Genome alignment Usman Roshan Applications Genome sequencing on


- Slides: 11

Genome alignment Usman Roshan

Applications • Genome sequencing on the rise • Whole genome comparison provides a deeper understanding of biology – Evolutionary history – Non-coding regions – Variant detection

Methods • General two-fold approach • 1. Find high scoring segments between pair of genomes. – Similar to BLAST like k-mer search using hashtables – Also done with suffix tree – Similar to short read mapping strategies • 2. Perform constrained alignment between high scoring segments

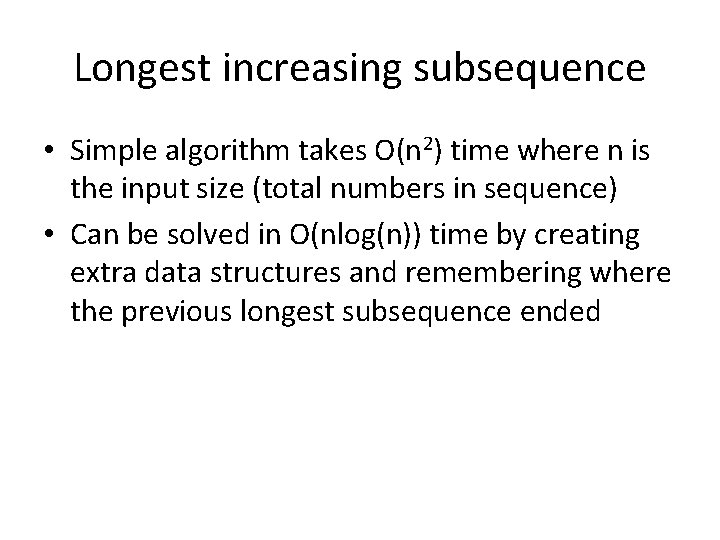
Longest increasing subsequence • Simple algorithm takes O(n 2) time where n is the input size (total numbers in sequence) • Can be solved in O(nlog(n)) time by creating extra data structures and remembering where the previous longest subsequence ended

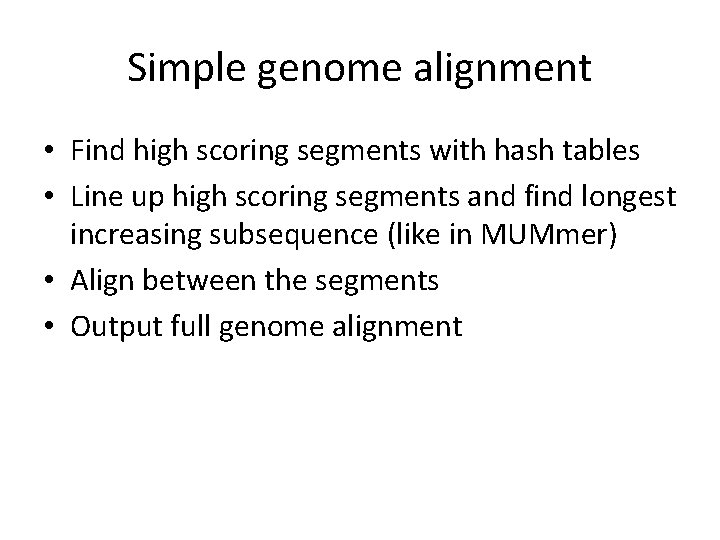
Simple genome alignment • Find high scoring segments with hash tables • Line up high scoring segments and find longest increasing subsequence (like in MUMmer) • Align between the segments • Output full genome alignment

Programs and experimental comparison • Alignathon: assessment of whole genome sequence alignment programs on simulated data • Several genome alignment programs were used • Since the data is simulated we know the true alignment and this can calculate the accuracy • See paper on website for overview

Exact genome alignment • Alignment of divergence sequences is challenging • How would an exact alignment method fare in comparison to traditional methods? • We compare a parallel GPU approach here to two popular methods.

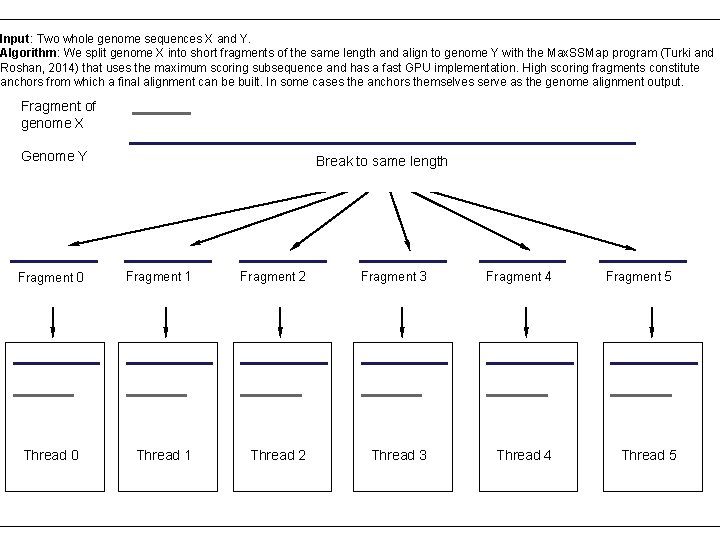
Input: Two whole genome sequences X and Y. Algorithm: We split genome X into short fragments of the same length and align to genome Y with the Max. SSMap program (Turki and Roshan, 2014) that uses the maximum scoring subsequence and has a fast GPU implementation. High scoring fragments constitute anchors from which a final alignment can be built. In some cases the anchors themselves serve as the genome alignment output. Fragment of genome X Genome Y Fragment 0 Thread 0 Break to same length Fragment 1 Fragment 2 Fragment 3 Fragment 4 Thread 1 Thread 2 Thread 3 Thread 4 Fragment 5 Thread 5

Experimental setup Simulated data: We use simulated pairwise alignments of divergent species from the Alignathon study (Earl et. al. 2014) Methods: LASTZ (Harris 2007), PECAN (Paten et. al. 2008), and GPU-EXACT (previous slide) All methods were applied without pre and post processing and with default parameters Accuracy:

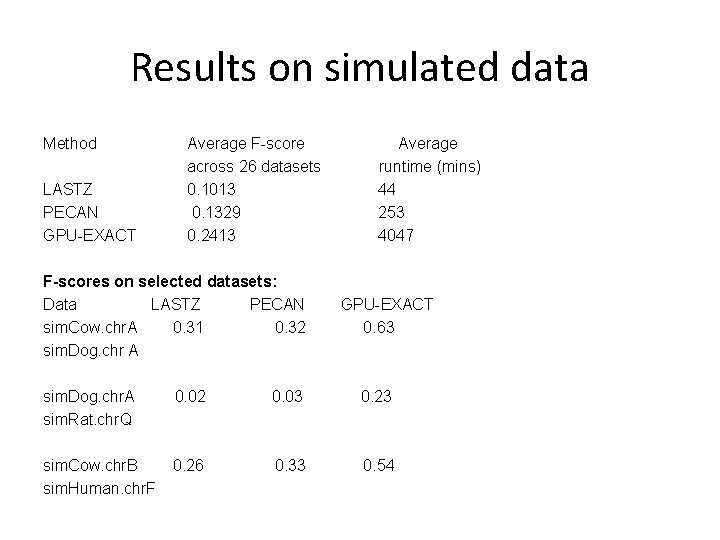
Results on simulated data Method LASTZ PECAN GPU-EXACT Average F-score across 26 datasets 0. 1013 0. 1329 0. 2413 F-scores on selected datasets: Data LASTZ PECAN sim. Cow. chr. A 0. 31 0. 32 sim. Dog. chr A Average runtime (mins) 44 253 4047 GPU-EXACT 0. 63 sim. Dog. chr. A sim. Rat. chr. Q 0. 02 0. 03 0. 23 sim. Cow. chr. B sim. Human. chr. F 0. 26 0. 33 0. 54

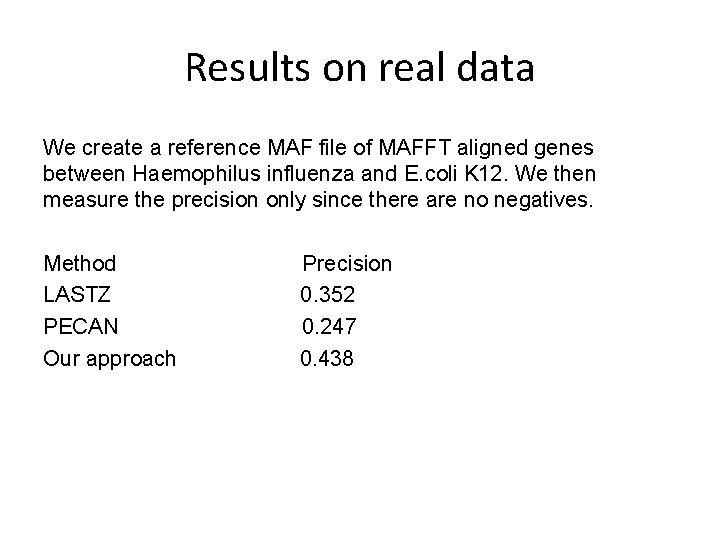
Results on real data We create a reference MAF file of MAFFT aligned genes between Haemophilus influenza and E. coli K 12. We then measure the precision only since there are no negatives. Method LASTZ PECAN Our approach Precision 0. 352 0. 247 0. 438