Sequence alignment with NeedlemanWunsch Usman Roshan Pairwise alignment



![NW pseudocode Initialization: for i = 1 to length of seq 1 { V[i][0] NW pseudocode Initialization: for i = 1 to length of seq 1 { V[i][0]](https://slidetodoc.com/presentation_image_h2/41c6723ac5fa54cdbe0a4576ac1d98e0/image-11.jpg)
- Slides: 12

Sequence alignment with Needleman-Wunsch Usman Roshan

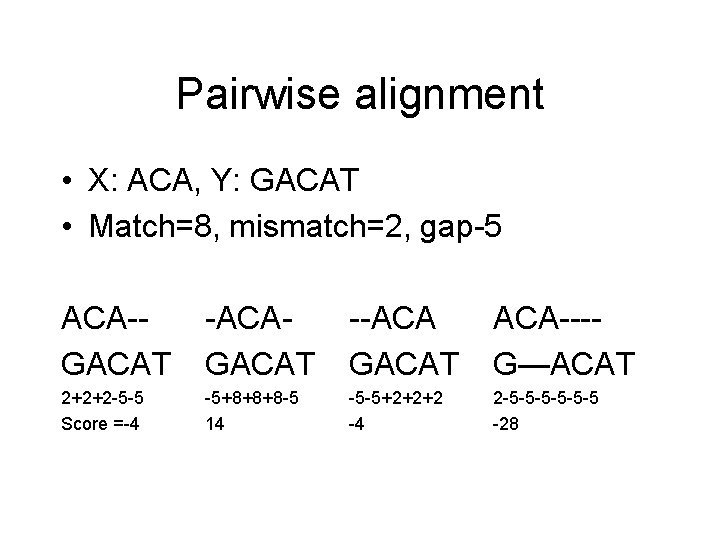
Pairwise alignment • X: ACA, Y: GACAT • Match=8, mismatch=2, gap-5 ACA-GACAT -ACAGACAT --ACA GACAT ACA---G—ACAT 2+2+2 -5 -5 Score =-4 -5+8+8+8 -5 14 -5 -5+2+2+2 -4 2 -5 -5 -5 -28

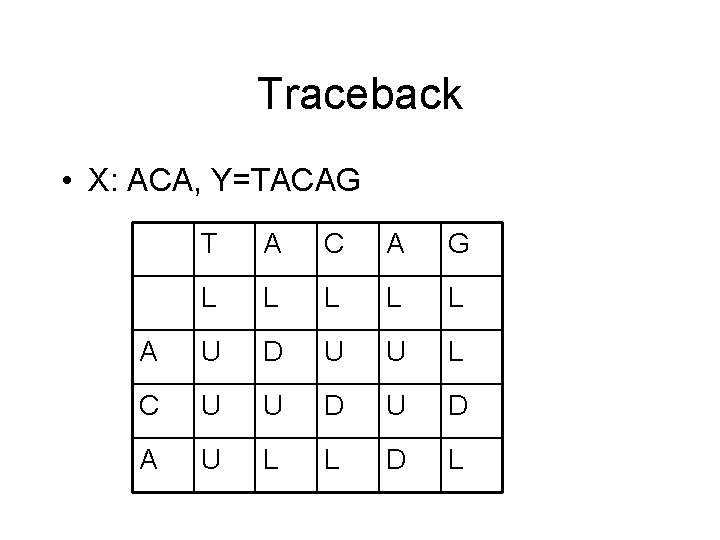
Traceback • We can compute an alignment of DNA (or protein or RNA) sequences X and Y with a traceback matrix T. • Sequence X is aligned along the rows and Y along the columns. • Each entry of the matrix T contains D, L, or U specifying diagonal, left or upper

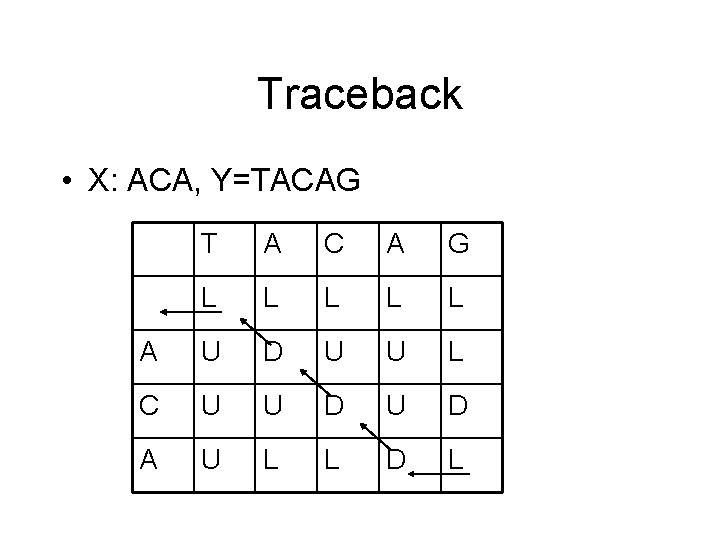
Traceback • X: ACA, Y=TACAG T A C A G L L L A U D U U L C U U D A U L L D L

Traceback • X: ACA, Y=TACAG T A C A G L L L A U D U U L C U U D A U L L D L

Traceback code aligned_seq 1 = "" aligned_seq 2 = "" i = len(seq 1) j = len(seq 2) while(i !=0 or j != 0): if(T[i][j] == “L”): aligned_seq 1 = “-” + aligned_seq 1 aligned_seq 2 = seq 2[j-1] + aligned_seq 2 j=j-1 elif(T[i][j] == "U"): aligned_seq 2 = "-" + aligned_seq 2 aligned_seq 1 = seq 1[i-1] + aligned_seq 1 i=i-1 else: aligned_seq 1 = seq 1[i-1] + aligned_seq 1 aligned_seq 2 = seq 2[j-1] + aligned_seq 2 i=i-1 j=j-1

Optimal alignment • An alignment can be specified by the traceback matrix. • How do we determine the traceback for the highest scoring alignment? • Needleman-Wunsch algorithm for global alignment – First proposed in 1970 – Widely used in genomics/bioinformatics – Dynamic programming algorithm

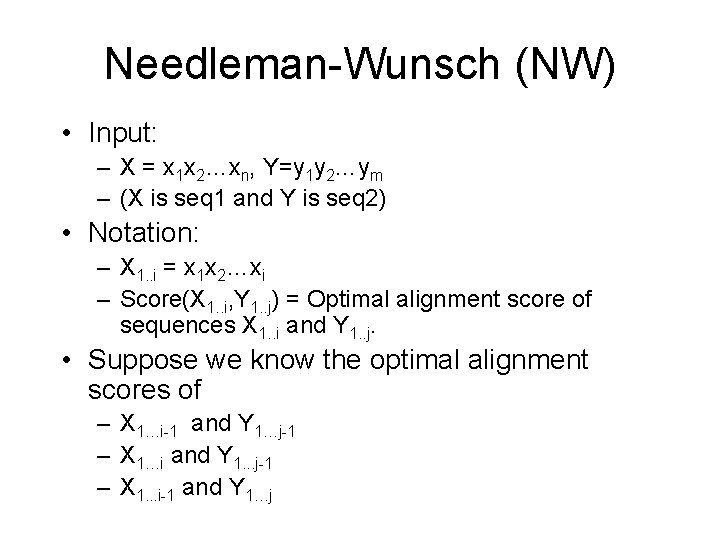
Needleman-Wunsch (NW) • Input: – X = x 1 x 2…xn, Y=y 1 y 2…ym – (X is seq 1 and Y is seq 2) • Notation: – X 1. . i = x 1 x 2…xi – Score(X 1. . i, Y 1. . j) = Optimal alignment score of sequences X 1. . i and Y 1. . j. • Suppose we know the optimal alignment scores of – X 1…i-1 and Y 1…j-1 – X 1…i and Y 1. . . j-1 – X 1. . . i-1 and Y 1…j

Needleman-Wunsch (NW) • Then the optimal alignment score of X 1…i and Y 1…j is the maximum of – Score(X 1…i-1, Y 1…j-1) + match/mismatch – Score(X 1…i, Y 1…j-1) + gap – Score(X 1…i-1, Y 1…j) + gap • We build on this observation to compute Score(Xn, Ym)

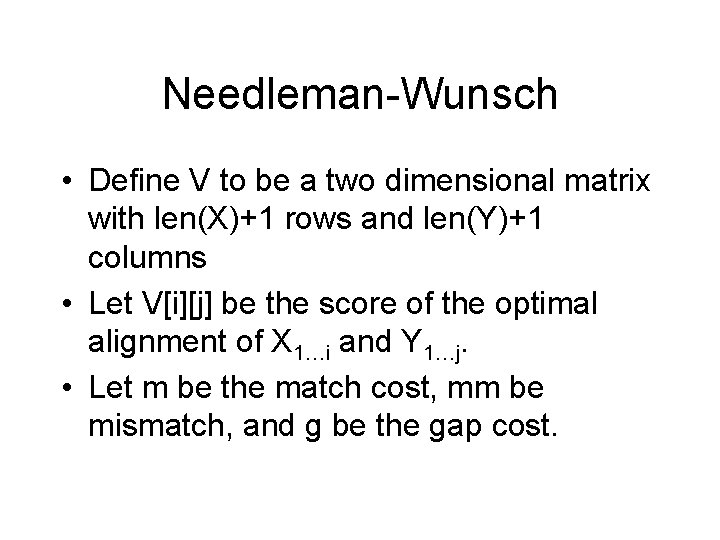
Needleman-Wunsch • Define V to be a two dimensional matrix with len(X)+1 rows and len(Y)+1 columns • Let V[i][j] be the score of the optimal alignment of X 1…i and Y 1…j. • Let m be the match cost, mm be mismatch, and g be the gap cost.
![NW pseudocode Initialization for i 1 to length of seq 1 Vi0 NW pseudocode Initialization: for i = 1 to length of seq 1 { V[i][0]](https://slidetodoc.com/presentation_image_h2/41c6723ac5fa54cdbe0a4576ac1d98e0/image-11.jpg)
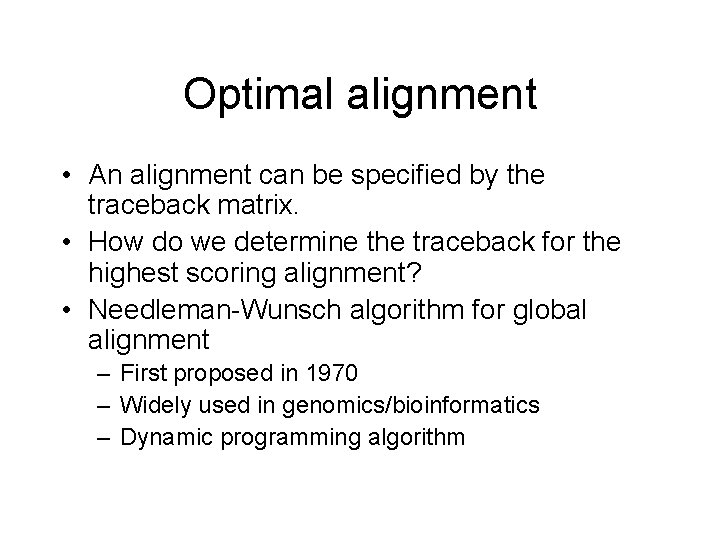
NW pseudocode Initialization: for i = 1 to length of seq 1 { V[i][0] = i*g; } For i = 1 to length of seq 2 { V[0][i] = i*g; } Recurrence: for i = 1 to length of seq 1{ for j = 1 to length of seq 2{ V[i-1][j-1] + m(or mm) V[i][j] = max { V[i-1][j] + g V[i][j-1] + g if(maximum is V[i-1][j-1] + m(or mm)) then T[i][j] = ‘D’ else if (maximum is V[i-1][j] + g) then T[i][j] = ‘U’ else then T[i][j] = ‘L’ } }

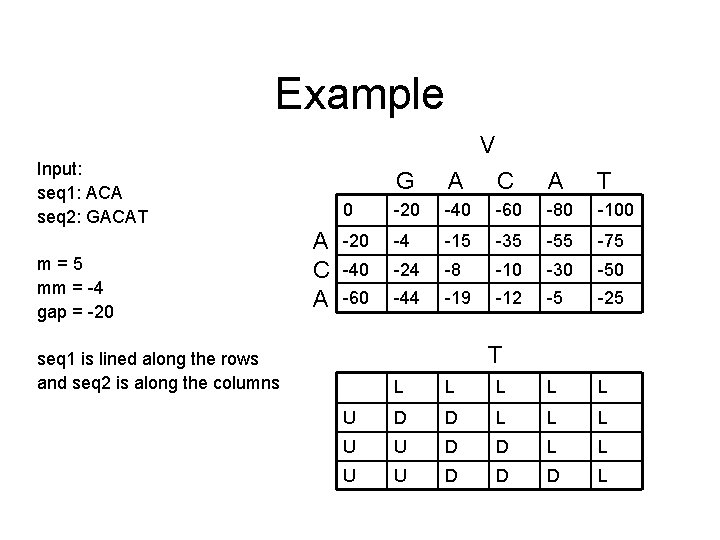
Example V Input: seq 1: ACA seq 2: GACAT m=5 mm = -4 gap = -20 A C A G A C A T 0 -20 -40 -60 -80 -100 -20 -4 -15 -35 -55 -75 -40 -24 -8 -10 -30 -50 -60 -44 -19 -12 -5 -25 T seq 1 is lined along the rows and seq 2 is along the columns L L L U D D L L L U U D D D L