Sequencing the Maize Genome Sequencing Consortium rwilsonwatson wustl










- Slides: 17


Sequencing the Maize Genome Sequencing Consortium rwilson@watson. wustl. edu

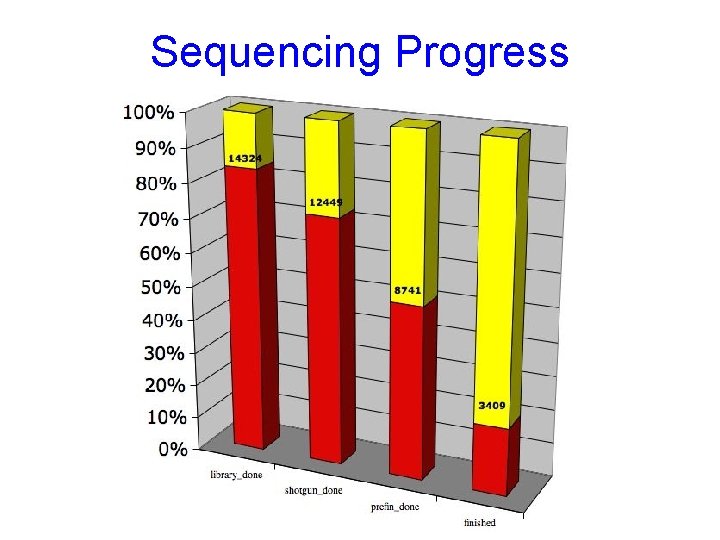
Sequencing Progress

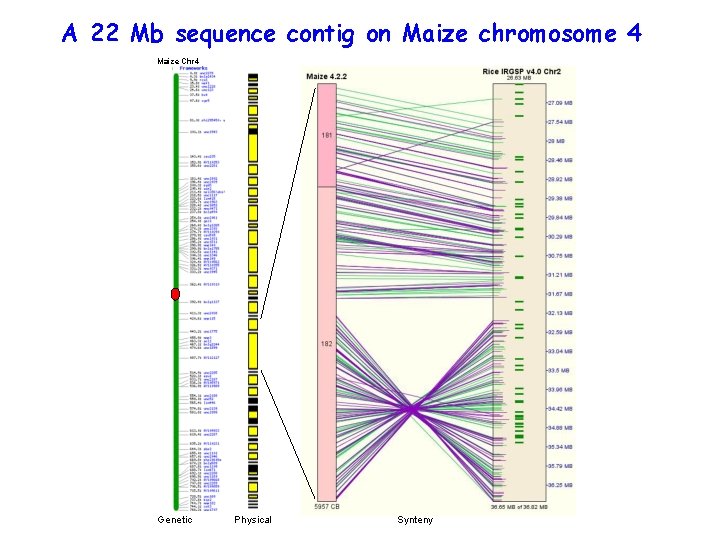
A 22 Mb sequence contig on Maize chromosome 4 Maize Chr 4 Genetic Physical Synteny

• Plans & Milestones 22 Mb contig on chromosome 4 - Analysis & publication • Draft sequence of the maize genome - All BACs: shotgun & pre-finishing (? ) - End of the calendar year - Announce at the Maize Meeting in D. C. • Completion of the maize genome sequence - Version 1. 0 - Analysis & Publication • Future Work - Secondary Annotation - Clean-up sequencing, maintenance

Today’s Agenda 9: 00 – 9: 15 am 9: 15 – 10: 30 am Introductions and Project Overview (Rick Wilson) Progress Updates – WU/AGI/CSHL/ISU Project - Map and Tile Path (Rod Wing) - Library Construction and Production (Lucinda Fulton) - Sequence Improvement (Bob Fulton, Dick Mc. Combie, Rod Wing) - Data Submission (Joanne Nelson) 10: 30 - 10: 45 am 10: 45 – Noon Break Progress Updates-continued - Assembly Improvement (Pat Schnable) - Annotation and Data Display (Doreen Ware) - DOE Project (Dan Rohksar) - Outreach (Dick Mc. Combie, Doreen Ware) Noon – 1: 00 pm 1: 00 – 2: 00 pm Executive Session-Working Lunch Discussion 2: 00 pm Depart for Airport

Maize Genome Sequencing at Arizona Rod A. Wing Arizona Genomics Institute BIO 5 Department of Plant Sciences University of Arizona

BAC by BAC Strategy to Sequence the Maize Genome Maize B 73 Genome (2300 Mb) Fingerprinting ~460, 000 BACs Genetic Anchoring in silico, overgo hybridization (19, 292) BAC End Sequencing ~800, 000 BAC physical maps (HICF & Agarose) FPC databases STC database (Agarose and HICF) Choose a seed BAC (800 Kb spacing) Shotgun sequencing and finishing STC database search, FP comparison Determine minimum overlap BACs Complete maize genome sequence Framework BAC library construction (Hind III, Eco. R I, Mbo. I ; 27 X genome coverage (~150 kb inserts)

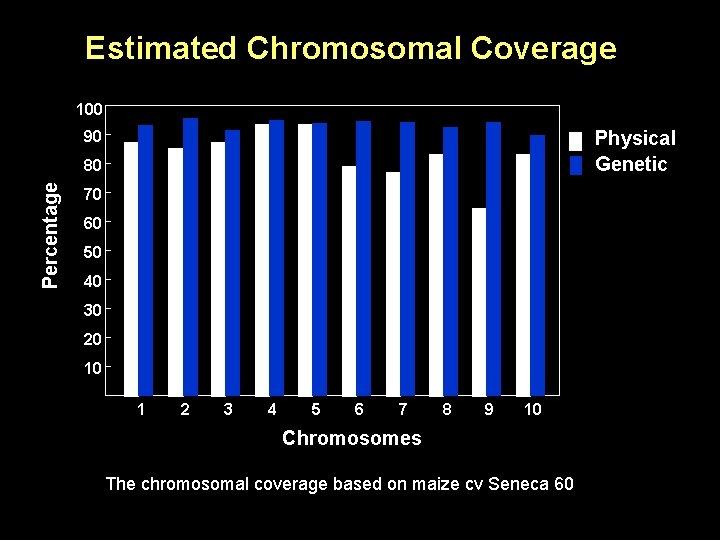
Estimated Chromosomal Coverage 100 Physical Genetic 90 Percentage 80 70 60 50 40 30 20 10 1 2 3 4 5 6 7 8 9 10 Chromosomes The chromosomal coverage based on maize cv Seneca 60

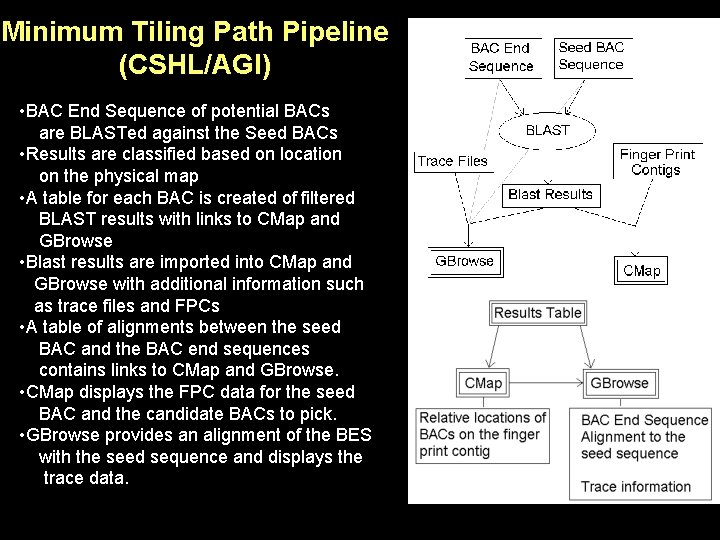
Minimum Tiling Path Pipeline (CSHL/AGI) • BAC End Sequence of potential BACs are BLASTed against the Seed BACs • Results are classified based on location on the physical map • A table for each BAC is created of filtered BLAST results with links to CMap and GBrowse • Blast results are imported into CMap and GBrowse with additional information such as trace files and FPCs • A table of alignments between the seed BAC and the BAC end sequences contains links to CMap and GBrowse. • CMap displays the FPC data for the seed BAC and the candidate BACs to pick. • GBrowse provides an alignment of the BES with the seed sequence and displays the trace data.

Clone Picking Progress • Seed BACs: 3, 400, complete • Clone Walking from Seed BACs: 12, 824 complete • Total clones picked = 16, 224 (169 96 -well plates) • 15, 400 successful • 7, 800 Year 1 • 7, 600 Year 2 • Gap-filling • ~600 Year 3, in progress

Clone Picking • Clone Walking – By sequence if seed BAC sequence was available – By fingerprints when no sequence was available • Clone verification – BAC end sequence – Seed BAC sequence

Library Picking • 60 cycles to look through 1, 221 384 -well plates for 16, 320 clones

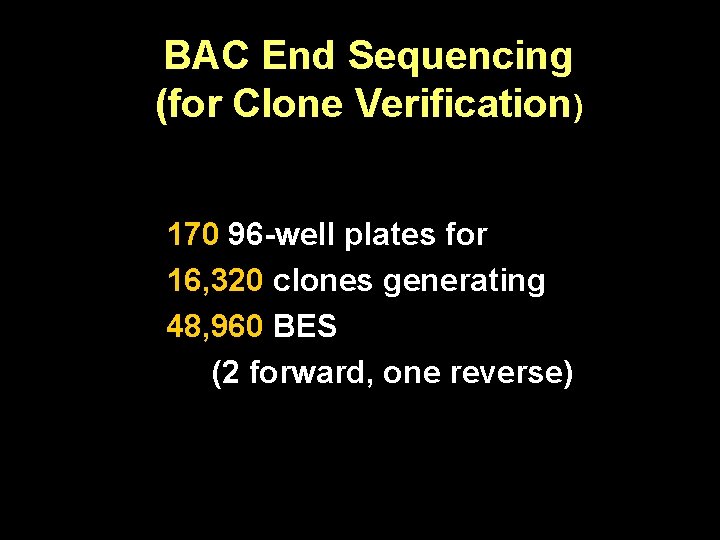
BAC End Sequencing (for Clone Verification) 170 96 -well plates for 16, 320 clones generating 48, 960 BES (2 forward, one reverse)

DNA Preparation and Shearing 170 96 -well plates for 16, 320 clones 10 plates each month 2. 5 plates person

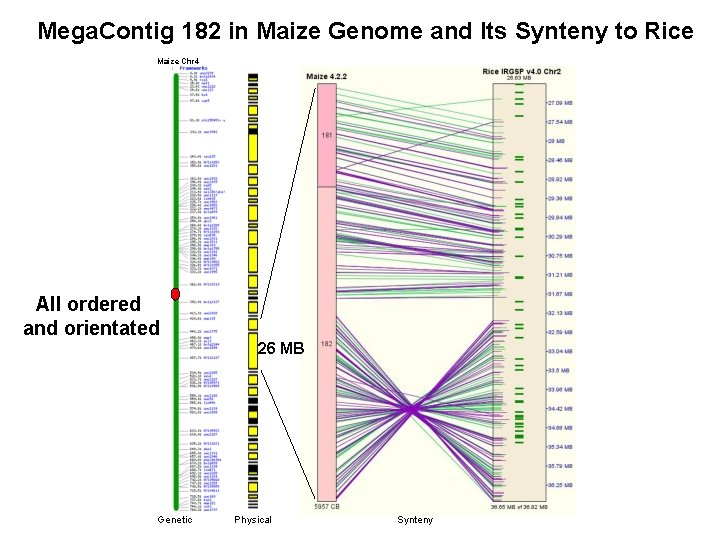
Mega. Contig 182 in Maize Genome and Its Synteny to Rice Maize Chr 4 All ordered and orientated 26 MB Genetic Physical Synteny

Maize Pseudomolecules for Rice Syntenic Chr 3 S 6. 9 Mb 7. 2 Mb (1. 5 gap/BAC) (1. 7 gap/BAC) Maize Chr 9 L Rice Chr 3 S Maize Chr 1 S