TTT TTC TTA TTG CTT CTC CTA CTG















































- Slides: 81






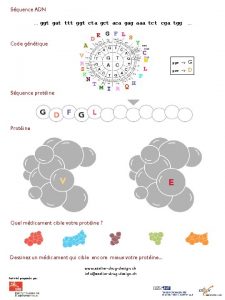
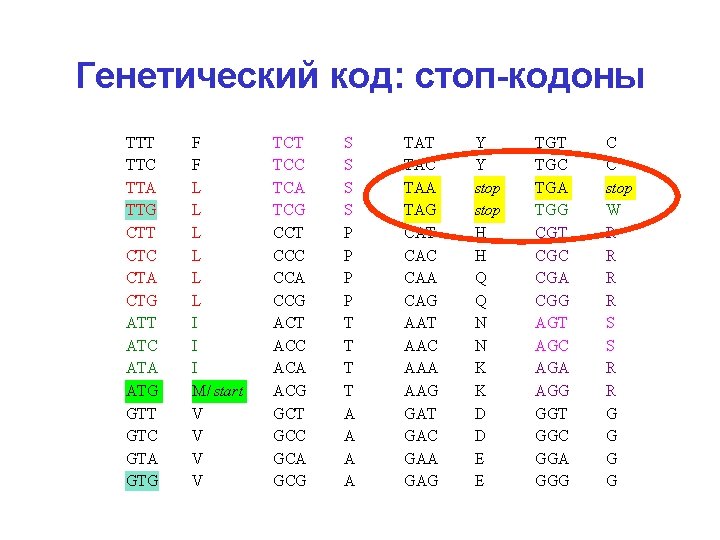
Генетический код: стоп-кодоны TTT TTC TTA TTG CTT CTC CTA CTG ATT ATC ATA ATG GTT GTC GTA GTG F F L L L I I I M/ start V V TCT TCC TCA TCG CCT CCC CCA CCG ACT ACC ACA ACG GCT GCC GCA GCG S S P P T T A A TAT TAC TAA TAG CAT CAC CAA CAG AAT AAC AAA AAG GАT GАC GАA GАG Y Y stop H H Q Q N N K K D D E E TGT TGC TGA TGG CGT CGC CGA CGG AGT AGC AGA AGG GGT GGC GGA GGG C C stop W R R S S R R G G



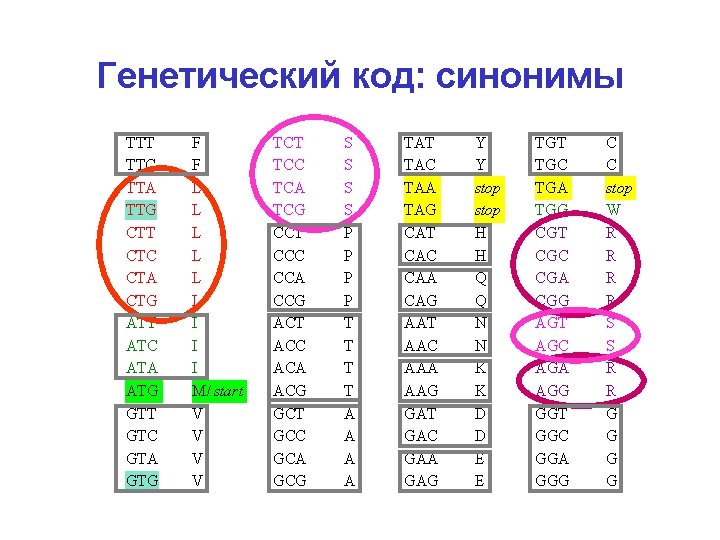
Генетический код: синонимы TTT TTC TTA TTG CTT CTC CTA CTG ATT ATC ATA ATG GTT GTC GTA GTG F F L L L I I I M/ start V V TCT TCC TCA TCG CCT CCC CCA CCG ACT ACC ACA ACG GCT GCC GCA GCG S S P P T T A A TAT TAC TAA TAG CAT CAC CAA CAG AAT AAC AAA AAG GАT GАC GАA GАG Y Y stop H H Q Q N N K K D D E E TGT TGC TGA TGG CGT CGC CGA CGG AGT AGC AGA AGG GGT GGC GGA GGG C C stop W R R S S R R G G


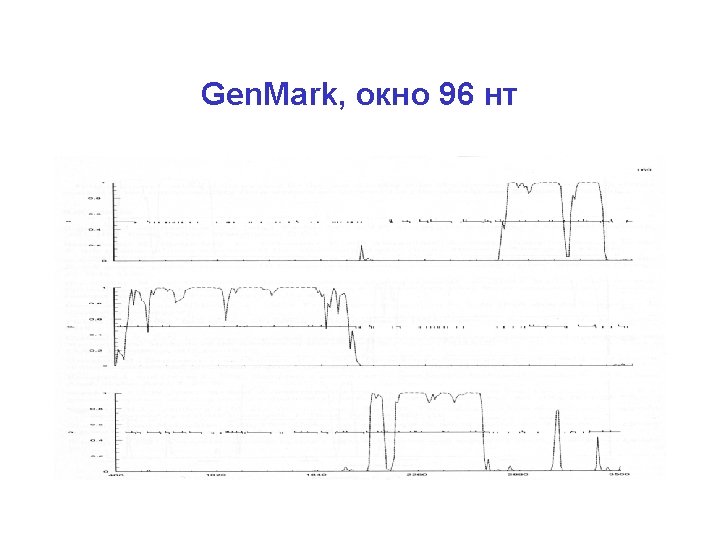
Gen. Mark, окно 96 нт

Генетический код: старт-кодоны TTT TTC TTA TTG CTT CTC CTA CTG ATT ATC ATA ATG GTT GTC GTA GTG F F L L L I I I M/ start V V TCT TCC TCA TCG CCT CCC CCA CCG ACT ACC ACA ACG GCT GCC GCA GCG S S P P T T A A TAT TAC TAA TAG CAT CAC CAA CAG AAT AAC AAA AAG GАT GАC GАA GАG Y Y stop H H Q Q N N K K D D E E TGT TGC TGA TGG CGT CGC CGA CGG AGT AGC AGA AGG GGT GGC GGA GGG C C stop W R R S S R R G G

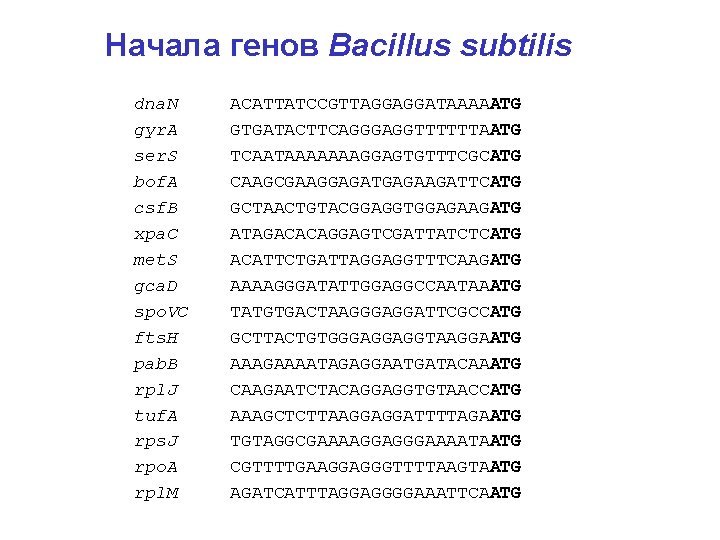
Начала генов Bacillus subtilis dna. N ACATTATCCGTTAGGAGGATAAAAATG gyr. A GTGATACTTCAGGGAGGTTTTTTAATG ser. S TCAATAAAAAAAGGAGTGTTTCGCATG bof. A CAAGCGAAGGAGATGAGAAGATTCATG csf. B GCTAACTGTACGGAGGTGGAGAAGATG xpa. C ATAGACACAGGAGTCGATTATCTCATG met. S ACATTCTGATTAGGAGGTTTCAAGATG gca. D AAAAGGGATATTGGAGGCCAATAAATG spo. VC TATGTGACTAAGGGAGGATTCGCCATG fts. H GCTTACTGTGGGAGGAGGTAAGGAATG pab. B AAAGAAAATAGAGGAATGATACAAATG rpl. J CAAGAATCTACAGGAGGTGTAACCATG tuf. A AAAGCTCTTAAGGAGGATTTTAGAATG rps. J TGTAGGCGAAAAGGAGGGAAAATAATG rpo. A CGTTTTGAAGGAGGGTTTTAAGTAATG rpl. M AGATCATTTAGGAGGGGAAATTCAATG

Участок связывания рибосом dna. N ACATTATCCGTTAGGAGGATAAAAATG gyr. A GTGATACTTCAGGGAGGTTTTTTAATG ser. S TCAATAAAAAAAGGAGTGTTTCGCATG bof. A CAAGCGAAGGAGATGAGAAGATTCATG csf. B GCTAACTGTACGGAGGTGGAGAAGATG xpa. C ATAGACACAGGAGTCGATTATCTCATG met. S ACATTCTGATTAGGAGGTTTCAAGATG gca. D AAAAGGGATATTGGAGGCCAATAAATG spo. VC TATGTGACTAAGGGAGGATTCGCCATG fts. H GCTTACTGTGGGAGGAGGTAAGGAATG pab. B AAAGAAAATAGAGGAATGATACAAATG rpl. J CAAGAATCTACAGGAGGTGTAACCATG tuf. A AAAGCTCTTAAGGAGGATTTTAGAATG rps. J TGTAGGCGAAAAGGAGGGAAAATAATG rpo. A CGTTTTGAAGGAGGGTTTTAAGTAATG rpl. M AGATCATTTAGGAGGGGAAATTCAATG

Сравнительный анализ (один и тот же ген в нескольких геномах) Гены консервативнее, чем межгенные области (точнее, особенности эволюции другие) Sty TCGCTCG--CAGCGGAAAGAGGATTACGCCCTTCGCCTGGAGGCTGTGCAGGGGC---GCCGGAGATGGGATGCATAATT Stm TCGCTCG--CAGCGGAAAGAGGATTACGCCCTTCGCCTGGAGGCTGTGCAGGGGC---GCCGGAGATGGGATGCATAATT Sen TCGCTCG--CAGCGGAAAGAGGATTACGCCCTTCGCCTGGAGGCTGTGCAGGGGC---GCCGGAGATGGGATGCATAATT Eco TTGCCCG--TGCCAGACGGCAGATTATCTCCCTGACCTGGTGGTTGCCCAGGAGGAGGGCCGGAAATAGGTTGTATCATT Kpn ----CGG--TGGCGCAGTGCCTGATGGG-CCTCGCCCTGGAGGACGGTCTGGCAT---ATCAGCAAGGGGGTGCGTCATG Ype TTGTTAGAACAGGGGAAAACGGTAAACAGTGTGGCATTAGATGTCGGTTATAGCT-----CCGCCTCTGCTTTTATCGCC * * * * * * * Sty AATTATCCTTTAAC-----CATAAATCTGAGCAATA-TATGCTTGGCGGCCAGATTATGGC--ACACTTGTCCGG Stm AATTATCCTTTAAC-----CATAAATCTGAGCAATA-TATGCCTGGCGGCCAGATTATGGC--ACACTTGTCCGG Sen AATTATCCTTTAAC-----CATAAATCTGAGCAATA-TATGCCTGGCGGCCAGATTATGGC--ACACTTGTCCGG Eco ACGTATCCTTATAC-----CTGAAATCTTCGCAAG--TATGCCTGGCCGCGAGATTATGGC--ACACTTGTCCGG Kpn ATTCATCCTTTCGATATCGCGGTGCTGGAACCAGGTGATGAGTATGCCTGGCGGCCAGATTATGGC--ACACTTCCCCAG Ype ATGTTTCAGCAAATAT----CGGGTACCA-CGCCTGAGCGTTTCCGGCGGGGCAATAGTGGCTTATACTAAGCCCC * ** * * *** * ** **** * *** Sty TTAACTCTCGTT-CTCAAACAG------GTACGACAGTC--GTGAAAATTCTCGTTGATGAAAATATGCCTTACGCCCGC Stm TTAACTCTCGTT-CTCAAACAG------GTACGACAGTC--GTGAAAATTCTCGTTGATGAAAATATGCCTTACGCCCGC Sen TTAACTCTCGTT-CTCAAACAG------GTACGACAGTC--GTGAAAATTCTCGTTGATGAAAATATGCCTTACGCCCGC Eco TTAACTCTCGT--CTCATACAG------GTAACACAAAC--GTGAAAATCCTTGTTGATGAAAATATGCCTTATGCCCGC Kpn TTAACTCTCGTT-CTCAGACAG------GTACTGAACT---GTGAAAATCCTCGTTGATGAAAATATGCCCGT Ype CTGTTTTTCATCTGTATGGCAGTTCGCTGTCGGAGAGTAAAGTGAAAATTCTGGTTGATGAAAATATGCCGTACGCTGAG * * ** * * *** **** ** ********* ** ** 123123123123123123123

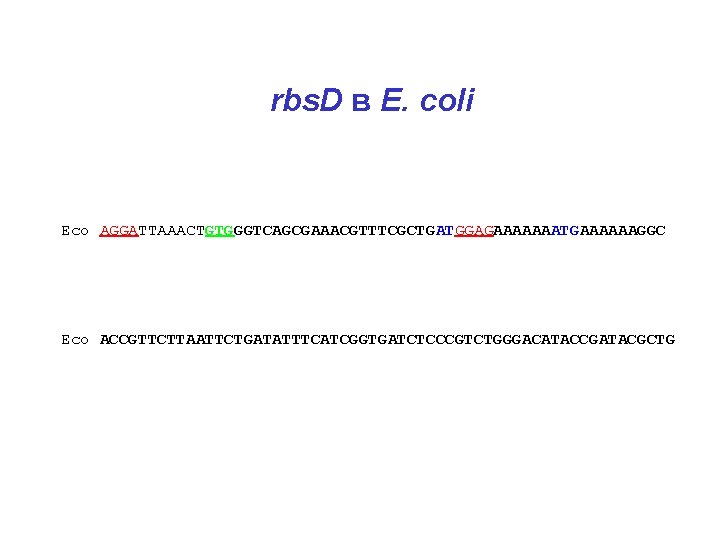
rbs. D в E. coli Eco AGGATTAAACTGTGGGTCAGCGAAACGTTTCGCTGATGGAGAAAAAAATGAAAAAAGGC Eco ACCGTTCTTAATTCTGATATTTCATCGGTGATCTCCCGTCTGGGACATACCGATACGCTG

rbs. D в энтеробактериях Sty AGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Sen AGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Stm GGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Eco AGGATTAAACTGTGGGTCAGCGAAACGTTTCGCTGATGGAGAA-AAAAATGAAAAAAGGC Ype TTTTCTAAACTCCTTGTTAGCGAAACGTTTCGCTCTTGGAGTA-GATCATGAAAAAAGGT ** ******** * * ***** Sty ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Sen ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Stm ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Eco ACCGTTCTTAATTCTGATATTTCATCGGTGATCTCCCGTCTGGGACATACCGATACGCTG Ype GTATTACTGAACGCTGATATTTCCGCGGTTATCTCCCGTCTGGGCCATACCGATCAGATT * ** ** ****** ***** *

rbs. D в энтеробактериях: ответ Sty AGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Sen AGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Stm GGGGTTACACTGCGGC-CAGCGAAACGTTTCGCTAGTGGAGCAGAAAAATGAAGAAAGGC Eco AGGATTAAACTGTGGGTCAGCGAAACGTTTCGCTGATGGAGAA-AAAAATGAAAAAAGGC Ype TTTTCTAAACTCCTTGTTAGCGAAACGTTTCGCTCTTGGAGTA-GATCATGAAAAAAGGT ** ******** * * ***** Sty ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Sen ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Stm ACCGTACTCAACTCTGAAATCTCGTCGGTCATTTCCCGTCTGGGGCATACTGATACTCTG Eco ACCGTTCTTAATTCTGATATTTCATCGGTGATCTCCCGTCTGGGACATACCGATACGCTG Ype GTATTACTGAACGCTGATATTTCCGCGGTTATCTCCCGTCTGGGCCATACCGATCAGATT * ** ** ****** ***** *



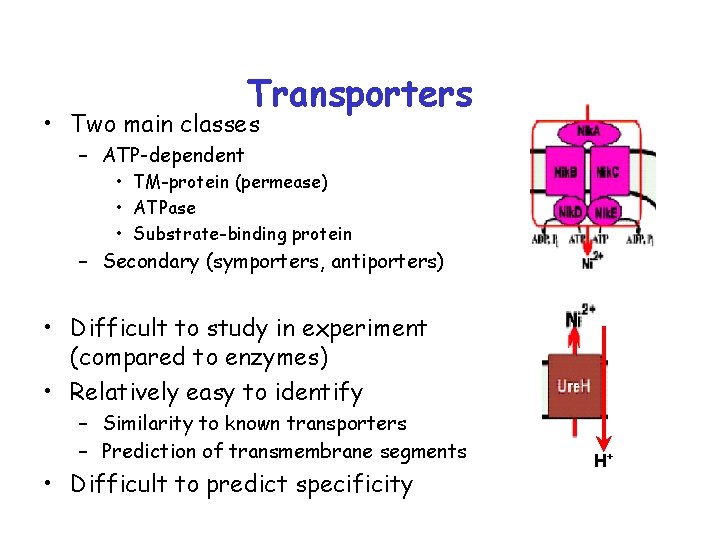
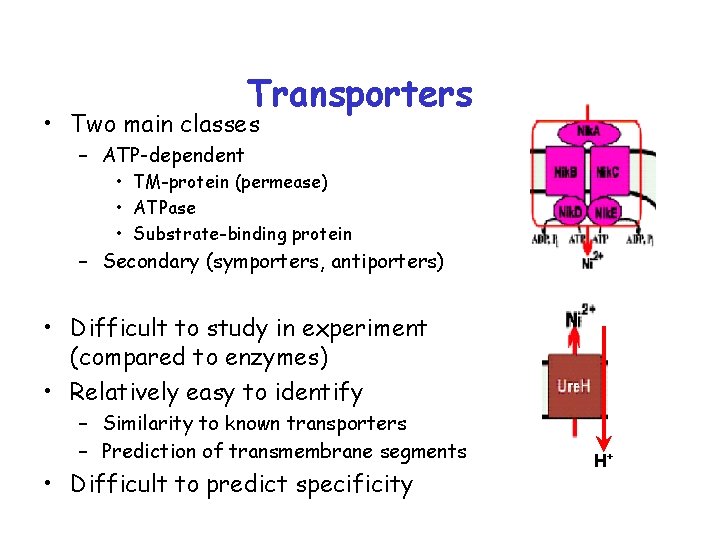
Transporters • Two main classes – ATP-dependent • TM-protein (permease) • ATPase • Substrate-binding protein – Secondary (symporters, antiporters) • Difficult to study in experiment (compared to enzymes) • Relatively easy to identify – Similarity to known transporters – Prediction of transmembrane segments • Difficult to predict specificity H+


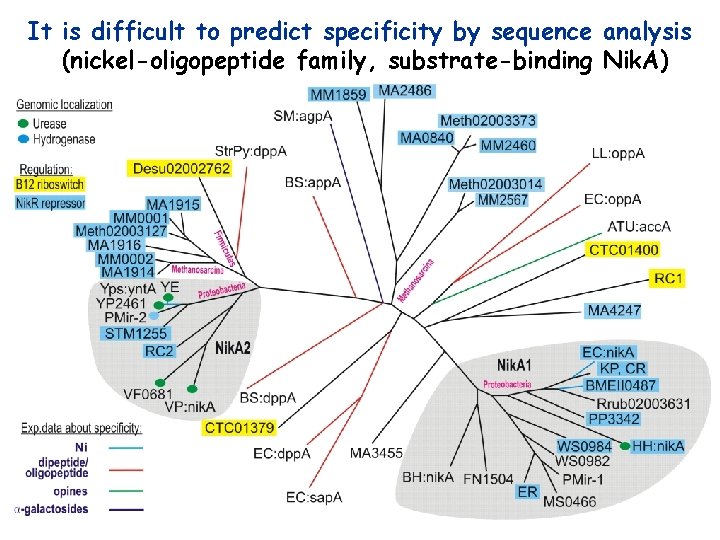
It is difficult to predict specificity by sequence analysis (nickel-oligopeptide family, substrate-binding Nik. A)

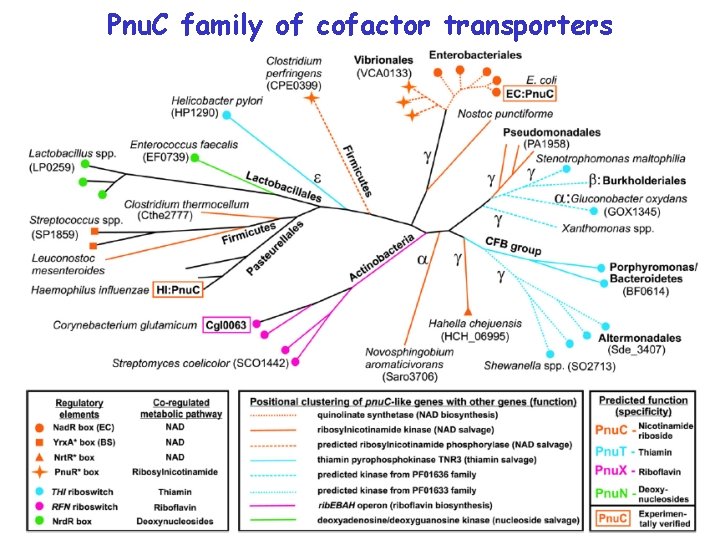
Pnu. C family of cofactor transporters

Riboflavin biosynthesis pathway

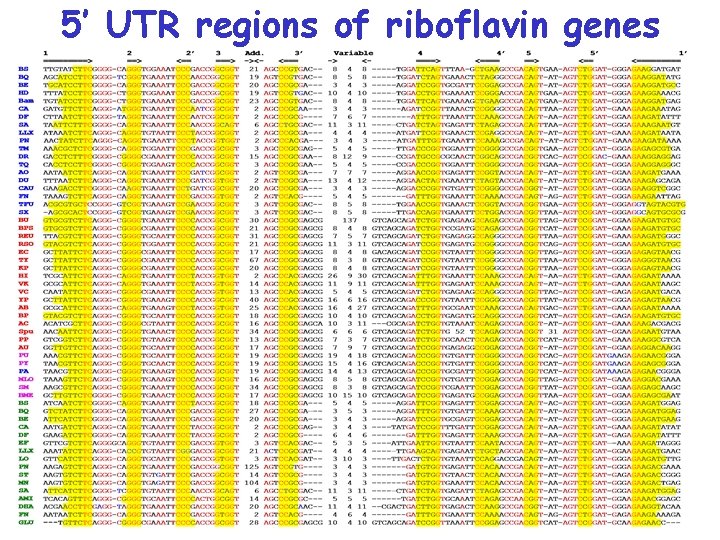
5’ UTR regions of riboflavin genes from various bacteria

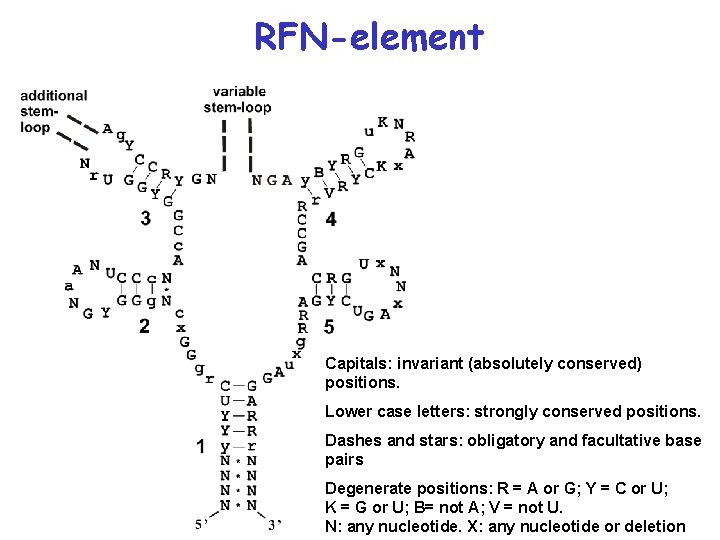
Conserved secondary structure of the RFN-element Capitals: invariant (absolutely conserved) positions. Lower case letters: strongly conserved positions. Dashes and stars: obligatory and facultative base pairs N: any nucleotide. X: any nucleotide or deletion

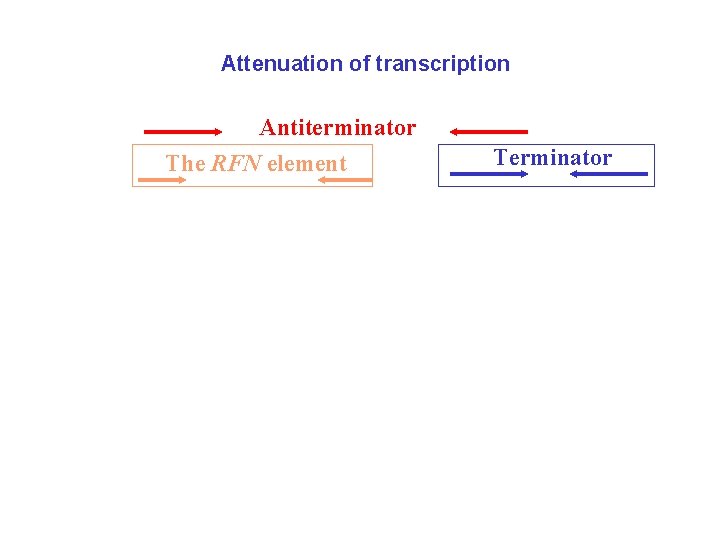
Attenuation of transcription Antiterminator The RFN element Antiterminator Terminator

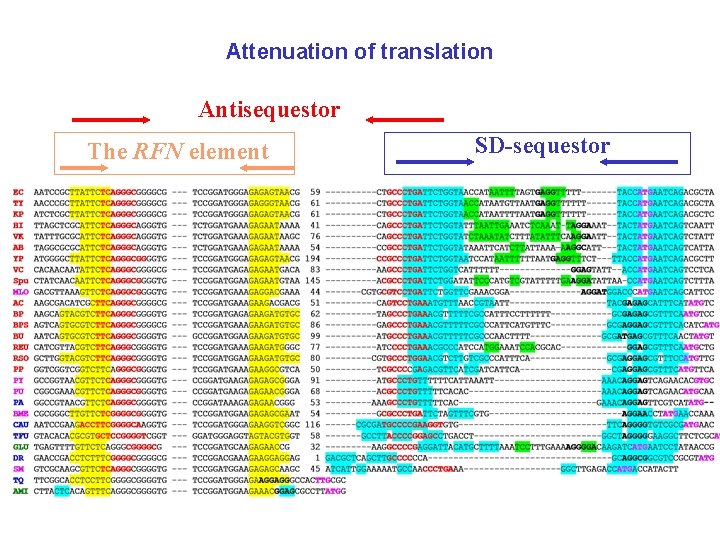
Attenuation of translation Antisequestor The RFN element SD-sequestor

Рибопереключатель RFN: регуляторный механизм Transcription attenuation Translation attenuation



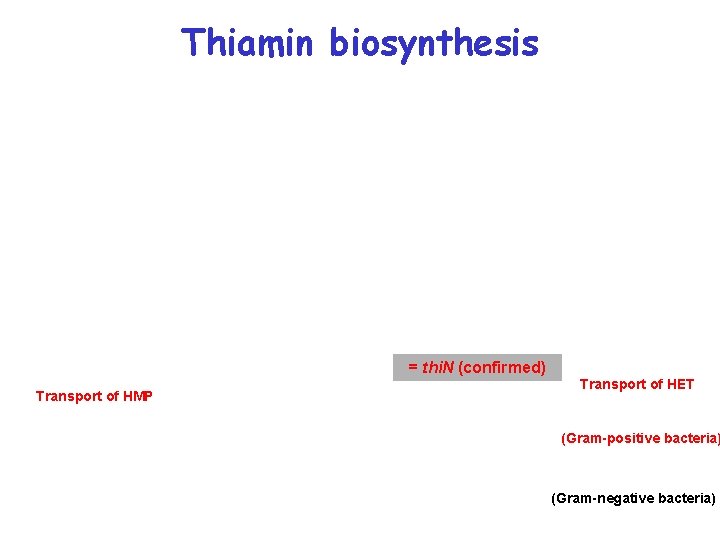
Метаболическая реконструкция тиаминового биосинтеза = thi. N (confirmed) Transport of HMP Transport of HET (Gram-positive bacteria) (Gram-negative bacteria)







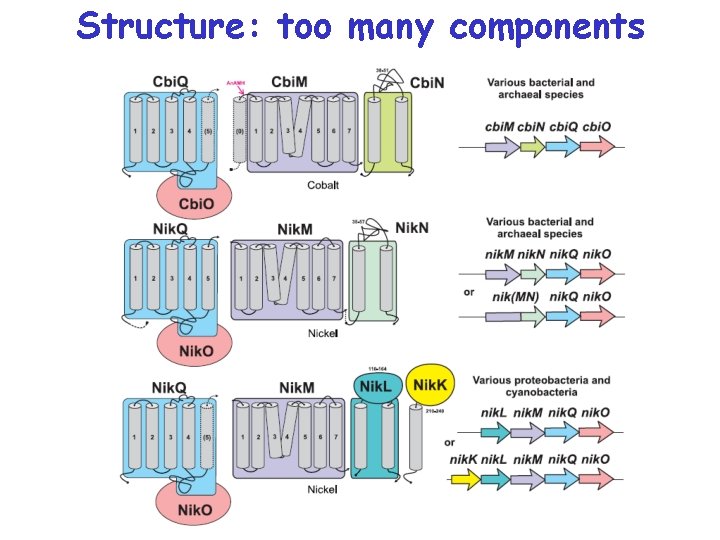
Структура cbi. O=Nik. O~bio. M cbi. Q=Nik. Q~bio. N









Transporters • Two main classes – ATP-dependent • TM-protein (permease) • ATPase • Substrate-binding protein – Secondary (symporters, antiporters) • Difficult to study in experiment (compared to enzymes) • Relatively easy to identify – Similarity to known transporters – Prediction of transmembrane segments • Difficult to predict specificity H+

It is difficult to predict specificity by sequence analysis (nickel-oligopeptide family, substrate-binding Nik. A)

Pnu. C family of cofactor transporters

Riboflavin biosynthesis pathway

5’ UTR regions of riboflavin genes

RFN-element Capitals: invariant (absolutely conserved) positions. Lower case letters: strongly conserved positions. Dashes and stars: obligatory and facultative base pairs Degenerate positions: R = A or G; Y = C or U; K = G or U; B= not A; V = not U. N: any nucleotide. X: any nucleotide or deletion

RFN: the mechanism of regulation • Transcription attenuation • Translation attenuation

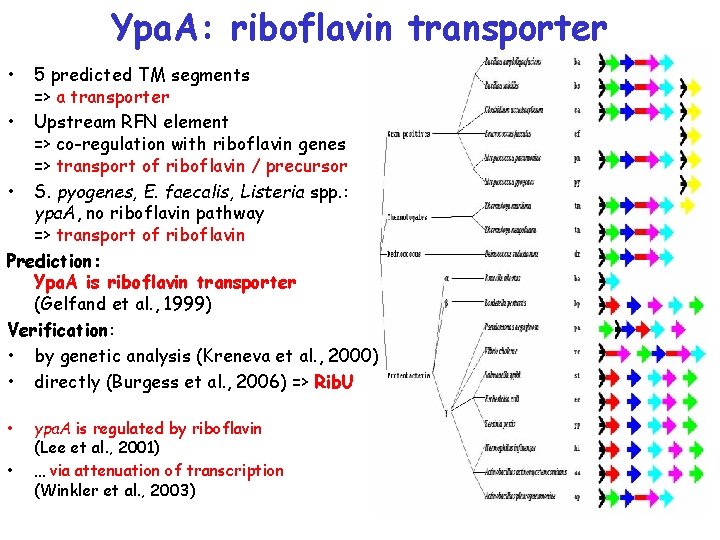
Ypa. A: riboflavin transporter • 5 predicted TM segments => a transporter • Upstream RFN element => co-regulation with riboflavin genes => transport of riboflavin / precursor • S. pyogenes, E. faecalis, Listeria spp. : ypa. A, no riboflavin pathway => transport of riboflavin Prediction: Ypa. A is riboflavin transporter (Gelfand et al. , 1999) Verification: • by genetic analysis (Kreneva et al. , 2000) • directly (Burgess et al. , 2006) => Rib. U • • ypa. A is regulated by riboflavin (Lee et al. , 2001) … via attenuation of transcription (Winkler et al. , 2003)

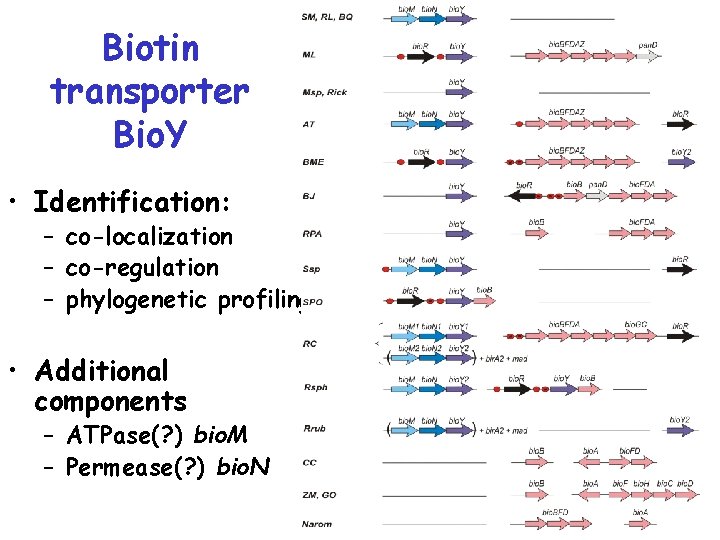
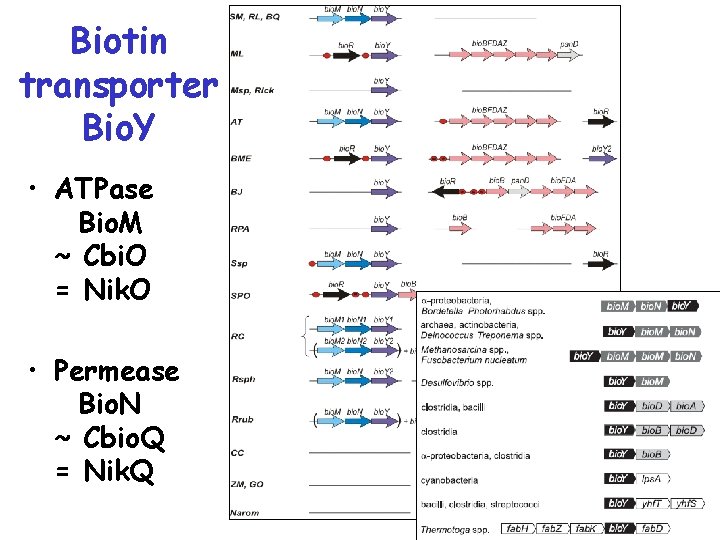
Biotin transporter Bio. Y • Identification: – co-localization – co-regulation – phylogenetic profiling • Additional components – ATPase(? ) bio. M – Permease(? ) bio. N

Thiamin biosynthesis = thi. N (confirmed) Transport of HMP Transport of HET (Gram-positive bacteria) (Gram-negative bacteria)

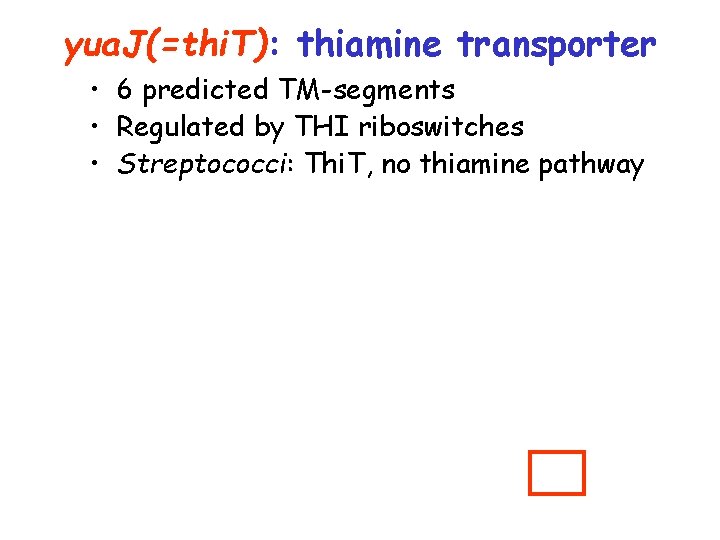
yua. J(=thi. T): thiamine transporter • 6 predicted TM-segments • Regulated by THI riboswitches • Streptococci: Thi. T, no thiamine pathway

yko. FEDC: ATP-dependent HMP transporter • • Regulated by THI riboswitches Newer occurs in genomes lacking thiamine pathway Always co-occurs with thi. D and thi. E Sometimes occurs without thi. C

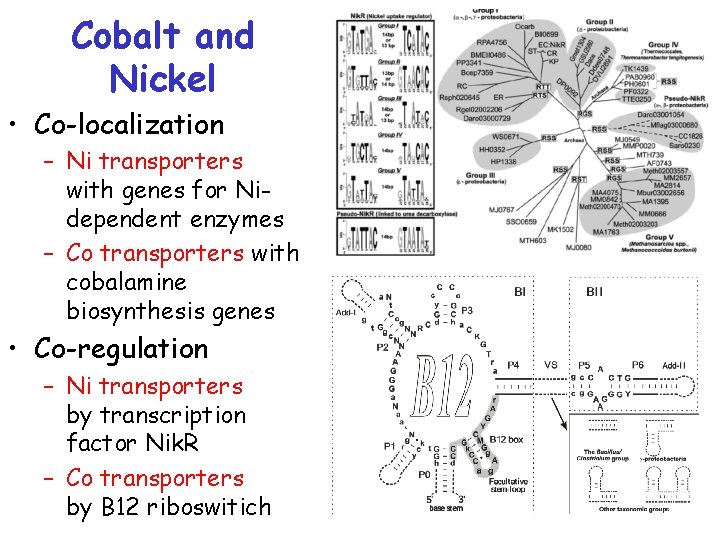
Cobalt and Nickel • Co-localization – Ni transporters with genes for Nidependent enzymes – Co transporters with cobalamine biosynthesis genes • Co-regulation – Ni transporters by transcription factor Nik. R – Co transporters by В 12 riboswitich

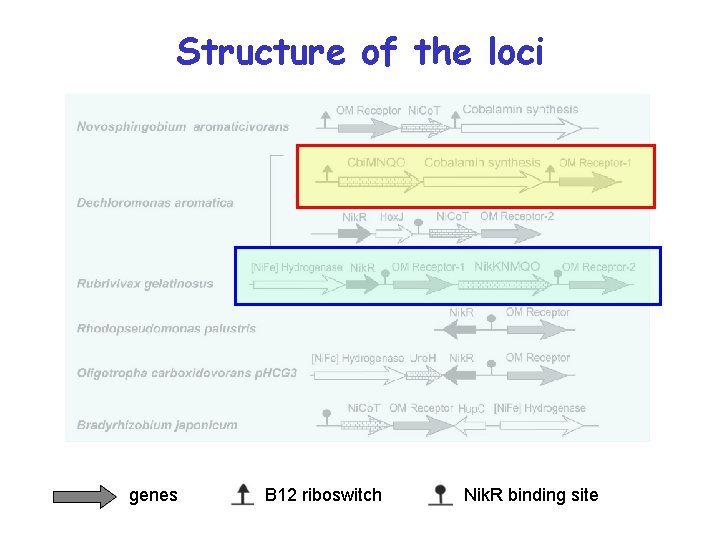
Structure of the loci genes B 12 riboswitch Nik. R binding site

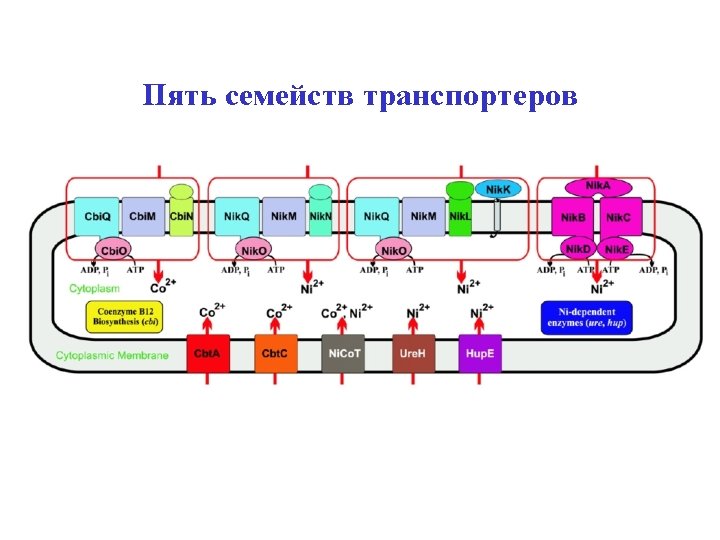
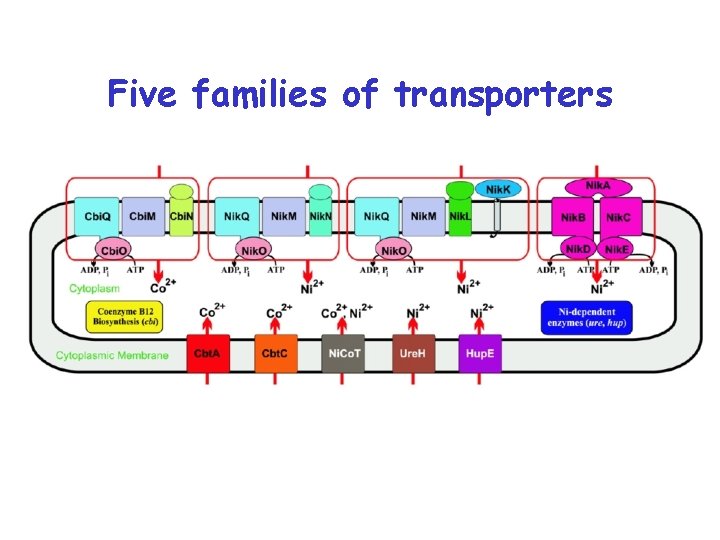
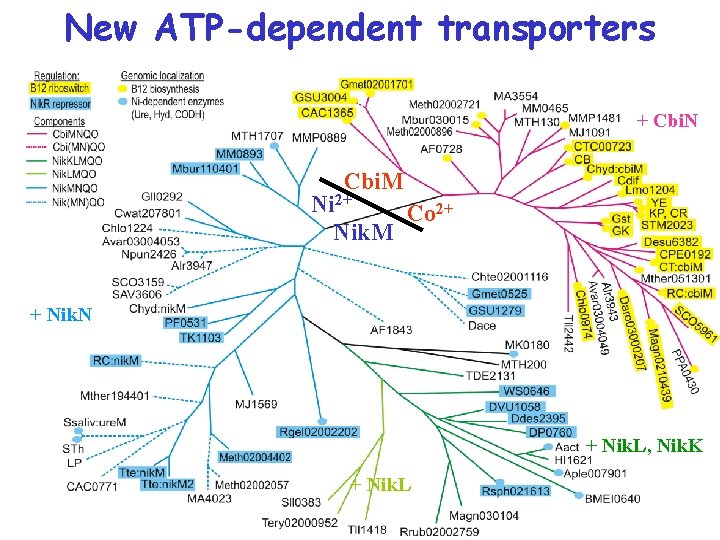
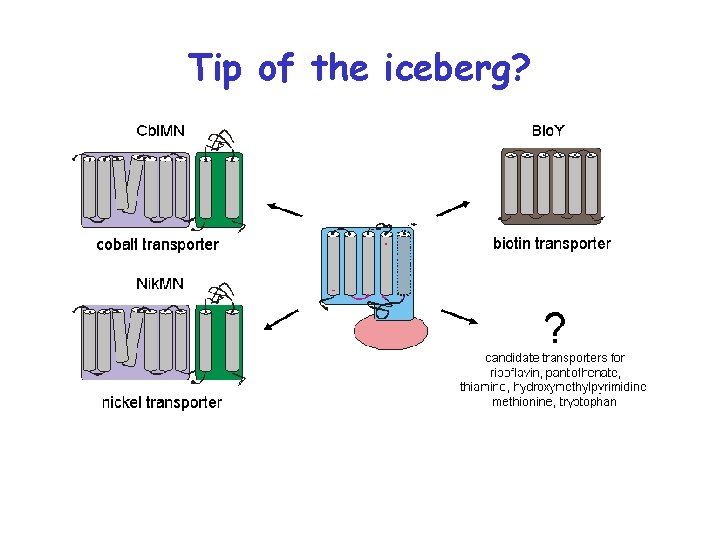
Five families of transporters

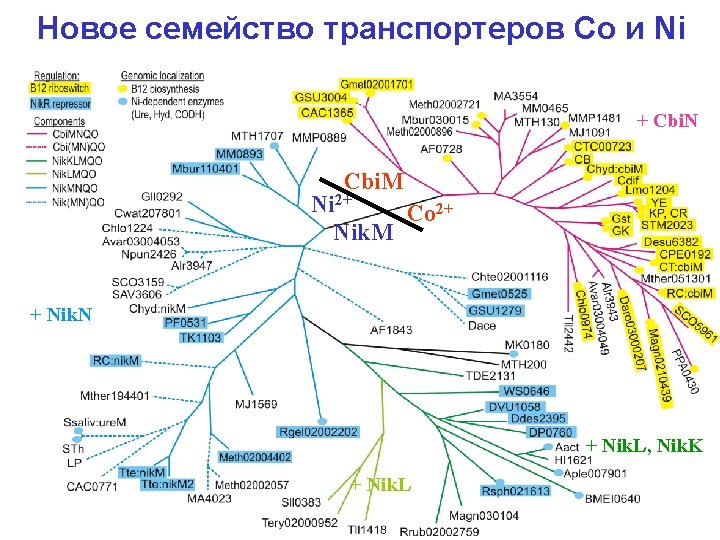
New ATP-dependent transporters + Cbi. N Cbi. M Ni 2+ Co 2+ Nik. M + Nik. N + Nik. L, Nik. K + Nik. L

Dmitry Rodionov Thomas Eitinger

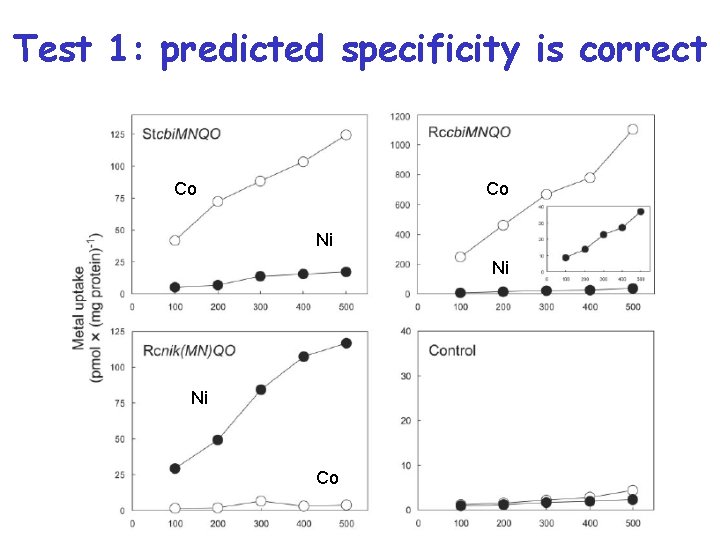
Test 1: predicted specificity is correct Co Co Ni Ni Ni Co

Structure: too many components

Biotin transporter Bio. Y • ATPase Bio. M ~ Cbi. O = Nik. O • Permease Bio. N ~ Cbio. Q = Nik. Q

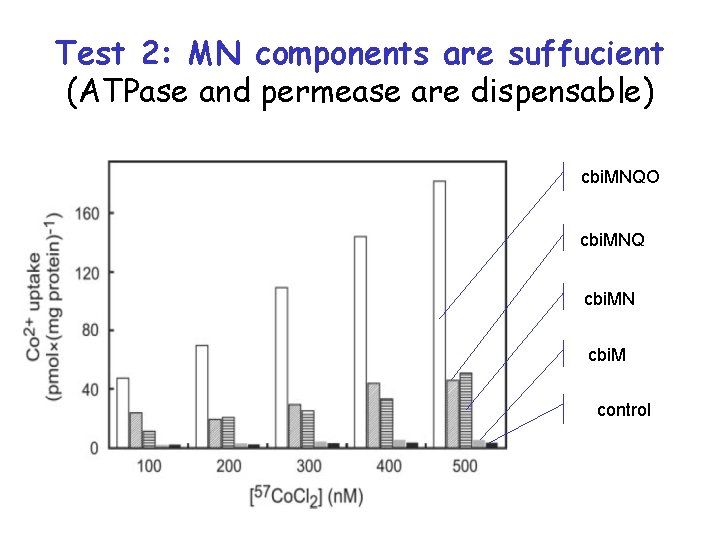
Test 2: MN components are suffucient (ATPase and permease are dispensable) cbi. MNQO cbi. MNQ cbi. MN cbi. M control

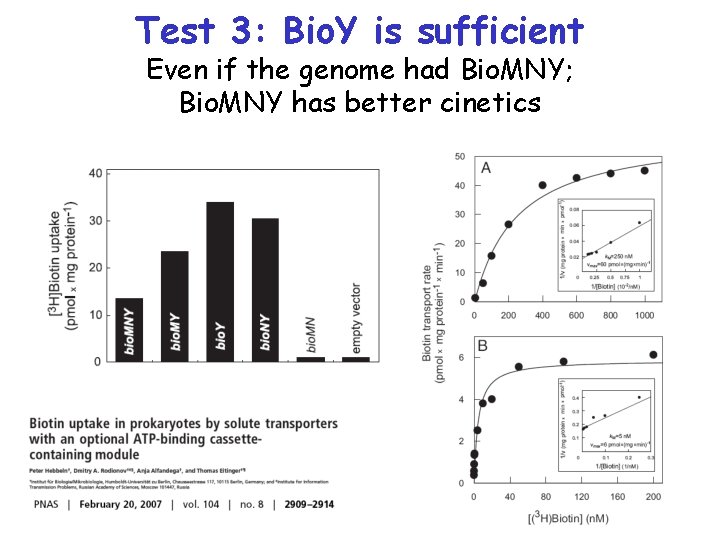
Test 3: Bio. Y is sufficient Even if the genome had Bio. MNY; Bio. MNY has better cinetics

Tip of the iceberg?




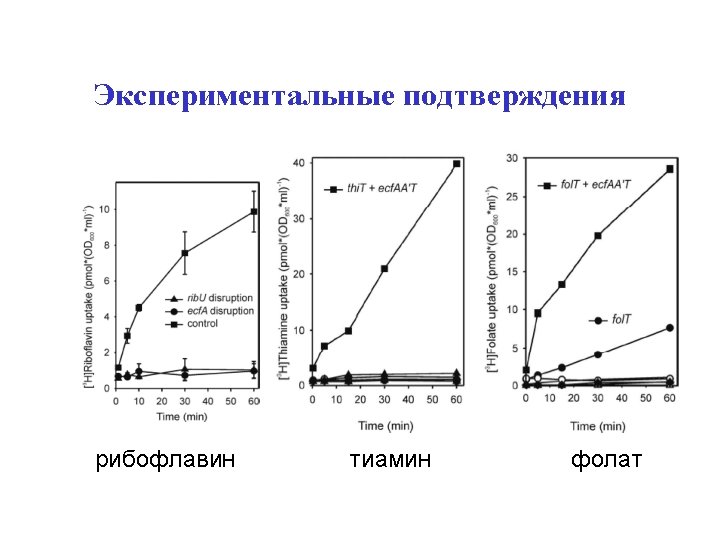
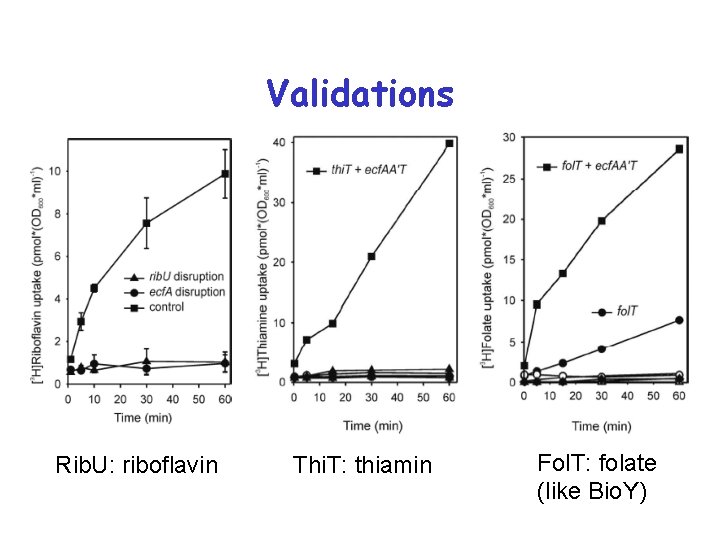
Validations Rib. U: riboflavin Thi. T: thiamin Fol. T: folate (like Bio. Y)

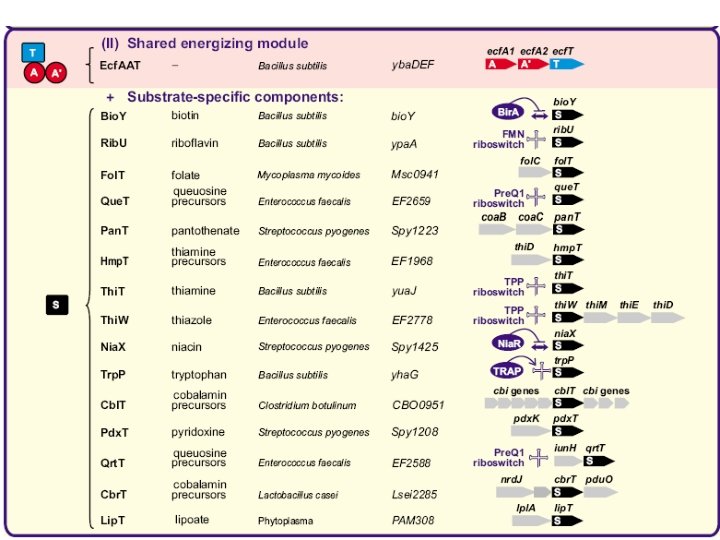
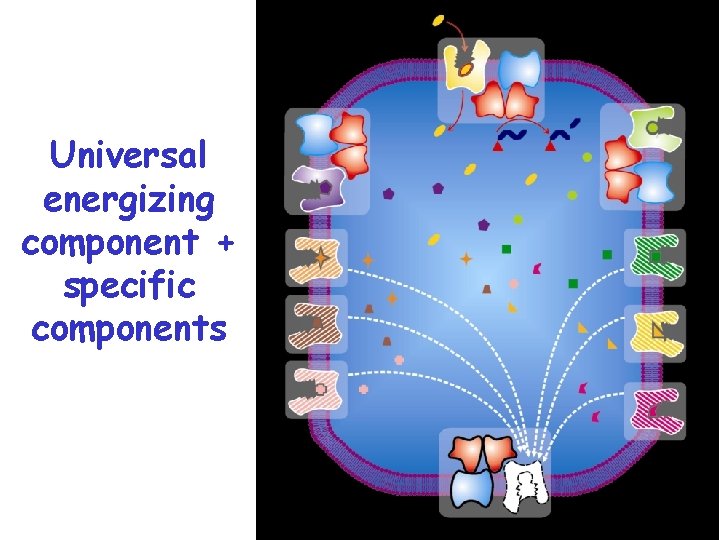
Universal energizing component + specific components





 Ctc cta
Ctc cta Qalqonsimon bez garmon preparatlari
Qalqonsimon bez garmon preparatlari Contoh kta dan tta
Contoh kta dan tta Tta adults
Tta adults Tta standards
Tta standards Seungtta
Seungtta Saigonctt
Saigonctt Saigon ctt
Saigon ctt Saigon ctt
Saigon ctt Saigonctt
Saigonctt Saigon ctt
Saigon ctt Ttt-järjestelmä
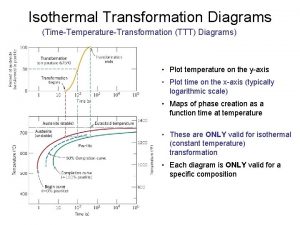
Ttt-järjestelmä Spheroidite ttt diagram
Spheroidite ttt diagram Diagrama ttt
Diagrama ttt Slidetodoc.com
Slidetodoc.com Diagram hardening
Diagram hardening Tell me and i forget teach me and i remember
Tell me and i forget teach me and i remember Diagrama ttt
Diagrama ttt Ttc methods
Ttc methods Look at the dna sequence gaa-ttc-gca
Look at the dna sequence gaa-ttc-gca Ttc birkenfeld
Ttc birkenfeld Ttc sunday service
Ttc sunday service Taux de marge
Taux de marge Normoxemia definicion
Normoxemia definicion Ttc electrical aptitude test
Ttc electrical aptitude test Ttc organization chart
Ttc organization chart Ace ttc
Ace ttc Ttc organization chart
Ttc organization chart Ttc organization chart
Ttc organization chart Satellite communication system
Satellite communication system California tamil academy books
California tamil academy books Cta goodman
Cta goodman Atlashr
Atlashr Agm salud cta
Agm salud cta Fir uir
Fir uir Cta templates
Cta templates Tipos de espacio aéreo
Tipos de espacio aéreo Cta download
Cta download Cta bus 4
Cta bus 4 Cta
Cta Cta freedom
Cta freedom Cta
Cta Control fisico
Control fisico Antigel cta
Antigel cta Cta liberty
Cta liberty Cta
Cta Mi cta
Mi cta Cta grading
Cta grading Autism authorization california online
Autism authorization california online Ctc en ctq
Ctc en ctq Ctc sheet
Ctc sheet Ctc
Ctc Ctc en ctq
Ctc en ctq Ctc link
Ctc link Ctc
Ctc Certido
Certido Ctc loss
Ctc loss Cambridge training college britain
Cambridge training college britain Ctc link
Ctc link Ctc anken
Ctc anken Ctc connectionist temporal classification
Ctc connectionist temporal classification Ctc registration
Ctc registration Ctc form 1
Ctc form 1 Autocontenido sinonimo
Autocontenido sinonimo Company profile with ctc
Company profile with ctc Ctc link sfcc
Ctc link sfcc Ctc
Ctc Saltatory variability
Saltatory variability Ctg como interpretar
Ctg como interpretar Saltatorni ctg
Saltatorni ctg Kaligrafi bismillah
Kaligrafi bismillah Grading of meconium stained liquor ppt
Grading of meconium stained liquor ppt Ctg akzeleration
Ctg akzeleration Next generation operating system
Next generation operating system Ctgs
Ctgs Uniforme deceleraties
Uniforme deceleraties Test ctg
Test ctg Ctg
Ctg Category 1 tracing
Category 1 tracing Ctg-1
Ctg-1 Classificazione ctg acog
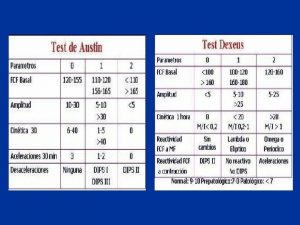
Classificazione ctg acog Normal ctg
Normal ctg