The Whole Genome Sequencing Revolution Martin Wiedmann Gellert
















- Slides: 32

The Whole Genome Sequencing Revolution Martin Wiedmann Gellert Family Professor of Food Safety Department of Food Science Cornell University, Ithaca, NY E-mail: mw 16@cornell. edu Phone: 607 -254 -2838

Outline • Subtyping for disease surveillance: from PFGE to WGS • WGS challenges: when are two isolates the same or different? Can we find identical isolates in different locations? • Looking in the future

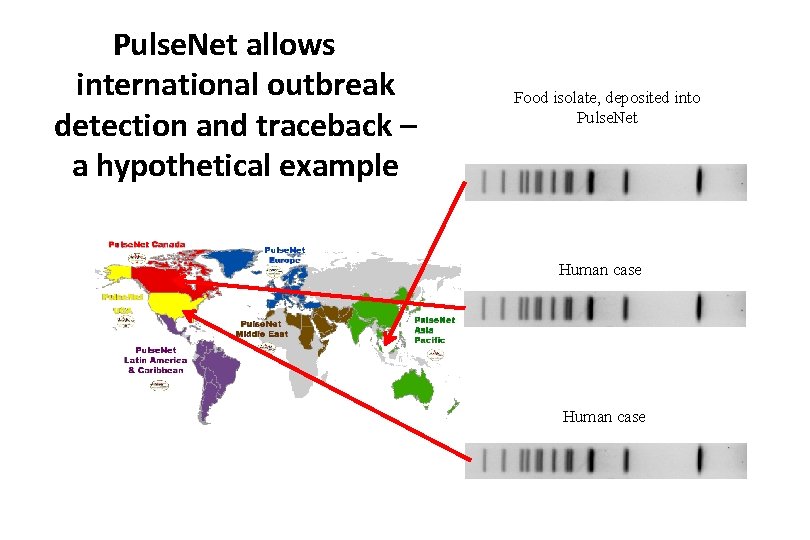
Pulse. Net allows international outbreak detection and traceback – a hypothetical example Food isolate, deposited into Pulse. Net Human case


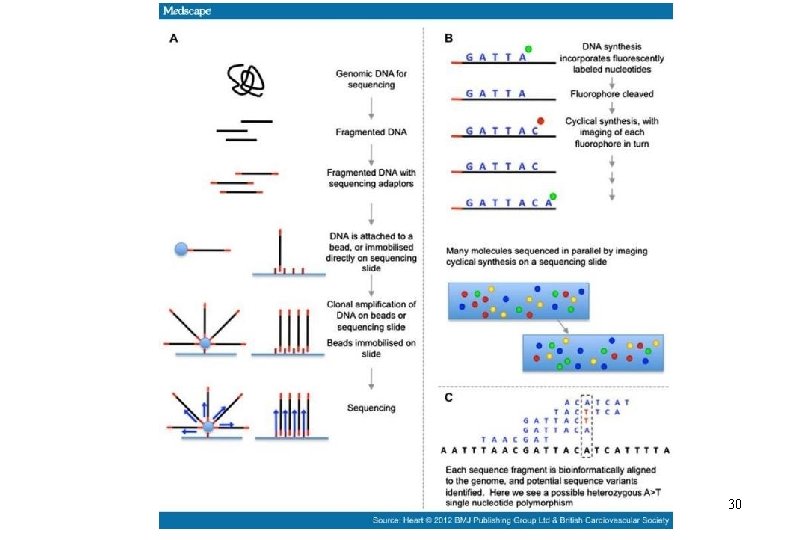
Whole Genome Sequencing • It all started with the human genome project • Sequencing of a bacterial genome is now feasible at costs of <$100/isolate • Costs will continue to drop • Commonly used platforms include • Roche 454 • Illumina Hi. Seq/Mi. Seq • Applied Biosystems SOLi. D Systems • Life Technologies/Thermofisher Ion Torrent; • Pac. Bio RS • Nanopore based systems (e. g. , Oxford Nanopore Min. ION)

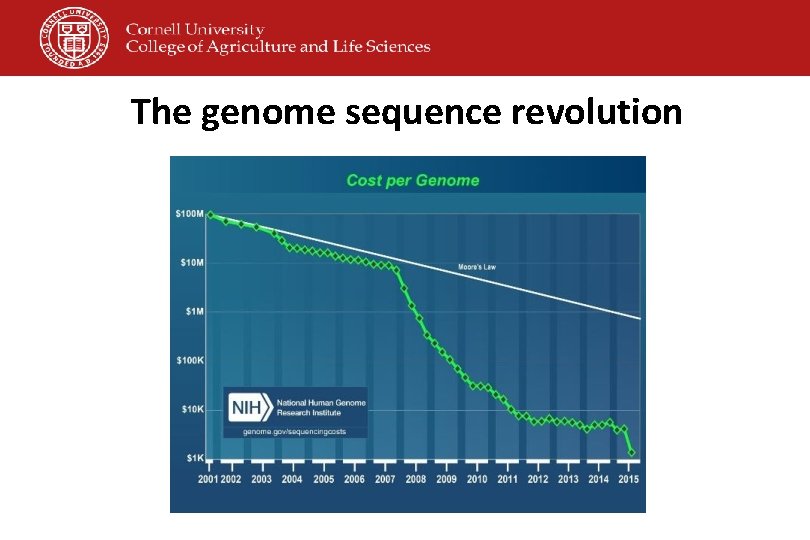
The genome sequence revolution

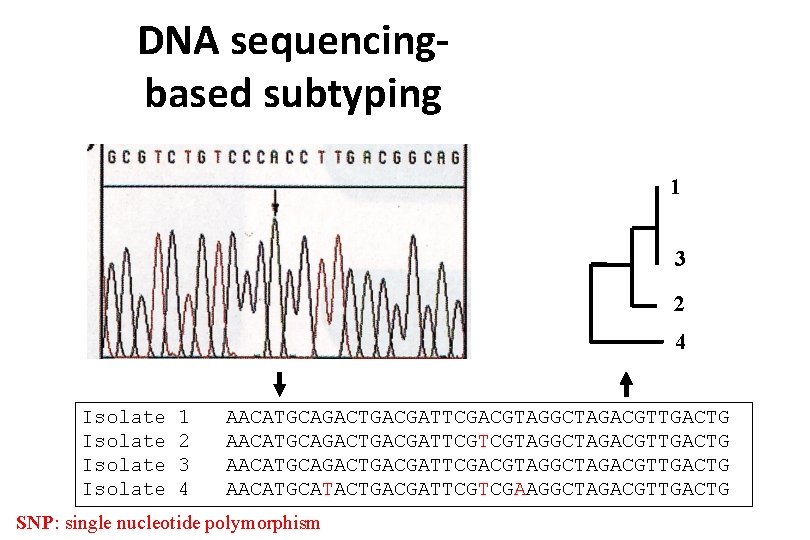
DNA sequencingbased subtyping 1 3 2 4 Isolate 1 2 3 4 AACATGCAGACTGACGATTCGACGTAGGCTAGACGTTGACTG AACATGCAGACTGACGATTCGTCGTAGGCTAGACGTTGACTG AACATGCAGACTGACGATTCGACGTAGGCTAGACGTTGACTG AACATGCATACTGACGATTCGTCGAAGGCTAGACGTTGACTG SNP: single nucleotide polymorphism

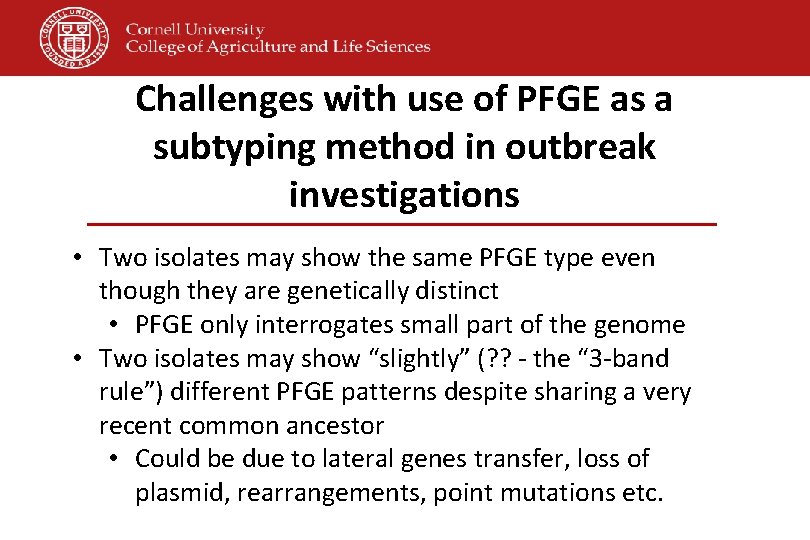
Challenges with use of PFGE as a subtyping method in outbreak investigations • Two isolates may show the same PFGE type even though they are genetically distinct • PFGE only interrogates small part of the genome • Two isolates may show “slightly” (? ? - the “ 3 -band rule”) different PFGE patterns despite sharing a very recent common ancestor • Could be due to lateral genes transfer, loss of plasmid, rearrangements, point mutations etc.

Xbal Spe. I Includes isolates form Salmonella outbreak linked to sausages (Rhode Island) and isolates from pistachios L Den Bakker et al. 2011. AEM.

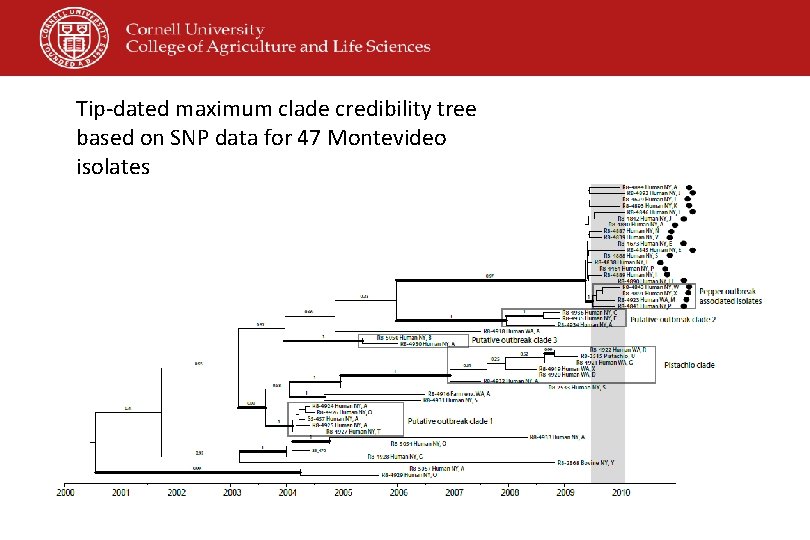
Tip-dated maximum clade credibility tree based on SNP data for 47 Montevideo isolates

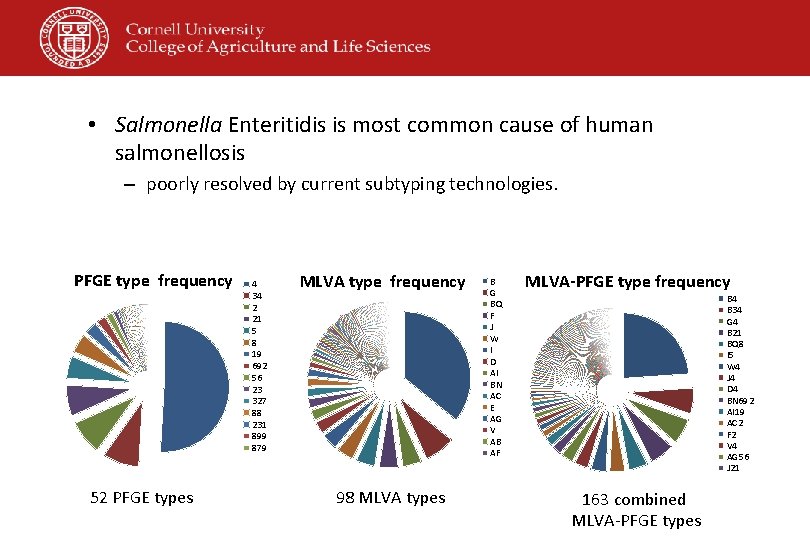
• Salmonella Enteritidis is most common cause of human salmonellosis – poorly resolved by current subtyping technologies. PFGE type frequency 52 PFGE types 4 34 2 21 5 8 19 692 56 23 327 88 231 899 879 MLVA type frequency 98 MLVA types B G BQ F J W I D AI BN AC E AG V AB AF MLVA-PFGE type frequency B 4 B 34 G 4 B 21 BQ 8 I 5 W 4 J 4 D 4 BN 692 AI 19 AC 2 F 2 V 4 AG 56 J 21 163 combined MLVA-PFGE types


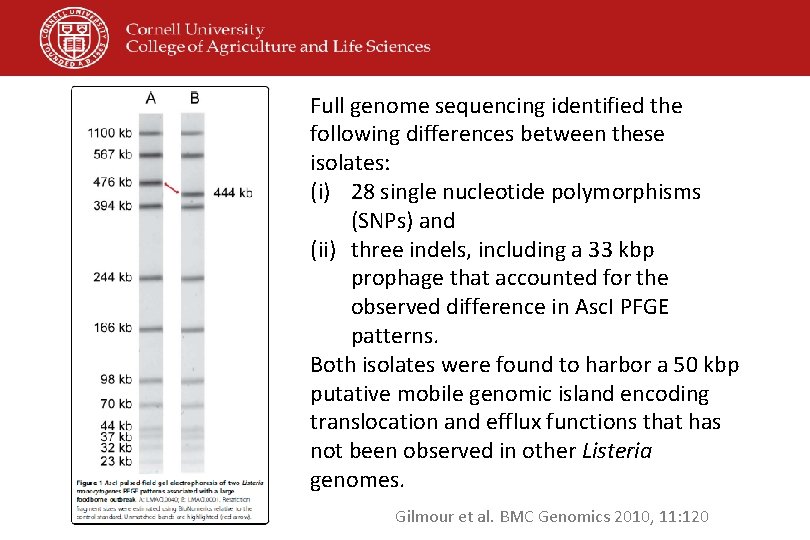
Full genome sequencing identified the following differences between these isolates: (i) 28 single nucleotide polymorphisms (SNPs) and (ii) three indels, including a 33 kbp prophage that accounted for the observed difference in Asc. I PFGE patterns. Both isolates were found to harbor a 50 kbp putative mobile genomic island encoding translocation and efflux functions that has not been observed in other Listeria genomes. Gilmour et al. BMC Genomics 2010, 11: 120


In addition, whole genome sequencing showed that 5 Listeria isolates collected in 2010 from the same facility were also closely related genetically to isolates from ill people.

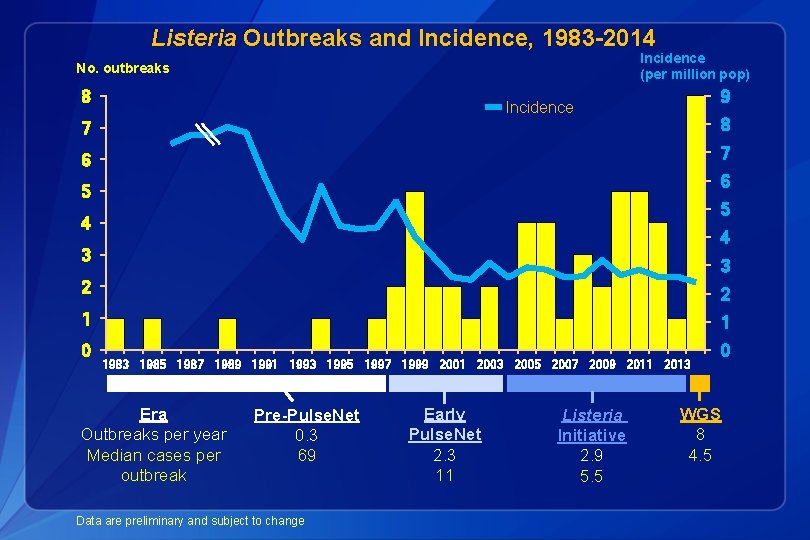
Listeria Outbreaks and Incidence, 1983 -2014 Incidence (per million pop) No. outbreaks 8 9 Incidence 7 8 6 7 6 5 5 4 4 3 3 2 2 1 1 0 1983 1985 1987 1989 1991 1993 1995 1997 1999 2001 2003 2005 2007 2009 2011 2013 Era Outbreaks per year Median cases per outbreak Pre-Pulse. Net 0. 3 69 Data are preliminary and subject to change Early Pulse. Net 2. 3 11 Listeria Initiative 2. 9 5. 5 0 WGS 8 4. 5

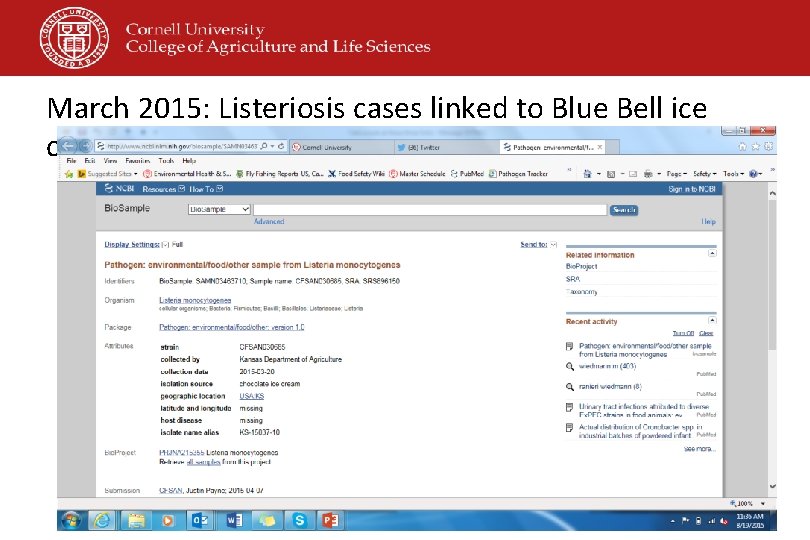
March 2015: Listeriosis cases linked to Blue Bell ice cream

Outline • Subtyping for disease surveillance: from PFGE to WGS • WGS challenges: when are two isolates the same or different? Can we find identical isolates in different locations? • Looking in the future

The challenge • Identical bacteria (100% match over the whole genome) can be found in different places that can be potential sources of foodborne disease outbreaks

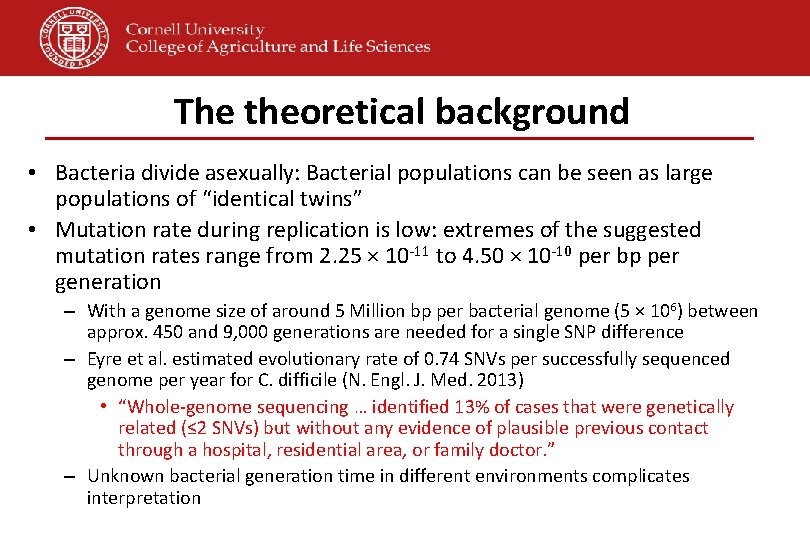
The theoretical background • Bacteria divide asexually: Bacterial populations can be seen as large populations of “identical twins” • Mutation rate during replication is low: extremes of the suggested mutation rates range from 2. 25 × 10 -11 to 4. 50 × 10 -10 per bp per generation – With a genome size of around 5 Million bp per bacterial genome (5 × 106) between approx. 450 and 9, 000 generations are needed for a single SNP difference – Eyre et al. estimated evolutionary rate of 0. 74 SNVs per successfully sequenced genome per year for C. difficile (N. Engl. J. Med. 2013) • “Whole-genome sequencing … identified 13% of cases that were genetically related (≤ 2 SNVs) but without any evidence of plausible previous contact through a hospital, residential area, or family doctor. ” – Unknown bacterial generation time in different environments complicates interpretation

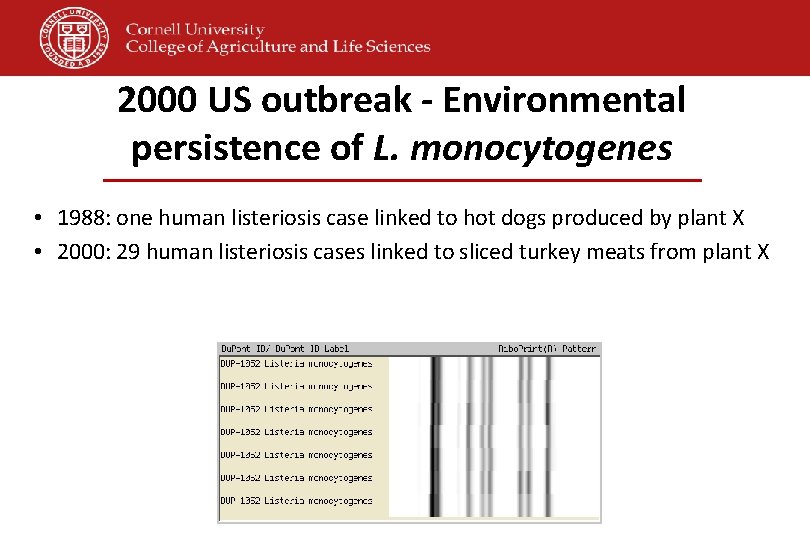
2000 US outbreak - Environmental persistence of L. monocytogenes • 1988: one human listeriosis case linked to hot dogs produced by plant X • 2000: 29 human listeriosis cases linked to sliced turkey meats from plant X

Real world observations


Real world observations In one case, isolates with < 3 SNP differences were found in retail delis in there different states

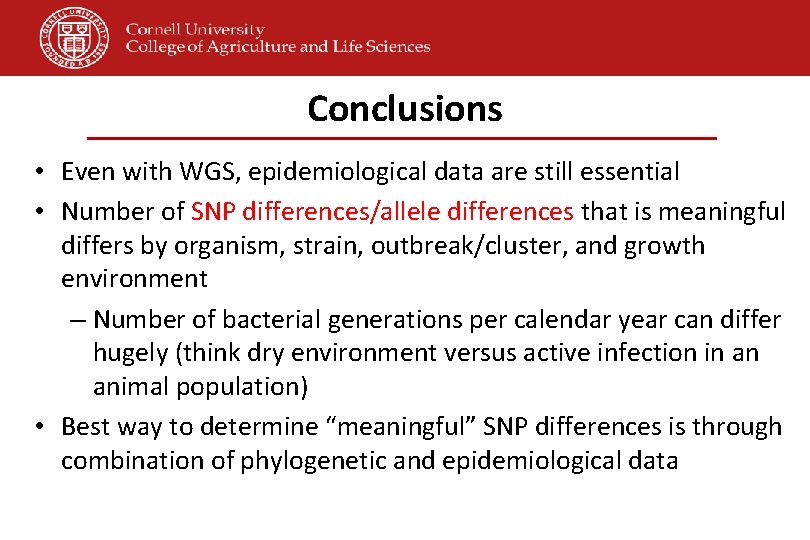
Conclusions • Even with WGS, epidemiological data are still essential • Number of SNP differences/allele differences that is meaningful differs by organism, strain, outbreak/cluster, and growth environment – Number of bacterial generations per calendar year can differ hugely (think dry environment versus active infection in an animal population) • Best way to determine “meaningful” SNP differences is through combination of phylogenetic and epidemiological data

Looking in the future • WGS will get cheaper and will be used more – STEC next, probably Salmonella Enteritidis after that – Detection of more clusters and outbreaks • WGS database will grow rapidly with inclusion of environmental isolates – More outbreak will be linked to source by using WGS matches between food or environmental isolates and human isolates as stating point • More broad application of WGS by private labs, maybe customers and consumers?


Conclusions • WGS is a game changer and will significantly improve detection of outbreaks, adulteration, etc. – False alarms will occur though • Pathogen detection in environments, by regulatory agencies, will lead to inclusion of WGS data in CDC/FDA/USDA databases (Genome. Trakr) – Environmental pathogen monitoring by industry will become even more important


30

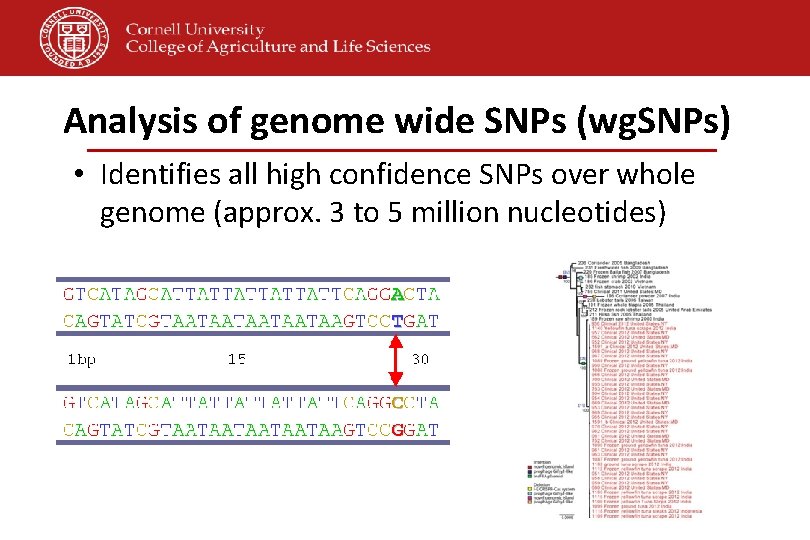
Analysis of genome wide SNPs (wg. SNPs) • Identifies all high confidence SNPs over whole genome (approx. 3 to 5 million nucleotides)

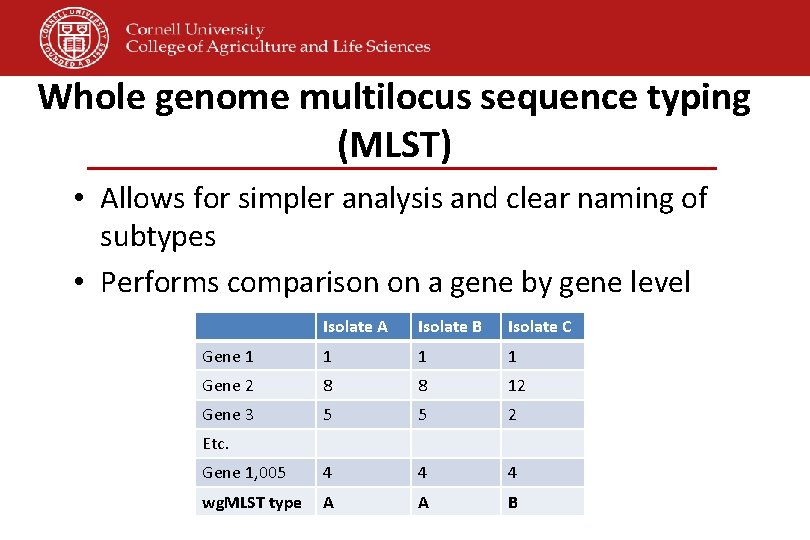
Whole genome multilocus sequence typing (MLST) • Allows for simpler analysis and clear naming of subtypes • Performs comparison on a gene by gene level Isolate A Isolate B Isolate C Gene 1 1 Gene 2 8 8 12 Gene 3 5 5 2 Gene 1, 005 4 4 4 wg. MLST type A A B Etc.
 Shotgun sequencing
Shotgun sequencing Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Perpartes
Perpartes Whole school whole community whole child model
Whole school whole community whole child model Genome-to-genome distance calculator
Genome-to-genome distance calculator Dna
Dna Genome sequencing
Genome sequencing Human genome project
Human genome project Franz josef gellert
Franz josef gellert Ziemi stengel
Ziemi stengel Dr gellért julianna
Dr gellért julianna Margaret wiedmann md
Margaret wiedmann md Massed practice
Massed practice Russian revolution vs french revolution
Russian revolution vs french revolution Modern commercial agriculture
Modern commercial agriculture You should hope that this game will be over soon
You should hope that this game will be over soon Savant genome browser
Savant genome browser Human genome project
Human genome project What is a genome
What is a genome Artemis genome
Artemis genome Yale university poster
Yale university poster Eukaryotic genome
Eukaryotic genome National human genome research institute
National human genome research institute Sickle cell karyotype
Sickle cell karyotype Genome modification ustaz auni
Genome modification ustaz auni Human genome size
Human genome size Epidna
Epidna Genome adalah
Genome adalah Genome mapping
Genome mapping Genome
Genome Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Satellite dna
Satellite dna Genome project
Genome project