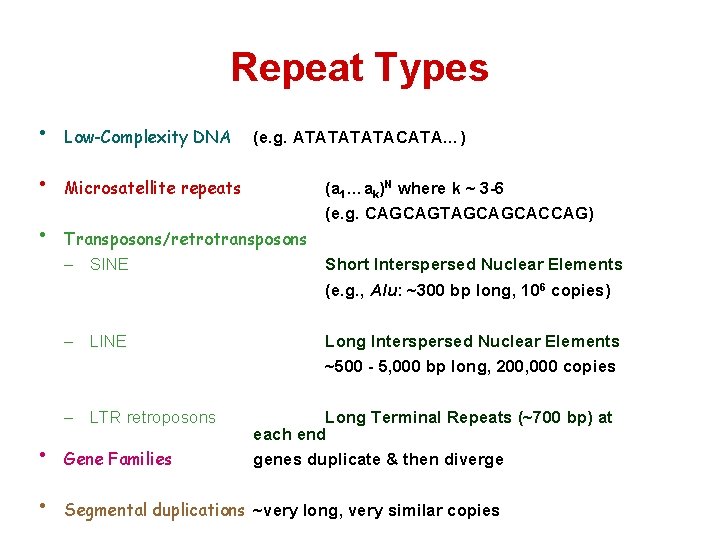
Repeat Types • Low-Complexity DNA • Microsatellite repeats • (e. g. ATATACATA…) (a 1…ak)N where k ~ 3 -6 (e. g. CAGCAGTAGCAGCACCAG) Transposons/retrotransposons – SINE Short Interspersed Nuclear Elements (e. g. , Alu: ~300 bp long, 106 copies) – LINE Long Interspersed Nuclear Elements ~500 - 5, 000 bp long, 200, 000 copies – LTR retroposons Long Terminal Repeats (~700 bp) at each end genes duplicate & then diverge • Gene Families • Segmental duplications ~very long, very similar copies

The sequencing errors, repeats, and the complexity of genomes make it necessary to use many heuristics in practice… The Shortest Superstring formulation is an over-simplification of the problem

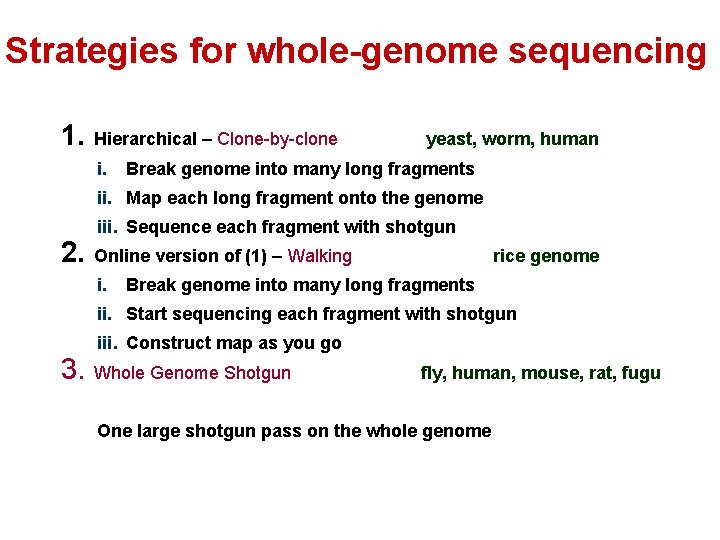
Strategies for whole-genome sequencing 1. Hierarchical – Clone-by-clone i. yeast, worm, human Break genome into many long fragments ii. Map each long fragment onto the genome iii. Sequence each fragment with shotgun 2. Online version of (1) – Walking i. rice genome Break genome into many long fragments ii. Start sequencing each fragment with shotgun iii. Construct map as you go 3. Whole Genome Shotgun fly, human, mouse, rat, fugu One large shotgun pass on the whole genome

Hierarchical Sequencing vs. Whole Genome Shotgun • Hierarchical Sequencing – Advantages: Easy assembly – Disadvantages: • Build library & physical map; • • Redundant sequencing Whole Genome Shotgun (WGS) – Advantages: No mapping, no redundant sequencing – Disadvantages: Difficult to assemble and resolve repeats Whole Genome Shotgun appears to get more popular…

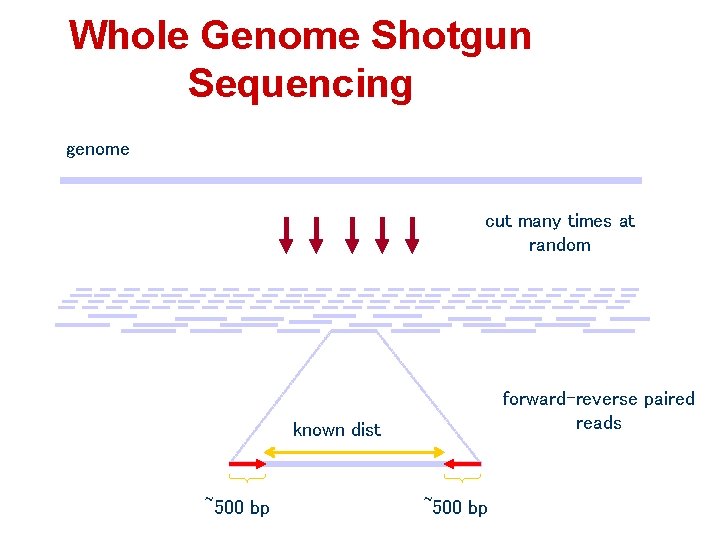
Whole Genome Shotgun Sequencing genome cut many times at random forward-reverse paired reads known dist ~500 bp

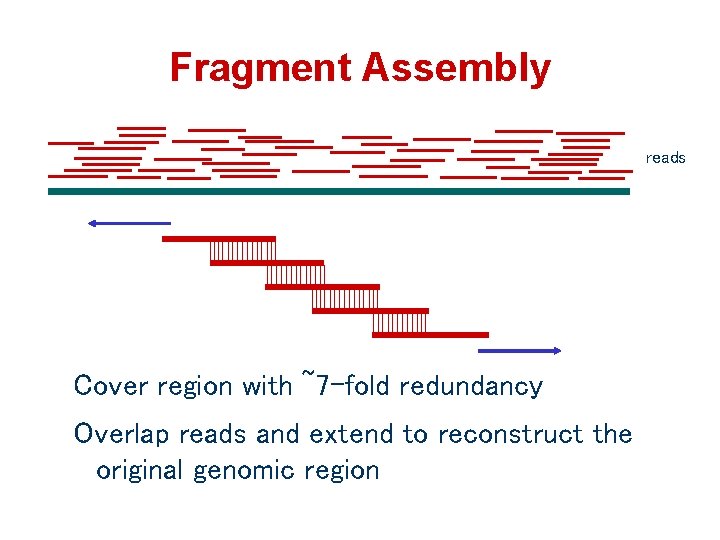
Fragment Assembly reads Cover region with ~7 -fold redundancy Overlap reads and extend to reconstruct the original genomic region

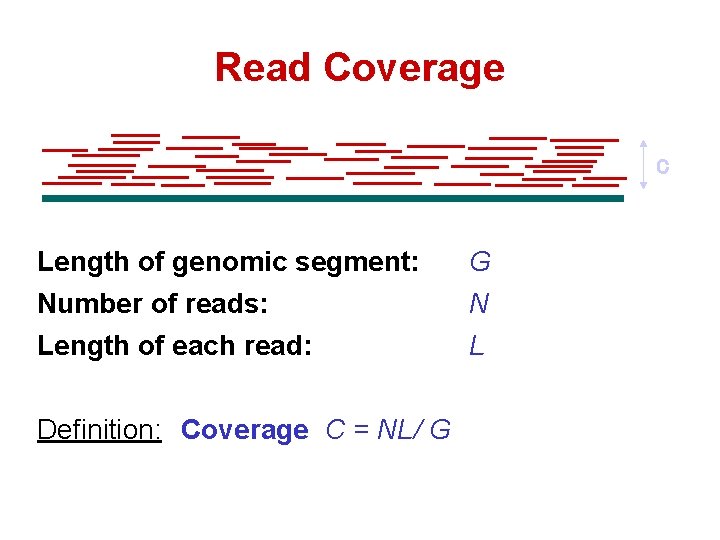
Read Coverage C Length of genomic segment: G Number of reads: N Length of each read: L Definition: Coverage C = NL/ G

Enough Coverage How much coverage is enough? According to the Lander-Waterman model: Assuming uniform distribution of reads, C=7 results in 1 gap per 1, 000 nucleotides

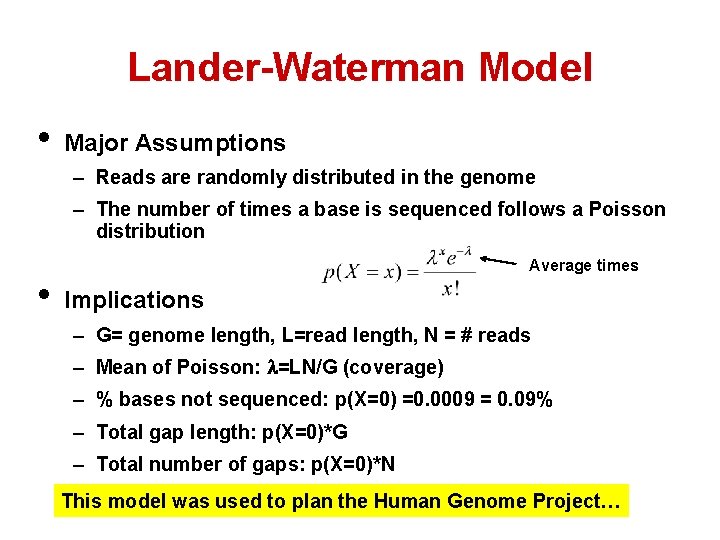
Lander-Waterman Model • Major Assumptions – Reads are randomly distributed in the genome – The number of times a base is sequenced follows a Poisson distribution • Average times Implications – G= genome length, L=read length, N = # reads – Mean of Poisson: =LN/G (coverage) – % bases not sequenced: p(X=0) =0. 0009 = 0. 09% – Total gap length: p(X=0)*G – Total number of gaps: p(X=0)*N This model was used to plan the Human Genome Project…

Repeats, Errors, and Read lengths • • • Repeats shorter than read length are OK Repeats with more base pair diffs than sequencing error rate are OK To make a smaller portion of the genome appear repetitive, try to: – Increase read length – Decrease sequencing error rate Role of error correction: Discards ~90% of single-letter sequencing errors decreases error rate decreases effective repeat content However, we have only limited read length. Many heuristics have been introduced to handle repeats…

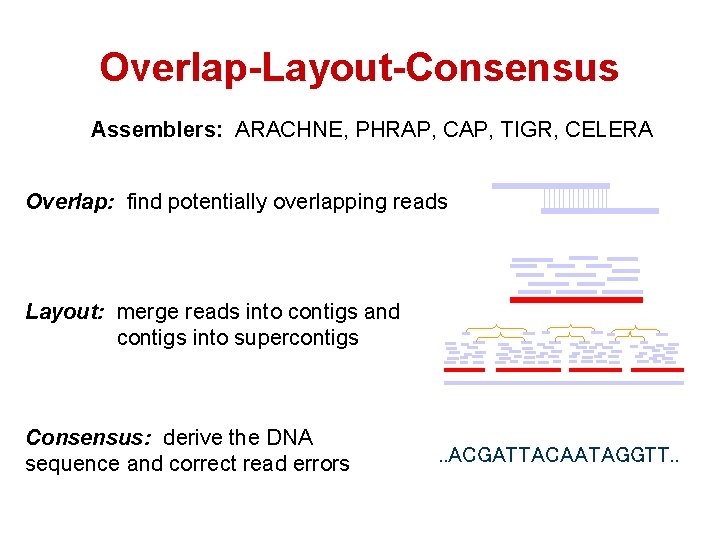
Overlap-Layout-Consensus Assemblers: ARACHNE, PHRAP, CAP, TIGR, CELERA Overlap: find potentially overlapping reads Layout: merge reads into contigs and contigs into supercontigs Consensus: derive the DNA sequence and correct read errors . . ACGATTACAATAGGTT. .

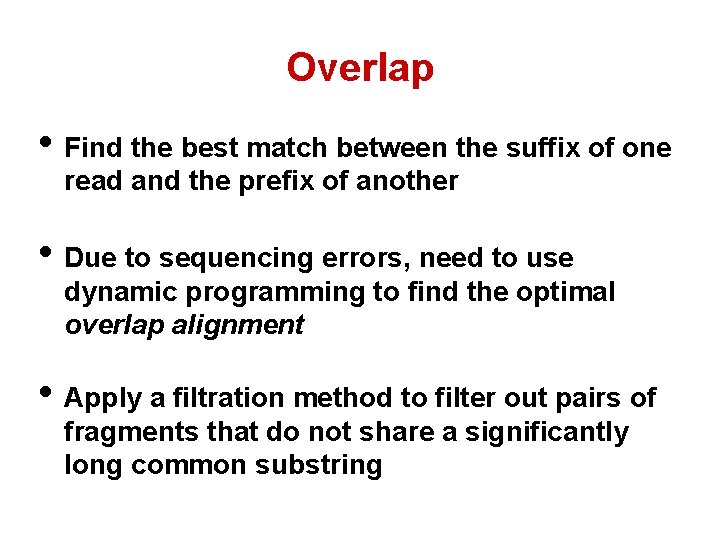
Overlap • Find the best match between the suffix of one read and the prefix of another • Due to sequencing errors, need to use dynamic programming to find the optimal overlap alignment • Apply a filtration method to filter out pairs of fragments that do not share a significantly long common substring

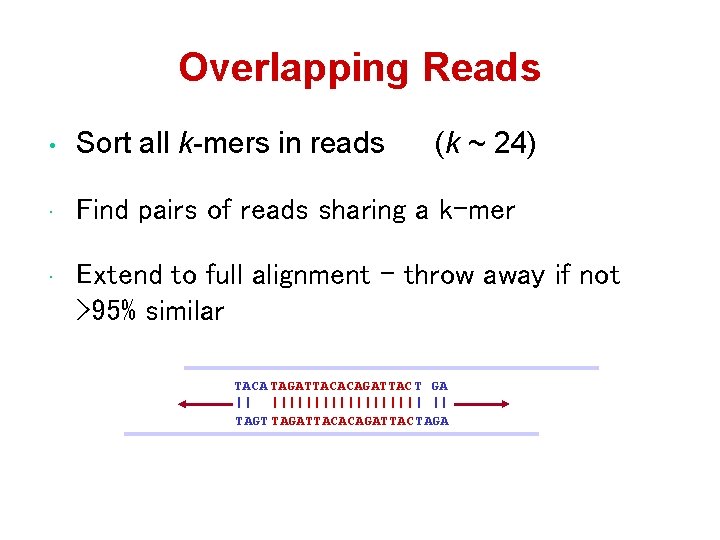
Overlapping Reads • Sort all k-mers in reads (k ~ 24) • Find pairs of reads sharing a k-mer • Extend to full alignment – throw away if not >95% similar TACA TAGATTACACAGATTAC T GA || ||||||||| | || TAGT TAGATTACACAGATTAC TAGA

Overlapping Reads and Repeats • A k-mer that appears N times, initiates N 2 comparisons • For an Alu that appears 106 times 1012 comparisons – too much • Solution: Discard all k-mers that appear more than t Coverage, (t ~ 10)

Finding Overlapping Reads Create local multiple alignments from the overlapping reads TAGATTACACAGATTACTGA TAG TTACACAGATTATTGA TAGATTACACAGATTACTGA

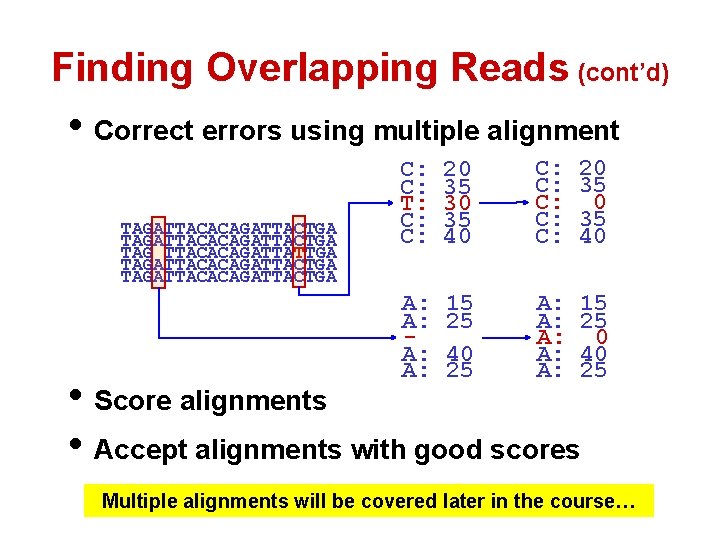
Finding Overlapping Reads (cont’d) • Correct errors using multiple alignment TAGATTACACAGATTACTGA TAG TTACACAGATTATTGA TAGATTACACAGATTACTGA C: C: T: C: C: 20 35 30 35 40 C: C: C: 20 35 40 A: A: 15 25 40 25 A: A: A: 15 25 0 40 25 • Score alignments • Accept alignments with good scores Multiple alignments will be covered later in the course…

Layout • Repeats are a major challenge • Do two aligned fragments really overlap, or are they from two copies of a repeat?

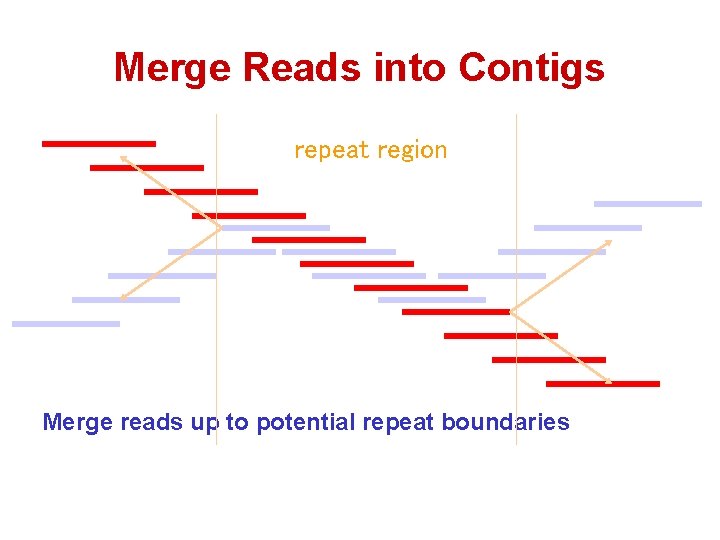
Merge Reads into Contigs repeat region Merge reads up to potential repeat boundaries

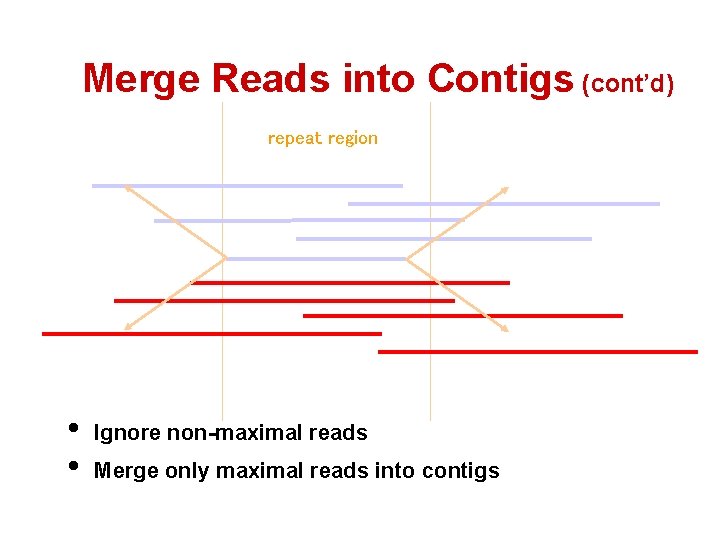
Merge Reads into Contigs (cont’d) repeat region • • Ignore non-maximal reads Merge only maximal reads into contigs

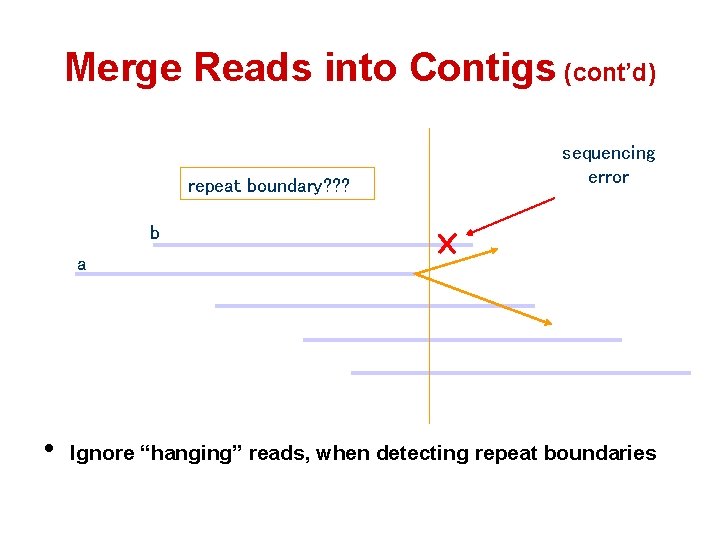
Merge Reads into Contigs (cont’d) repeat boundary? ? ? sequencing error b a • Ignore “hanging” reads, when detecting repeat boundaries

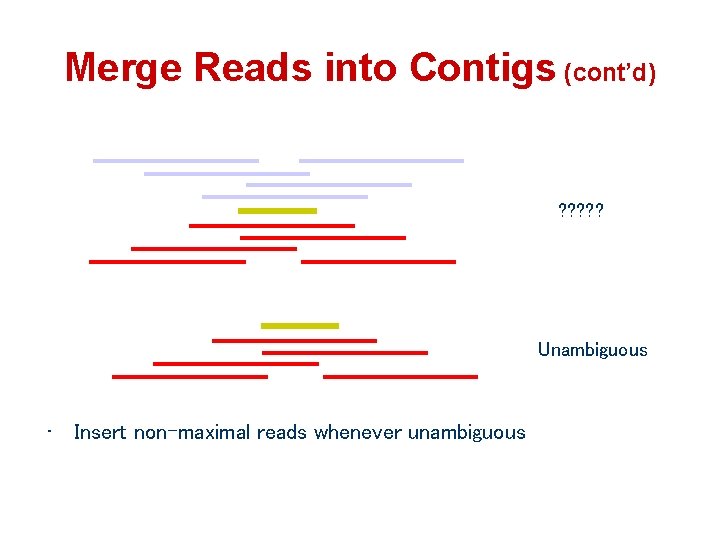
Merge Reads into Contigs (cont’d) ? ? ? Unambiguous • Insert non-maximal reads whenever unambiguous

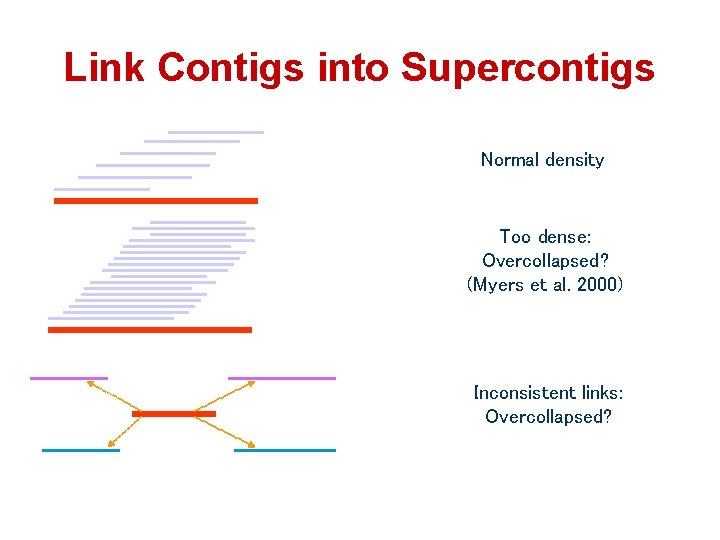
Link Contigs into Supercontigs Normal density Too dense: Overcollapsed? (Myers et al. 2000) Inconsistent links: Overcollapsed?

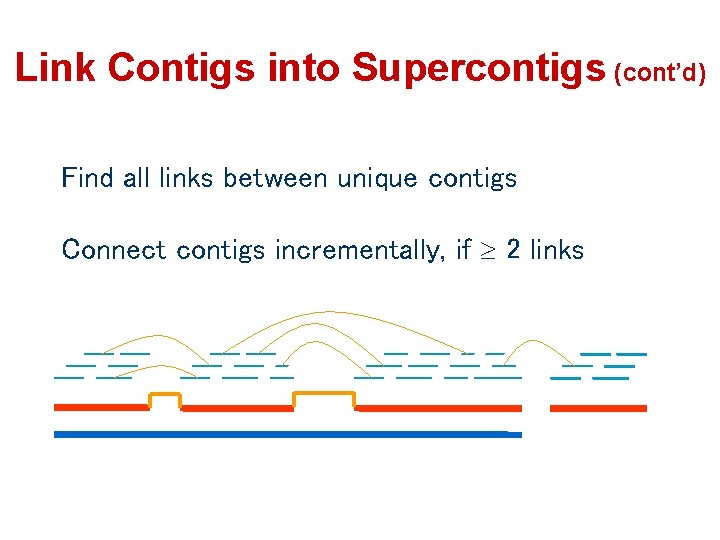
Link Contigs into Supercontigs (cont’d) Find all links between unique contigs Connect contigs incrementally, if 2 links

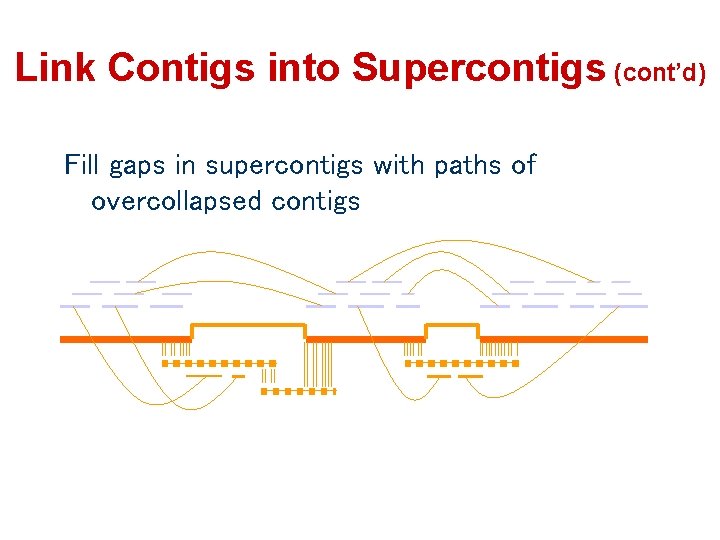
Link Contigs into Supercontigs (cont’d) Fill gaps in supercontigs with paths of overcollapsed contigs

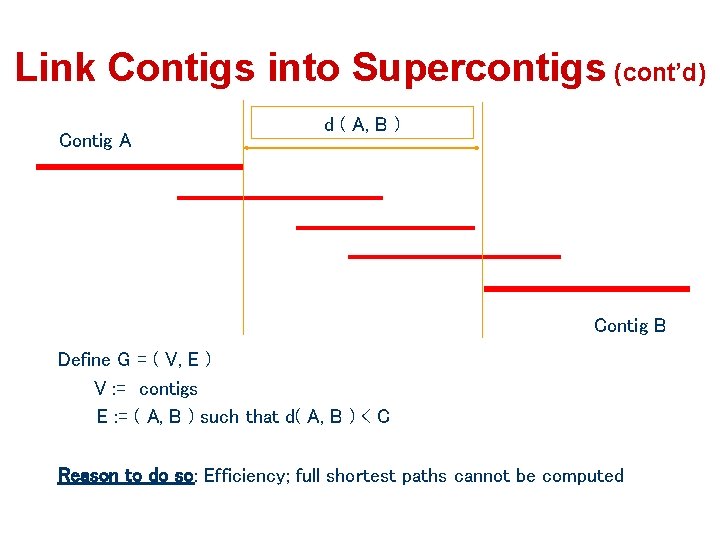
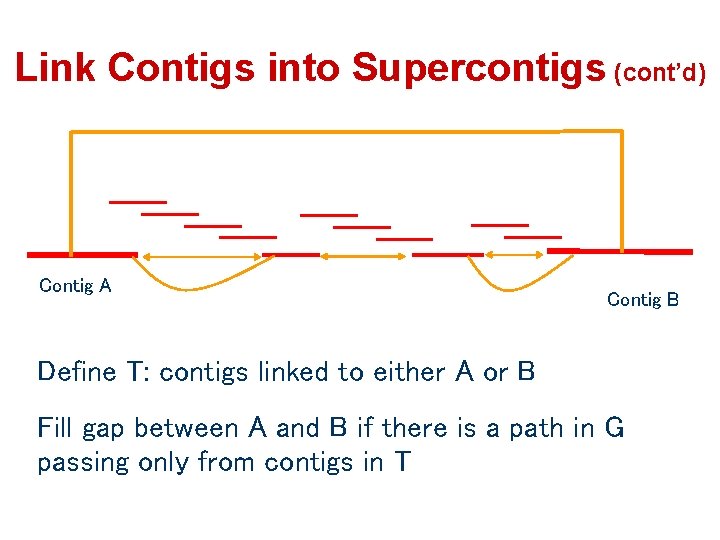
Link Contigs into Supercontigs (cont’d) Contig A d ( A, B ) Contig B Define G = ( V, E ) V : = contigs E : = ( A, B ) such that d( A, B ) < C Reason to do so: Efficiency; full shortest paths cannot be computed

Link Contigs into Supercontigs (cont’d) Contig A Contig B Define T: contigs linked to either A or B Fill gap between A and B if there is a path in G passing only from contigs in T

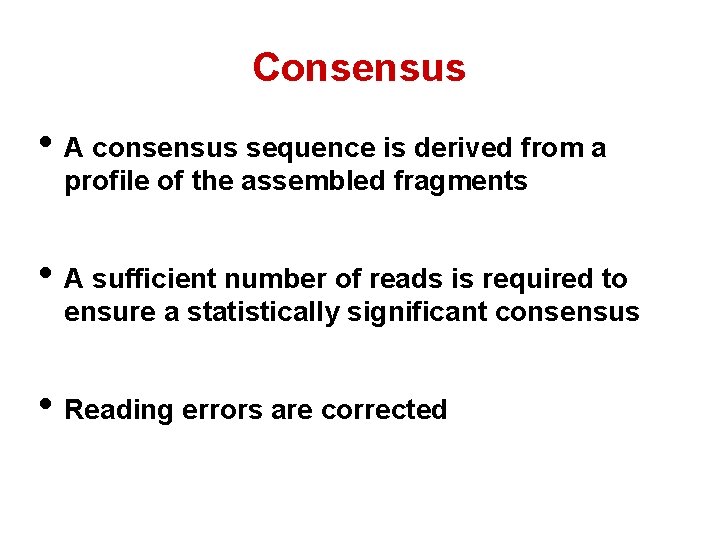
Consensus • A consensus sequence is derived from a profile of the assembled fragments • A sufficient number of reads is required to ensure a statistically significant consensus • Reading errors are corrected

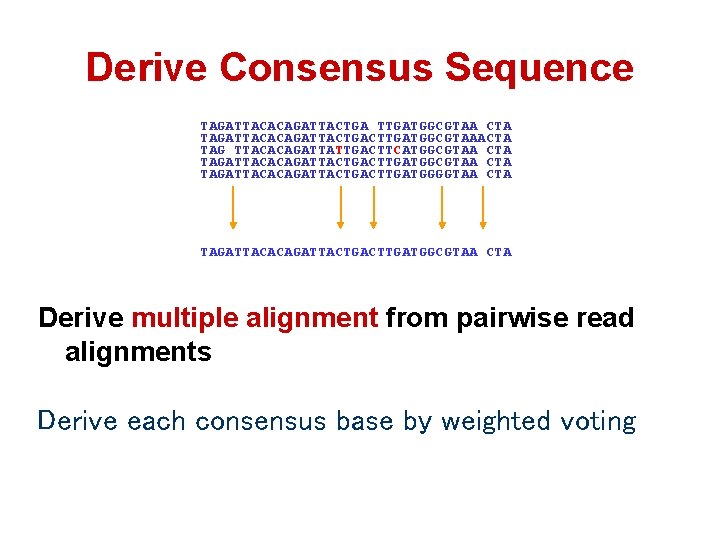
Derive Consensus Sequence TAGATTACACAGATTACTGA TTGATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAAACTA TAG TTACACAGATTATTGACTTCATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGGGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAA CTA Derive multiple alignment from pairwise read alignments Derive each consensus base by weighted voting

What You Should Know • The challenges in assembling fragments for whole genome shotgun sequencing • Lander-Waterman model • Main heuristics used in Overlap-Layout. Consensus
 Pyrosequencing animation
Pyrosequencing animation Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Shotgun sequencing
Shotgun sequencing Cdc whole school whole community whole child
Cdc whole school whole community whole child Genome-to-genome distance calculator
Genome-to-genome distance calculator Sequencing human genome
Sequencing human genome Genome sequencing
Genome sequencing Genome sequencing
Genome sequencing Massed practice
Massed practice 01:640:244 lecture notes - lecture 15: plat, idah, farad
01:640:244 lecture notes - lecture 15: plat, idah, farad Quem pode o mais pode o menos
Quem pode o mais pode o menos 498
498 Eecs 498
Eecs 498 Cs 498 vr
Cs 498 vr Ece 498
Ece 498 Umich eecs 498
Umich eecs 498 Cs 498 vr
Cs 498 vr Eecs 498
Eecs 498 +1 (432) 498-0251
+1 (432) 498-0251 Cs 498 cloud computing applications
Cs 498 cloud computing applications Embedded systems
Embedded systems Ece 498
Ece 498 498
498 Cs 498 vr
Cs 498 vr Engr 498 uvic
Engr 498 uvic Cs 498 vr
Cs 498 vr Human genome project source code
Human genome project source code Savant genome browser
Savant genome browser What is a genome
What is a genome Section 14-3 human molecular genetics
Section 14-3 human molecular genetics Innovation genome project
Innovation genome project Consensus sequence in prokaryotes
Consensus sequence in prokaryotes Ucsc genome browser tutorial
Ucsc genome browser tutorial