Spatial Localization and Multinuclear MR Spectroscopy Techniques Navin













- Slides: 64

Spatial Localization and Multinuclear MR Spectroscopy Techniques Navin Bansal, Ph. D. Associate Professor and Director of MR Research

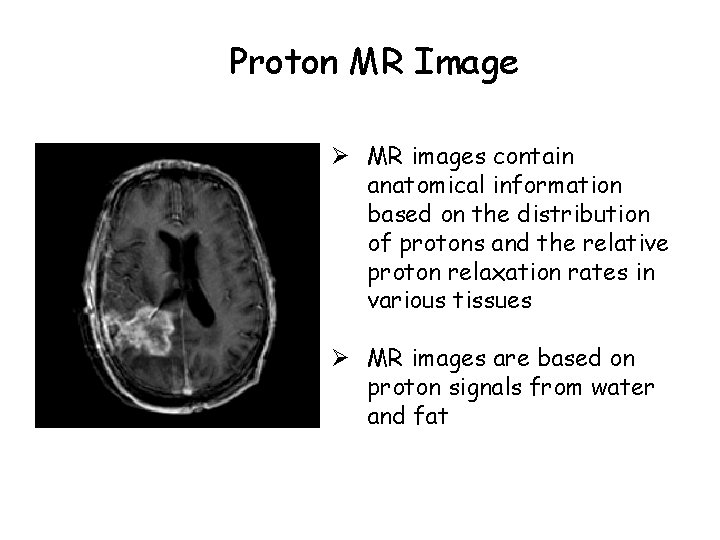
Proton MR Image Ø MR images contain anatomical information based on the distribution of protons and the relative proton relaxation rates in various tissues Ø MR images are based on proton signals from water and fat

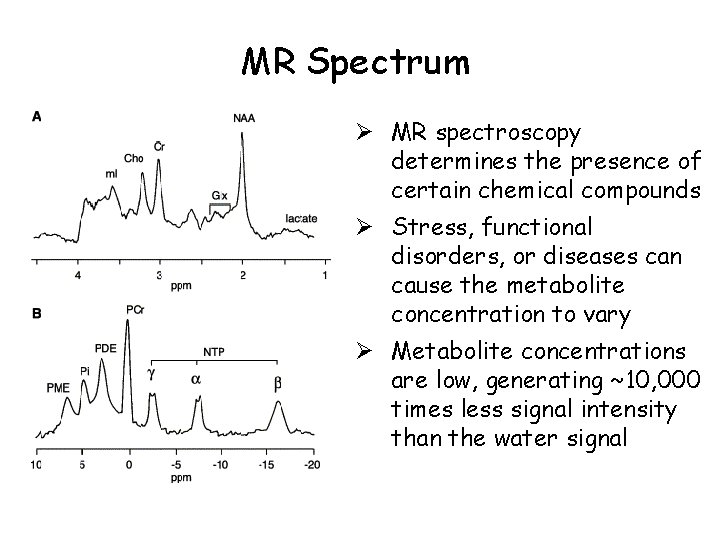
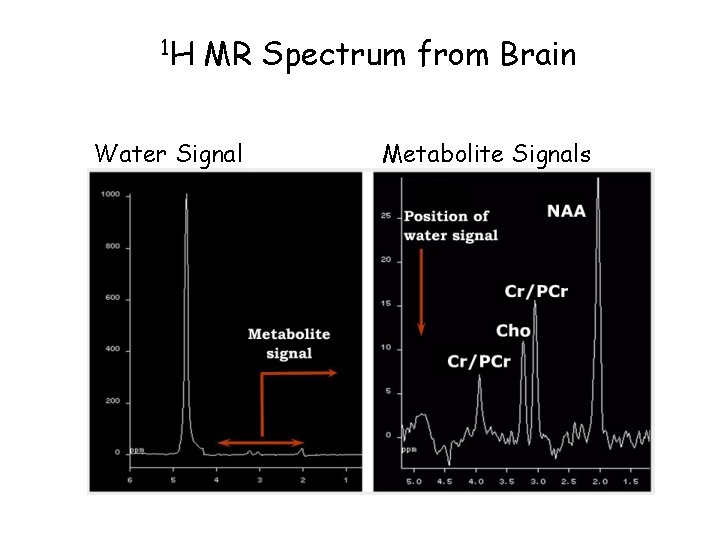
MR Spectrum Ø MR spectroscopy determines the presence of certain chemical compounds Ø Stress, functional disorders, or diseases can cause the metabolite concentration to vary Ø Metabolite concentrations are low, generating ~10, 000 times less signal intensity than the water signal

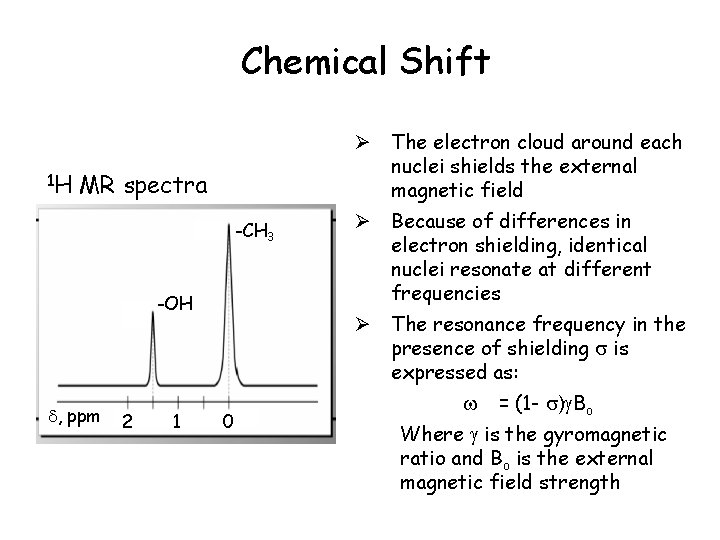
Chemical Shift 1 H Ø The electron cloud around each nuclei shields the external magnetic field MR spectra -CH 3 -OH , ppm 2 1 Ø Because of differences in electron shielding, identical nuclei resonate at different frequencies Ø The resonance frequency in the presence of shielding is expressed as: 0 = (1 - ) Bo Where is the gyromagnetic ratio and Bo is the external magnetic field strength

Chemical Shift Ø The frequency shift increases with field strength. For example, shift difference between water and fat ( water - fat) at 1. 5 T is 255 Hz at 3. 0 T is 510 Hz = ( water - fat) 106/ Bo, in ppm units water-fat is 3. 5 ppm independent of field strength Ø By convention o Signals of weakly shielded nuclei with higher frequency are on the left o Signals of more heavily shielded nuclei with lower frequency are on the right Ø Chemical shift of water is set to 4. 7 ppm at body temperature

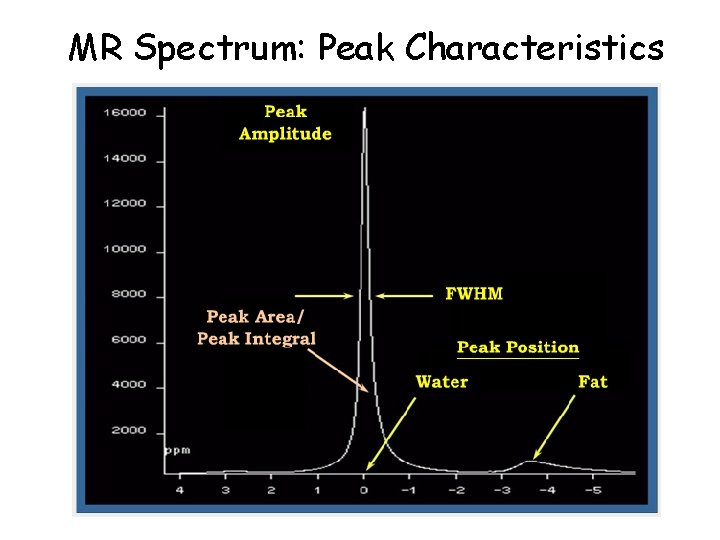
MR Spectrum: Peak Characteristics

1 H MR Spectrum from Brain Water Signal Metabolite Signals

Spatial Localization Surface Coil Localization Ø Simple surface coil acquisition Ø Depth Resolved Surface Coil Spectroscopy, DRESS Single Volume Localization Ø Image Selected In Vivo Spectroscopy, ISIS Ø Point Resolved Spectroscopy, PRESS Ø Stimulated Echo Acquisition Mode, STEAM Multiple Volume Acquisition Ø Chemical Shift Imaging, CSI

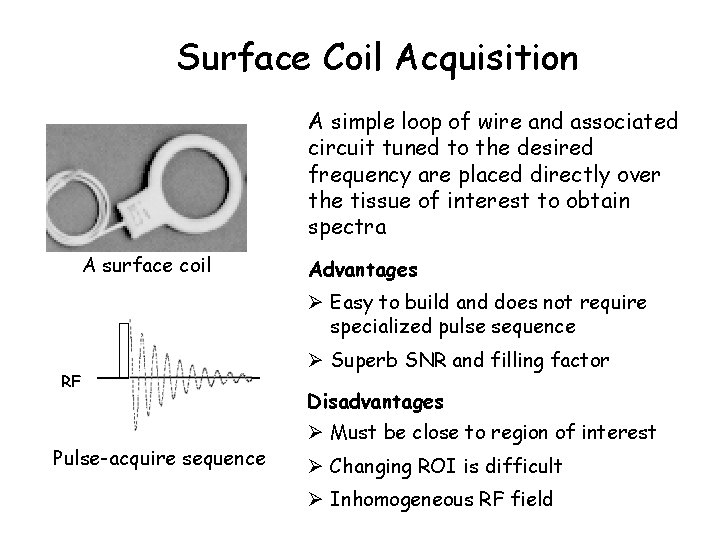
Surface Coil Acquisition A simple loop of wire and associated circuit tuned to the desired frequency are placed directly over the tissue of interest to obtain spectra A surface coil Advantages Ø Easy to build and does not require specialized pulse sequence RF Pulse-acquire sequence Ø Superb SNR and filling factor Disadvantages Ø Must be close to region of interest Ø Changing ROI is difficult Ø Inhomogeneous RF field

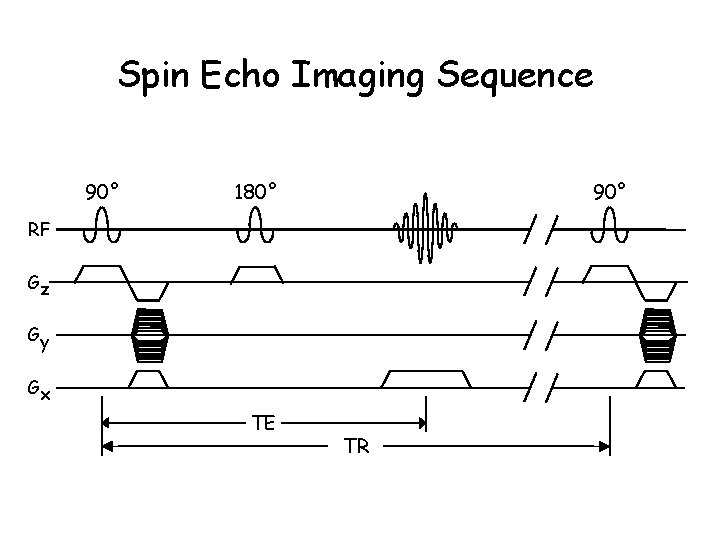
Spin Echo Imaging Sequence 90° 180° 90° RF Gz Gy Gx TE TR

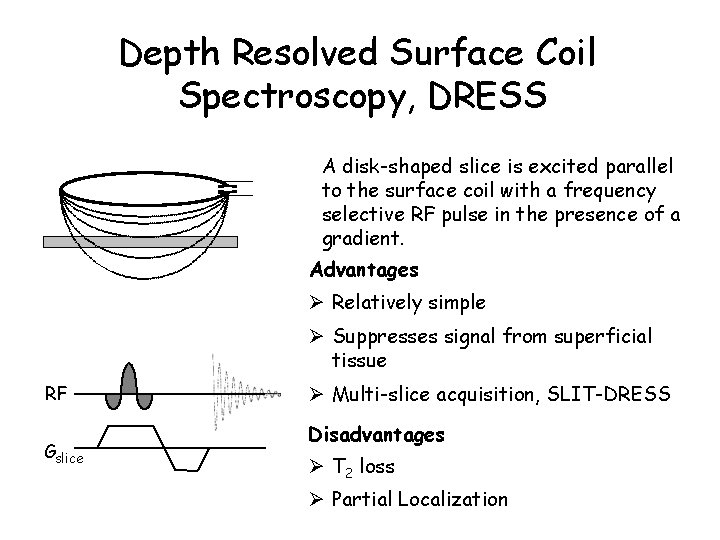
Depth Resolved Surface Coil Spectroscopy, DRESS A disk-shaped slice is excited parallel to the surface coil with a frequency selective RF pulse in the presence of a gradient. Advantages Ø Relatively simple Ø Suppresses signal from superficial tissue RF Gslice Ø Multi-slice acquisition, SLIT-DRESS Disadvantages Ø T 2 loss Ø Partial Localization

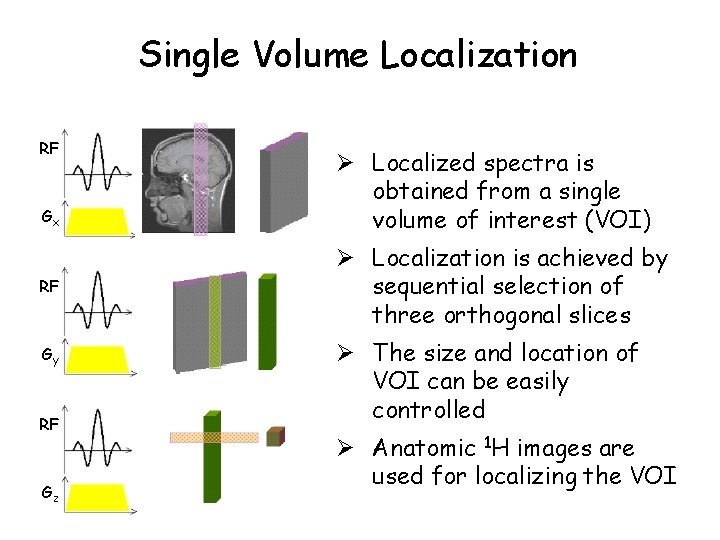
Single Volume Localization RF Gx RF Gy RF Gz Ø Localized spectra is obtained from a single volume of interest (VOI) Ø Localization is achieved by sequential selection of three orthogonal slices Ø The size and location of VOI can be easily controlled Ø Anatomic 1 H images are used for localizing the VOI

Single Volume Localization ØImage selected in vivo spectroscopy, ISIS ØPoint resolved spectroscopy, PRESS ØStimulated echo acquisition mode, STEAM

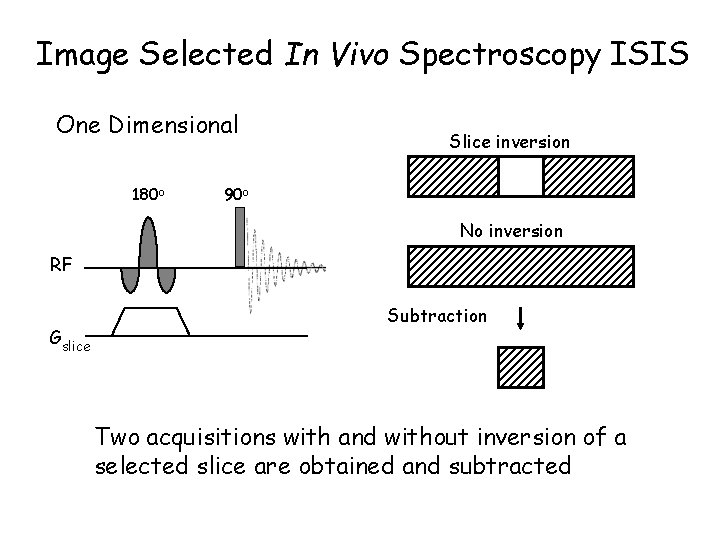
Image Selected In Vivo Spectroscopy ISIS One Dimensional 180 o Slice inversion 90 o No inversion RF Gslice Subtraction Two acquisitions with and without inversion of a selected slice are obtained and subtracted

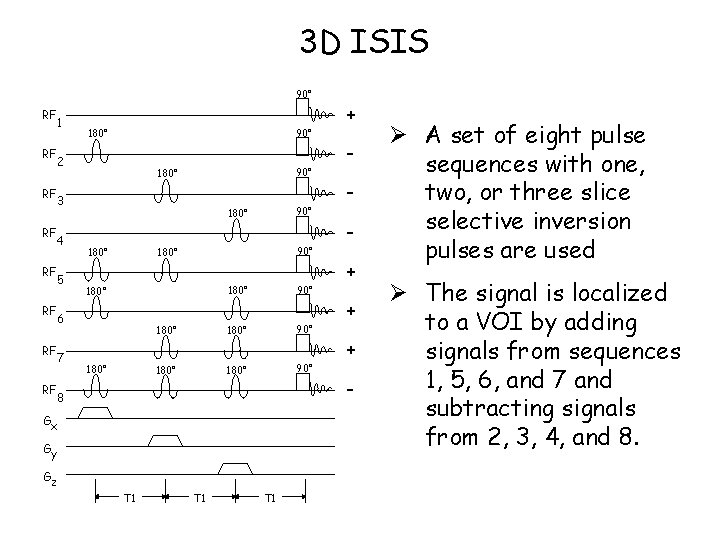
3 D ISIS 90° RF RF 1 90° 180° 2 90° 180° 3 4 5 180° 90° 180° 6 7 90° 180° 8 Gx Gy Gz T 1 T 1 + + - Ø A set of eight pulse sequences with one, two, or three slice selective inversion pulses are used Ø The signal is localized to a VOI by adding signals from sequences 1, 5, 6, and 7 and subtracting signals from 2, 3, 4, and 8.

Image Selected In Vivo Spectroscopy, ISIS Advantages Ø No T 2 loss – 31 P MRS Ø Less sensitive to gradient imperfections Ø Can be used with a surface coil Disadvantages Ø Dynamic range Ø Subtraction error due to motion

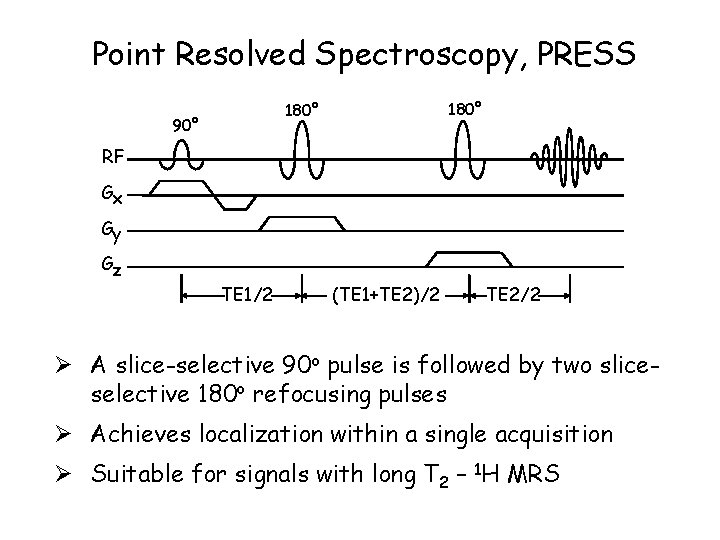
Point Resolved Spectroscopy, PRESS 180° 90° RF Gx Gy Gz TE 1/2 (TE 1+TE 2)/2 TE 2/2 Ø A slice-selective 90 o pulse is followed by two sliceselective 180 o refocusing pulses Ø Achieves localization within a single acquisition Ø Suitable for signals with long T 2 – 1 H MRS

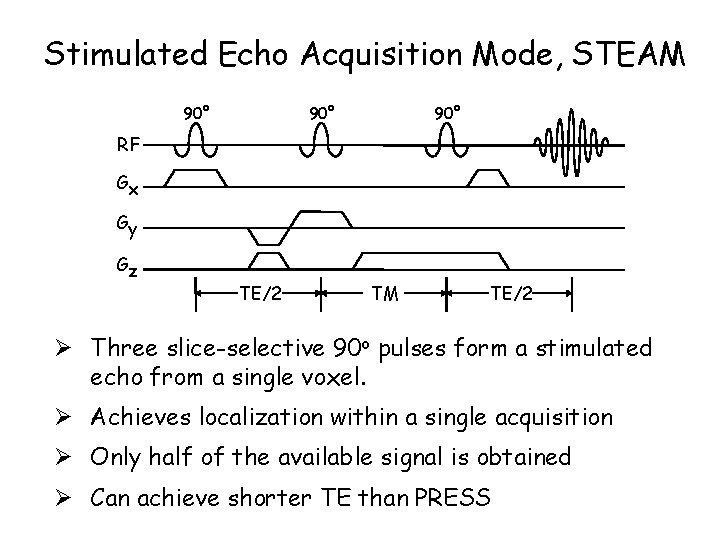
Stimulated Echo Acquisition Mode, STEAM 90° 90° RF Gx Gy Gz TE/2 TM TE/2 Ø Three slice-selective 90 o pulses form a stimulated echo from a single voxel. Ø Achieves localization within a single acquisition Ø Only half of the available signal is obtained Ø Can achieve shorter TE than PRESS

Effects of MR Parameters on PRESS spectra Ø Repetition Time, TR Ø Number of Signal Averages Ø Echo Time, TE Ø Voxel Size

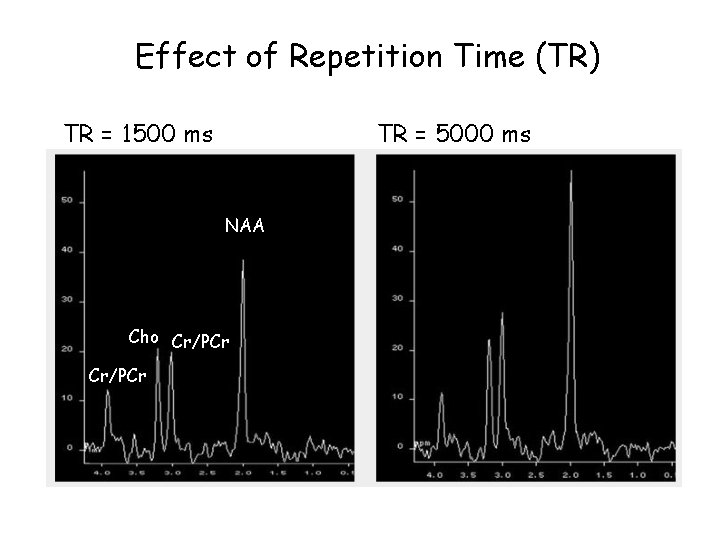
Effect of Repetition Time (TR) TR = 1500 ms TR = 5000 ms NAA Cho Cr/PCr

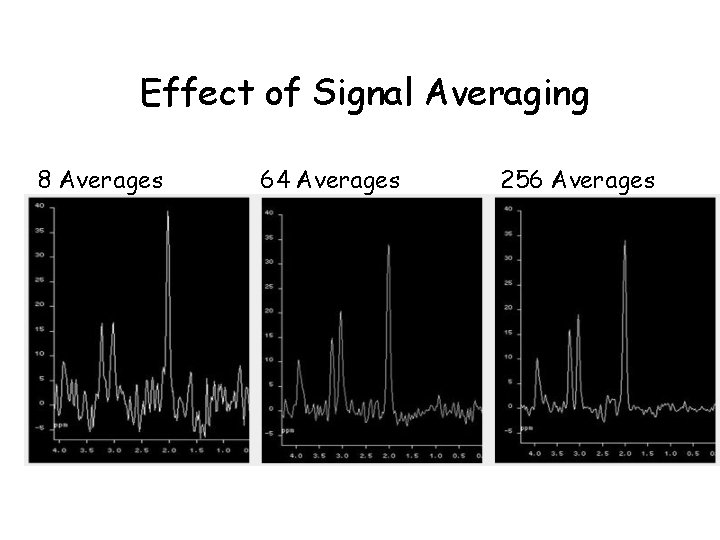
Effect of Signal Averaging 8 Averages 64 Averages 256 Averages

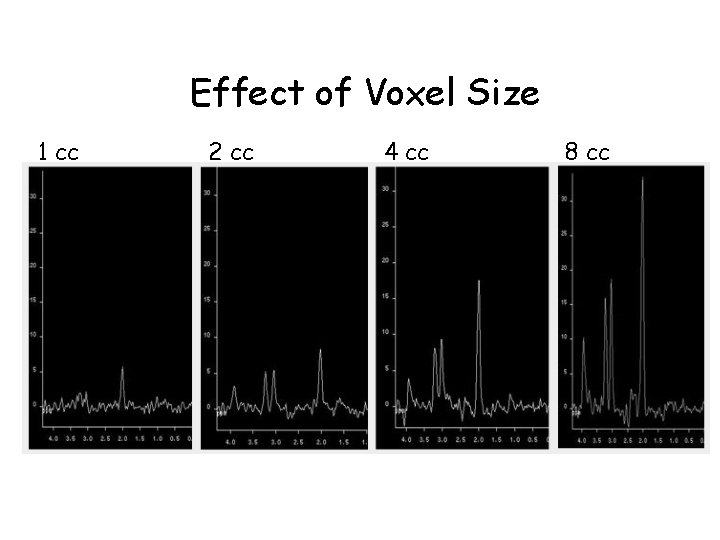
Effect of Voxel Size 1 cc 2 cc 4 cc 8 cc

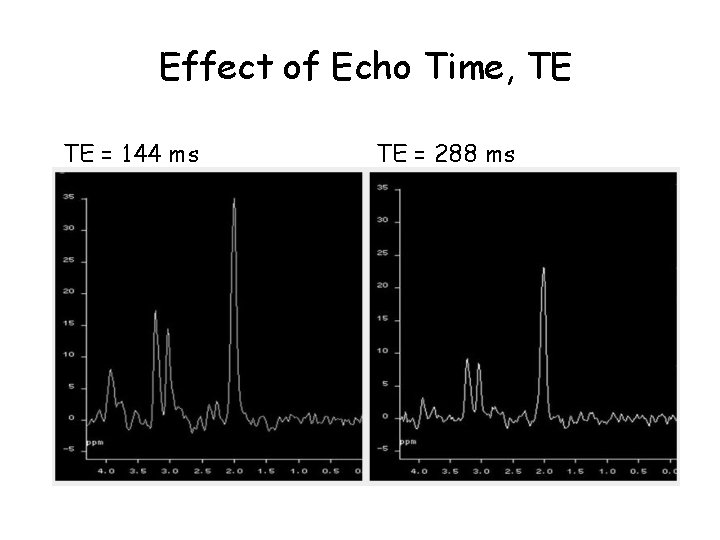
Effect of Echo Time, TE TE = 144 ms TE = 288 ms

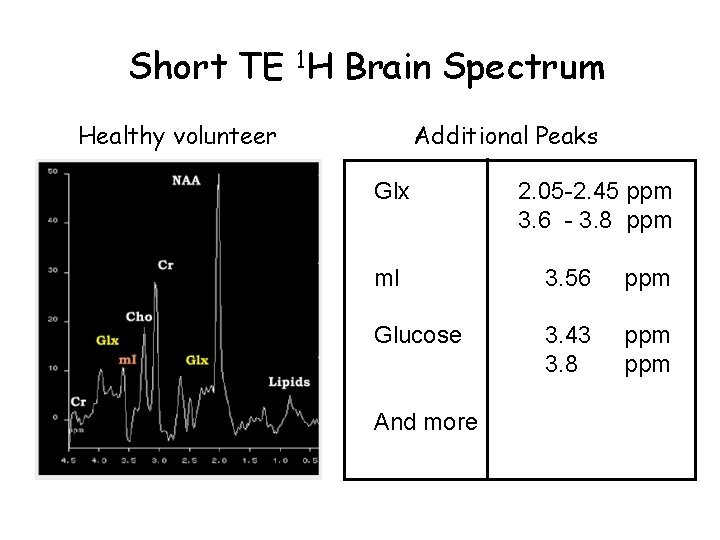
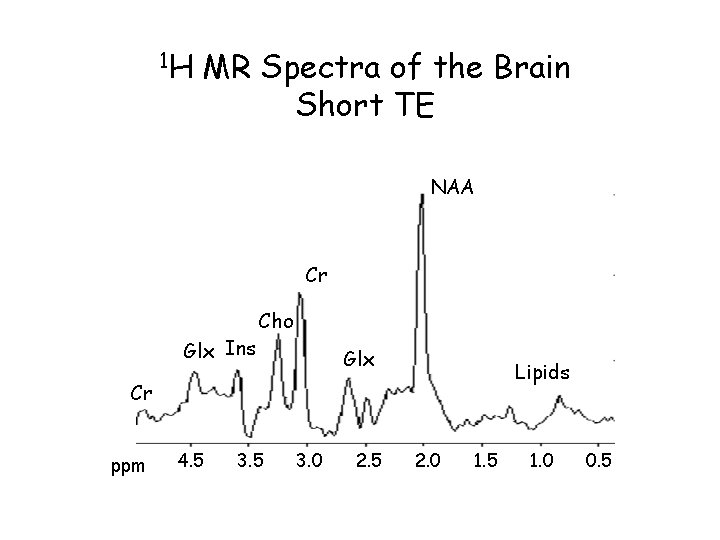
Short TE 1 H Brain Spectrum Healthy volunteer Additional Peaks Glx 2. 05 -2. 45 ppm 3. 6 - 3. 8 ppm m. I 3. 56 ppm Glucose 3. 43 3. 8 ppm And more

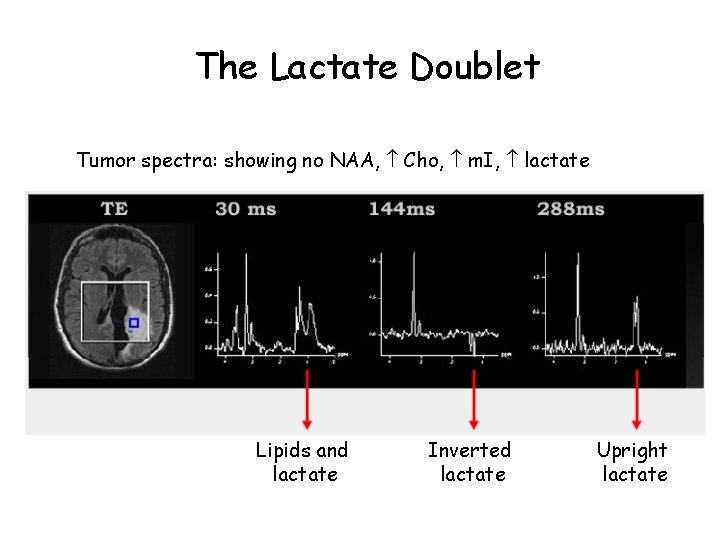
The Lactate Doublet Tumor spectra: showing no NAA, Cho, m. I, lactate Lipids and lactate Inverted lactate Upright lactate

Single Voxel Spectroscopy: Overview Ø Simplicity Ø Flexibility in voxel size and position Ø Accurate definition of VOI Ø Excellent shim and spectral resolution Ø Many voxels within the same dataset

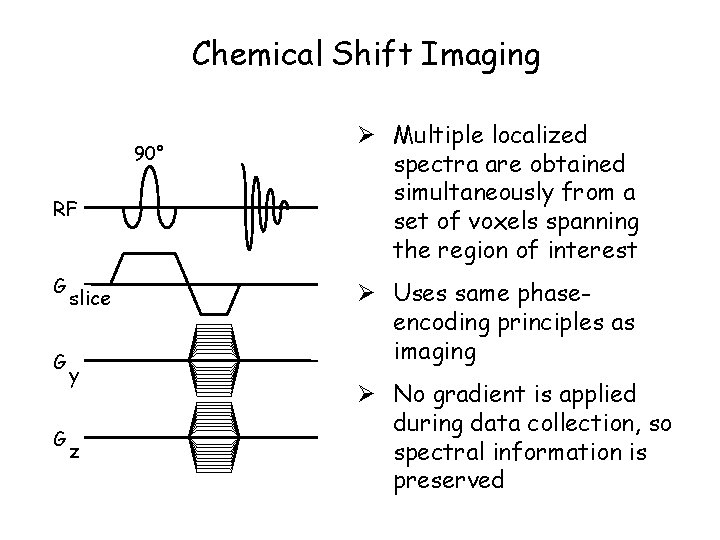
Chemical Shift Imaging 90° RF G G G slice y z Ø Multiple localized spectra are obtained simultaneously from a set of voxels spanning the region of interest Ø Uses same phaseencoding principles as imaging Ø No gradient is applied during data collection, so spectral information is preserved

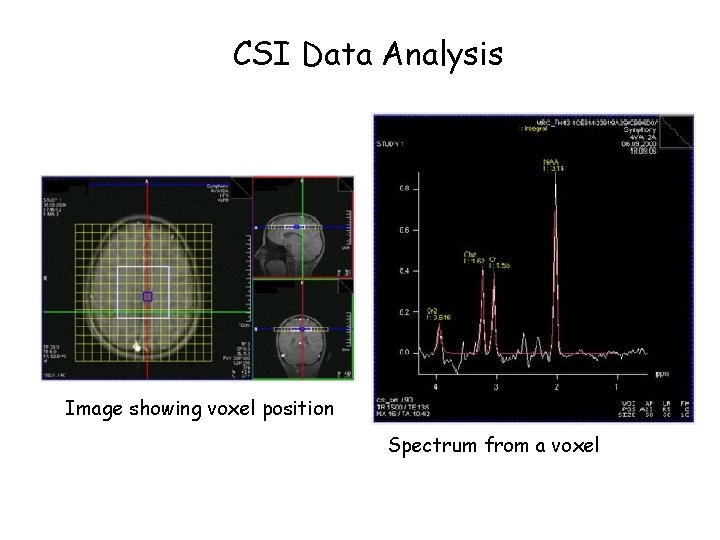
CSI Spectral Map Ø Display of all spectra Ø Underlying reference image shows voxel position Ø Individual spectra can be displayed enlarged Ø Spectral map can be archived together with the reference image and the CSI grid

CSI Data Analysis Image showing voxel position Spectrum from a voxel

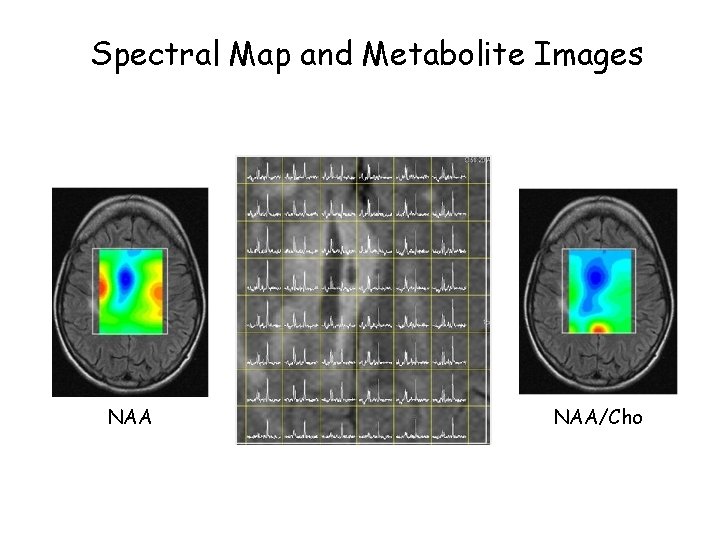
Spectral Map and Metabolite Images NAA/Cho

CSI: Overview Advantages Ø Acquisition of multiple voxels Ø Metabolite images, spectral maps, peak information maps, and results table Ø Many voxels within the same dataset Disadvantages Ø Large volume – more difficult to shim Ø Voxel bleeding Ø Large datasets

Multinuclear MR Spectroscopy

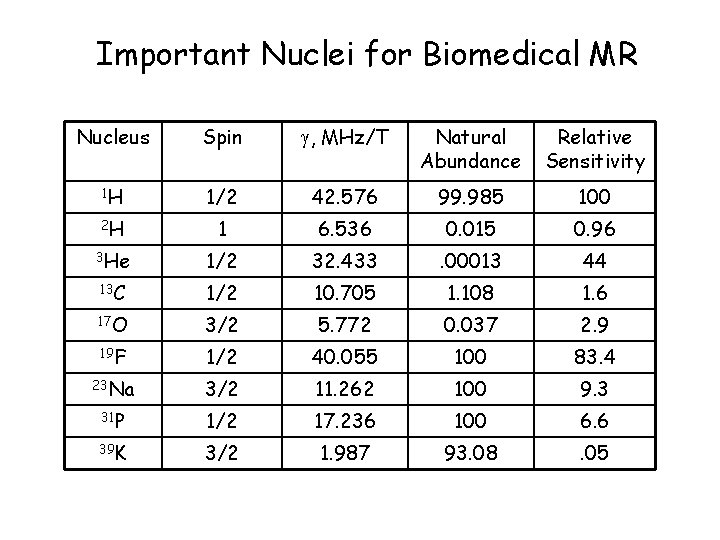
Important Nuclei for Biomedical MR Nucleus Spin , MHz/T Natural Abundance Relative Sensitivity 1 H 1/2 42. 576 99. 985 100 2 H 1 6. 536 0. 015 0. 96 3 He 1/2 32. 433 . 00013 44 13 C 1/2 10. 705 1. 108 1. 6 17 O 3/2 5. 772 0. 037 2. 9 19 F 1/2 40. 055 100 83. 4 23 Na 3/2 11. 262 100 9. 3 31 P 1/2 17. 236 100 6. 6 39 K 3/2 1. 987 93. 08 . 05

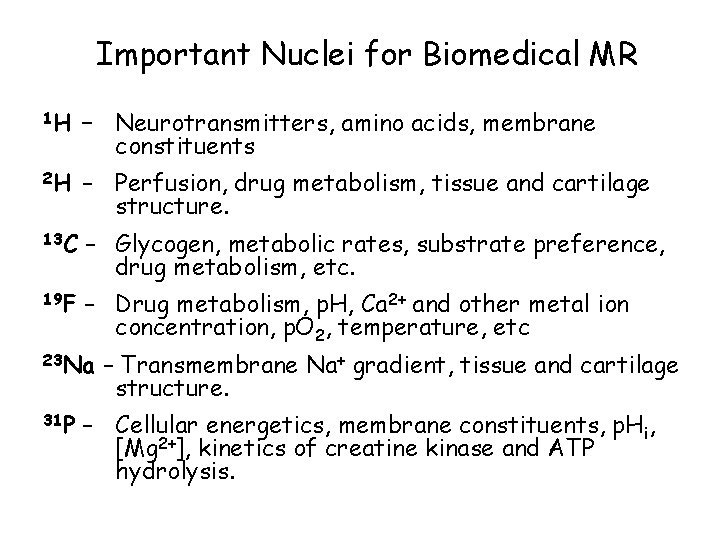
Important Nuclei for Biomedical MR 1 H – Neurotransmitters, amino acids, membrane constituents 2 H – Perfusion, drug metabolism, tissue and cartilage structure. 13 C – Glycogen, metabolic rates, substrate preference, drug metabolism, etc. 19 F – Drug metabolism, p. H, Ca 2+ and other metal ion concentration, p. O 2, temperature, etc 23 Na 31 P – Transmembrane Na+ gradient, tissue and cartilage structure. – Cellular energetics, membrane constituents, p. Hi, [Mg 2+], kinetics of creatine kinase and ATP hydrolysis.

1 H MR Spectroscopy

1 H MR Spectra of the Brain Short TE NAA Cr Glx Ins Cho Glx Lipids Cr ppm 4. 5 3. 0 2. 5 2. 0 1. 5 1. 0 0. 5

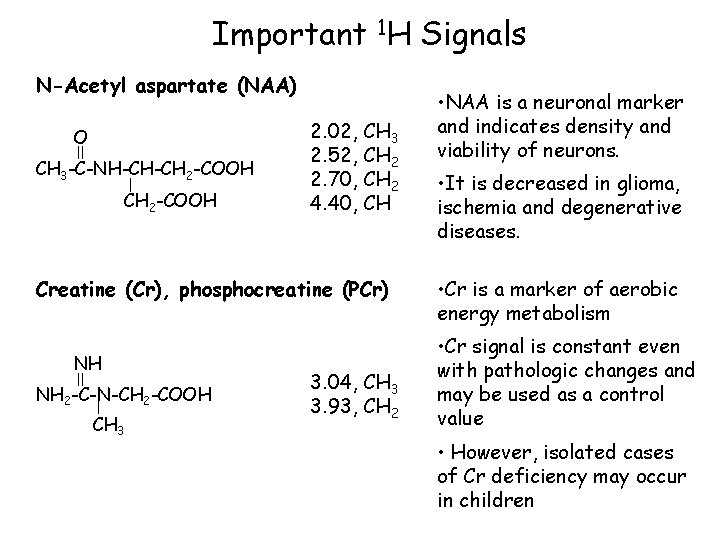
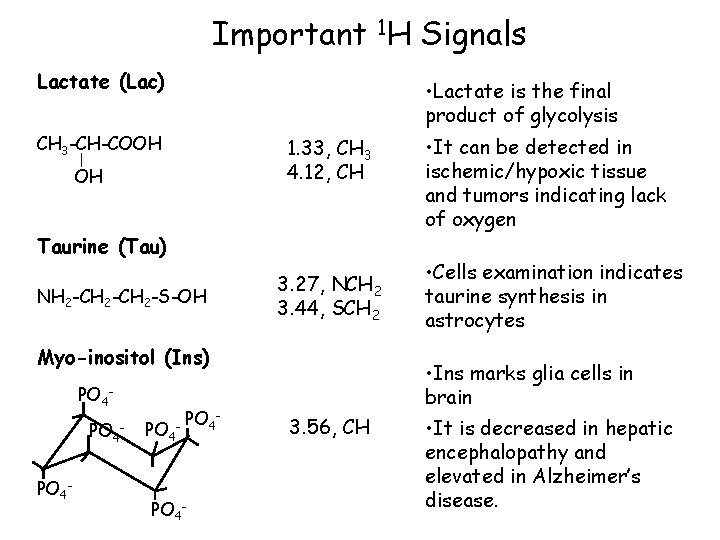
Important 1 H Signals N-Acetyl aspartate (NAA) O CH 3 -C-NH-CH-CH 2 -COOH 2. 02, CH 3 2. 52, CH 2 2. 70, CH 2 4. 40, CH Creatine (Cr), phosphocreatine (PCr) NH NH 2 -C-N-CH 2 -COOH CH 3 3. 04, CH 3 3. 93, CH 2 • NAA is a neuronal marker and indicates density and viability of neurons. • It is decreased in glioma, ischemia and degenerative diseases. • Cr is a marker of aerobic energy metabolism • Cr signal is constant even with pathologic changes and may be used as a control value • However, isolated cases of Cr deficiency may occur in children

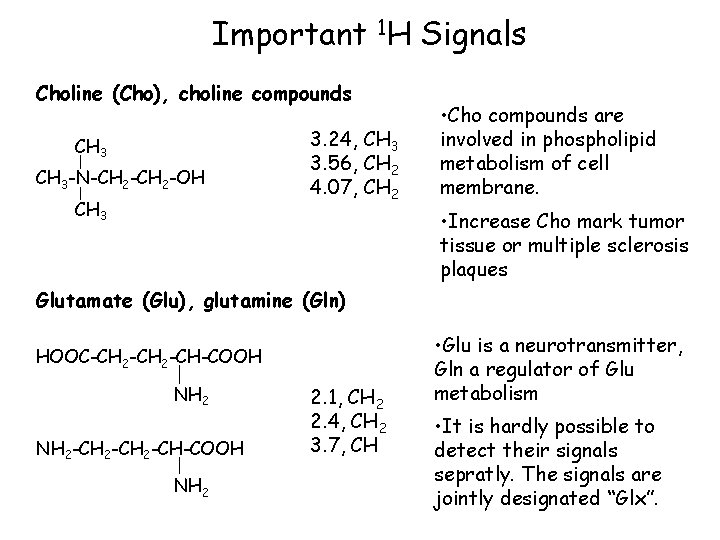
Important 1 H Signals Choline (Cho), choline compounds CH 3 -N-CH 2 -OH CH 3 3. 24, CH 3 3. 56, CH 2 4. 07, CH 2 • Cho compounds are involved in phospholipid metabolism of cell membrane. • Increase Cho mark tumor tissue or multiple sclerosis plaques Glutamate (Glu), glutamine (Gln) HOOC-CH 2 -CH-COOH NH 2 -CH-COOH NH 2 2. 1, CH 2 2. 4, CH 2 3. 7, CH • Glu is a neurotransmitter, Gln a regulator of Glu metabolism • It is hardly possible to detect their signals sepratly. The signals are jointly designated “Glx”.

Important 1 H Signals Lactate (Lac) • Lactate is the final product of glycolysis CH 3 -CH-COOH 1. 33, CH 3 4. 12, CH OH Taurine (Tau) NH 2 -CH 2 -S-OH 3. 27, NCH 2 3. 44, SCH 2 Myo-inositol (Ins) PO 4 - - PO 4 - • It can be detected in ischemic/hypoxic tissue and tumors indicating lack of oxygen • Cells examination indicates taurine synthesis in astrocytes • Ins marks glia cells in brain 3. 56, CH • It is decreased in hepatic encephalopathy and elevated in Alzheimer’s disease.

31 P MR Spectroscopy

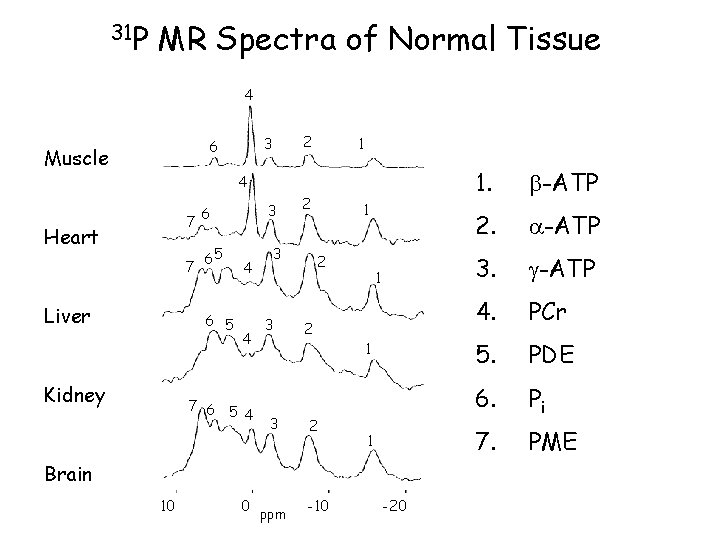
31 P MR Spectra of Normal Tissue 4 Muscle 2 3 6 1 4 7 Heart 7 Liver 65 6 5 Kidney 3 6 4 4 7 6 5 4 2 3 3 3 1 2 2 2 1 1 1 Brain 10 0 ppm -10 -20 1. -ATP 2. -ATP 3. -ATP 4. PCr 5. PDE 6. Pi 7. PME

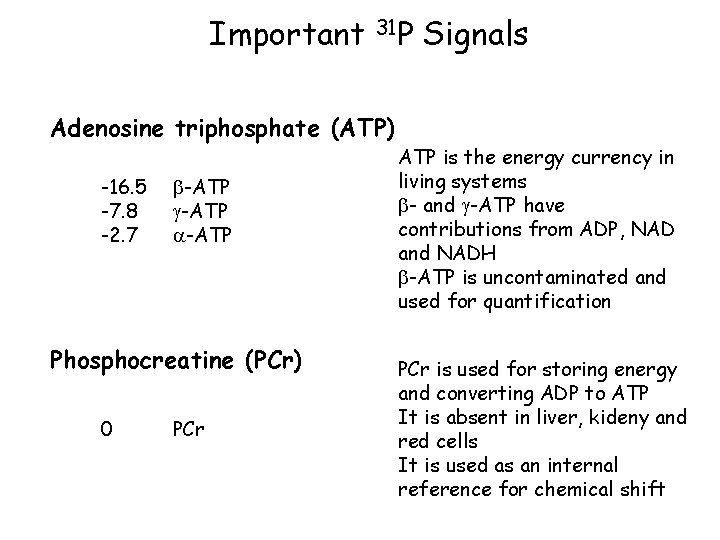
Important 31 P Adenosine triphosphate (ATP) -16. 5 -7. 8 -2. 7 -ATP Phosphocreatine (PCr) 0 PCr Signals ATP is the energy currency in living systems - and -ATP have contributions from ADP, NAD and NADH -ATP is uncontaminated and used for quantification PCr is used for storing energy and converting ADP to ATP It is absent in liver, kideny and red cells It is used as an internal reference for chemical shift

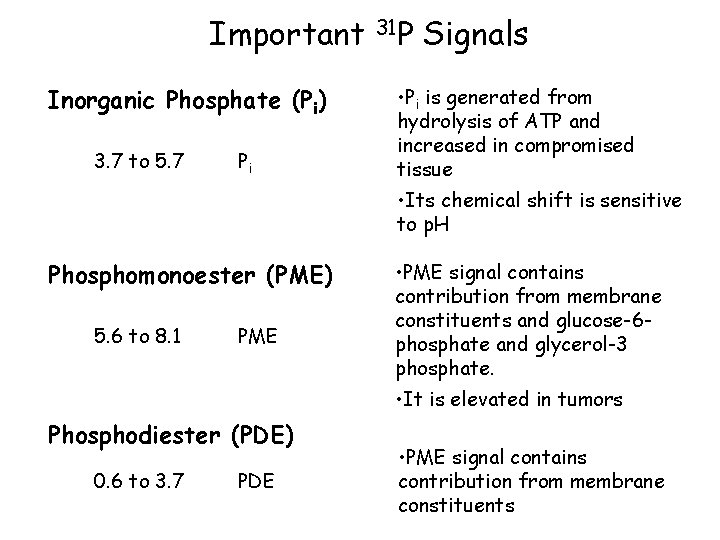
Important Inorganic Phosphate (Pi) 3. 7 to 5. 7 Pi 31 P Signals • Pi is generated from hydrolysis of ATP and increased in compromised tissue • Its chemical shift is sensitive to p. H Phosphomonoester (PME) 5. 6 to 8. 1 PME • PME signal contains contribution from membrane constituents and glucose-6 phosphate and glycerol-3 phosphate. • It is elevated in tumors Phosphodiester (PDE) 0. 6 to 3. 7 PDE • PME signal contains contribution from membrane constituents

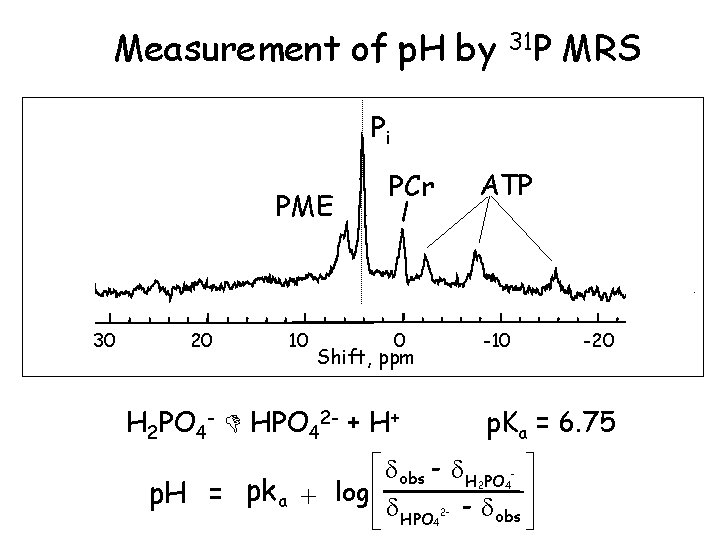
Measurement of p. H by 31 P MRS Pi PME 30 20 10 PCr ATP 0 -10 Shift, ppm H 2 PO 4 - HPO 42 - + H+ -20 p. Ka = 6. 75 é obs - H PO ù ú p. H = pka + log ê êë HPO - obs úû 2 4 2 - 4

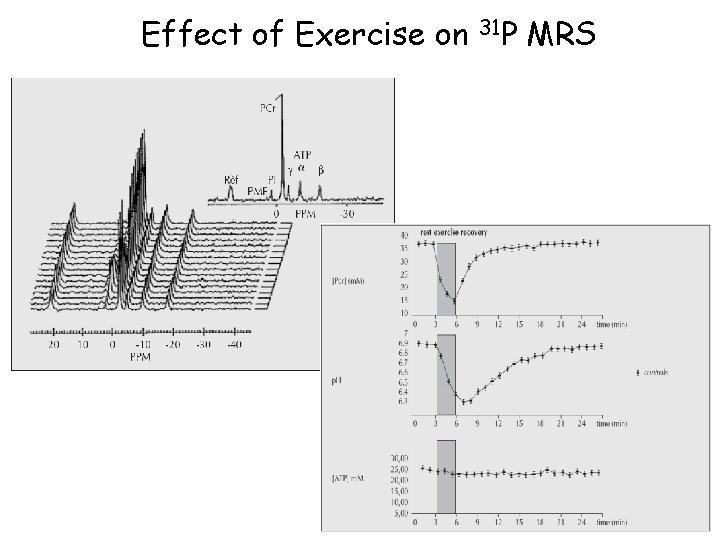
Effect of Exercise on 31 P MRS

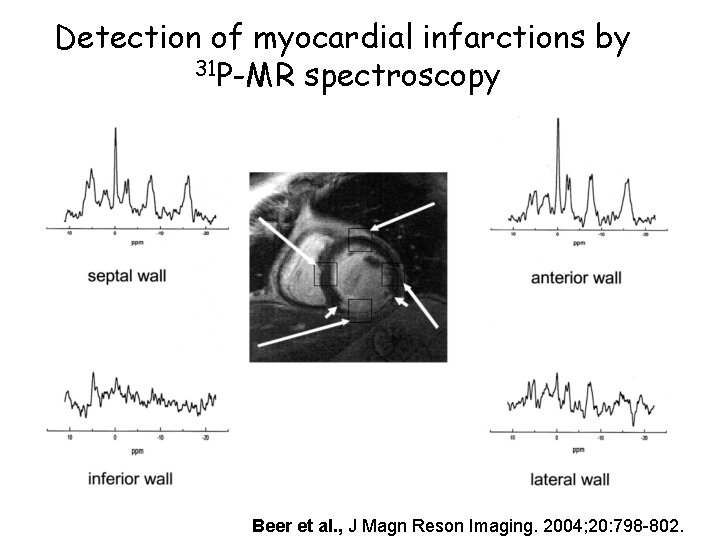
Detection of myocardial infarctions by 31 P-MR spectroscopy Beer et al. , J Magn Reson Imaging. 2004; 20: 798 -802.

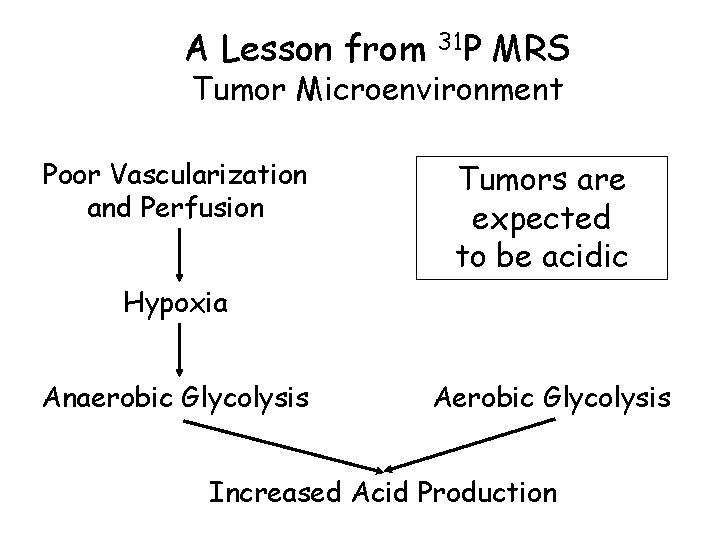
A Lesson from 31 P MRS Tumor Microenvironment Poor Vascularization and Perfusion Tumors are expected to be acidic Hypoxia Anaerobic Glycolysis Aerobic Glycolysis Increased Acid Production

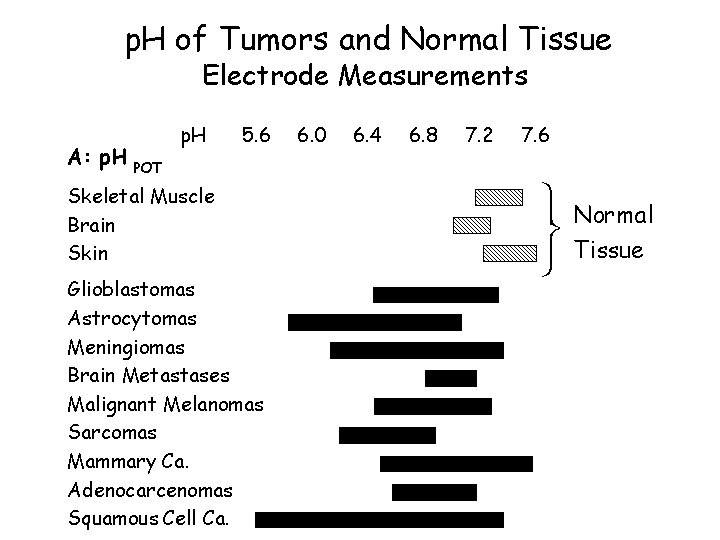
p. H of Tumors and Normal Tissue Electrode Measurements A: p. H POT p. H 5. 6 Skeletal Muscle Brain Skin Glioblastomas Astrocytomas Meningiomas Brain Metastases Malignant Melanomas Sarcomas Mammary Ca. Adenocarcenomas Squamous Cell Ca. 6. 0 6. 4 6. 8 7. 2 7. 6 ü Normal ý Tissue þ

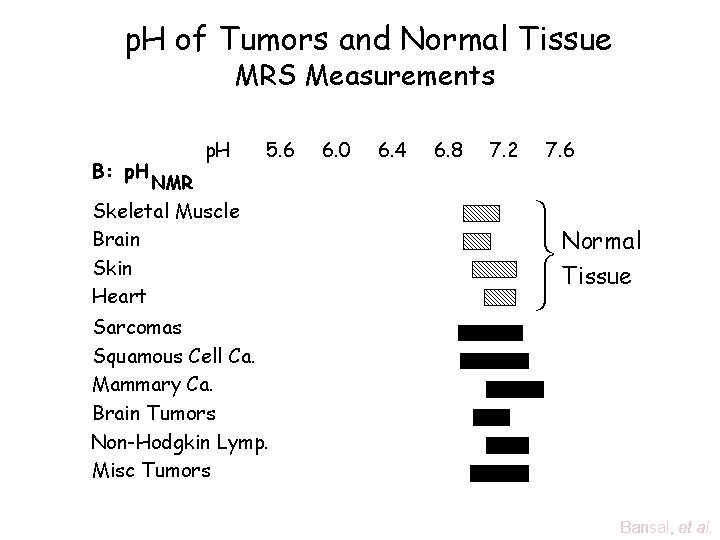
p. H of Tumors and Normal Tissue MRS Measurements B: p. H 5. 6 6. 0 6. 4 6. 8 7. 2 7. 6 NMR Skeletal Muscle Brain Skin Heart Sarcomas Squamous Cell Ca. Mammary Ca. Brain Tumors Non-Hodgkin Lymp. Misc Tumors ü Normal ý Tissue þ Bansal, et al.

23 Na MR Spectroscopy and Imaging

Biological Importance of Sodium Ø Sodium and other ions are inhomogeneously distributed across the cell membrane. Ø A transmembrane sodium gradient reflects a dynamic equilibrium between Na+-K+ ATPase versus passive or mediated flux. Ø The sodium gradient may be altered in certain diseased states. Bansal, et al.

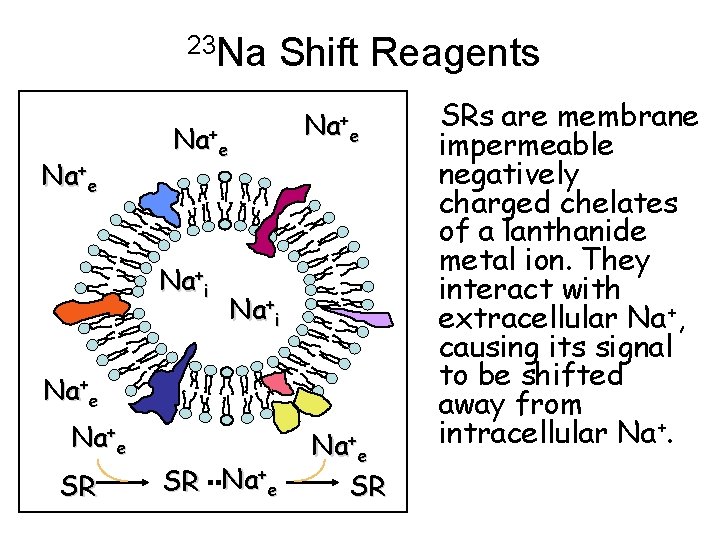
Biomedical 23 Na NMR Ø 23 Na is the second most sensitive nucleus for biomedical NMR. Ø Intra- and extracellular sodium resonate at the same frequency. Ø Two approaches to distinguish between different sodium pools: Paramagnetic Shift Reagents Multiple Quantum Filters Bansal, et al.

23 Na Na+e Shift Reagents Na+e Na+i Na+e SR SRs are membrane impermeable negatively charged chelates of a lanthanide metal ion. They interact with extracellular Na+, causing its signal to be shifted away from intracellular Na+.

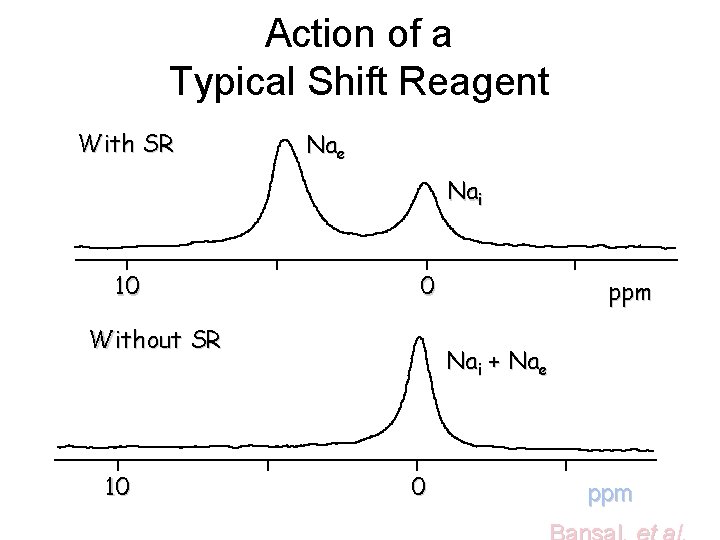
Action of a Typical Shift Reagent With SR Nae Nai 10 0 Without SR 10 ppm Nai + Nae 0 ppm

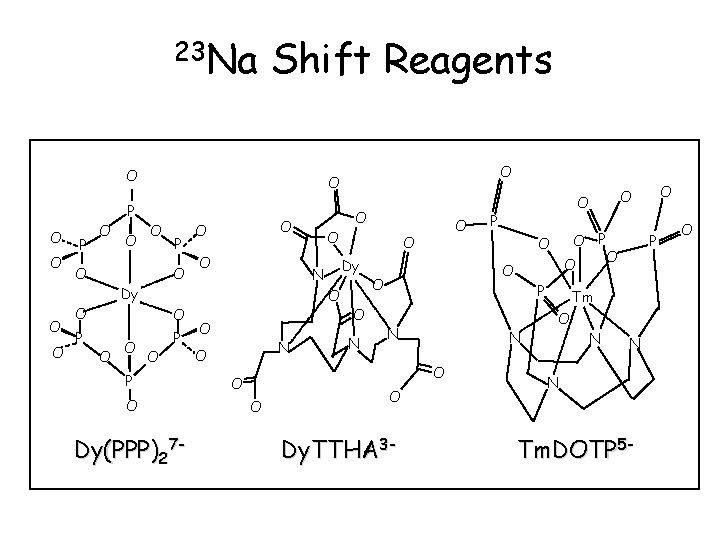
23 Na Shift Reagents O P O O O O Dy O O P O Dy(PPP)27 - N O O O P O Dy N O O O O P N O Dy. TTHA 3 - P O O N O O P O Tm O N N N Tm. DOTP 5 - O O O

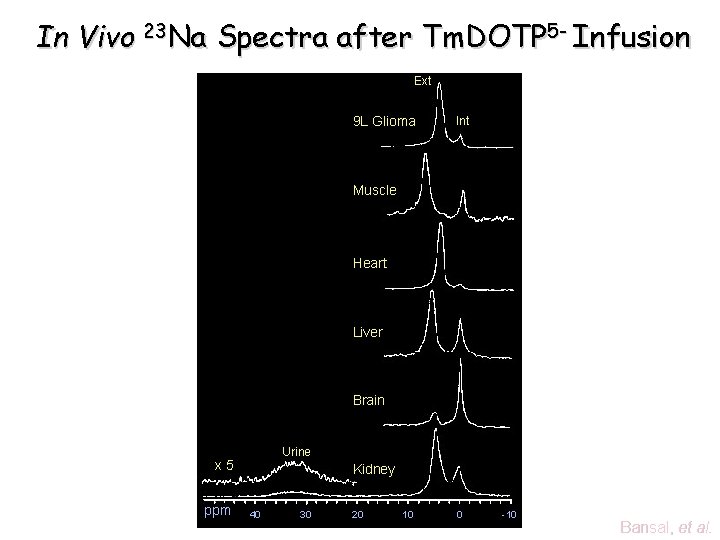
In Vivo 23 Na Spectra after Tm. DOTP 5 - Infusion Ext 9 L Glioma Int Muscle Heart Liver Brain Urine x 5 ppm Kidney 40 30 20 10 0 -10 Bansal, et al.

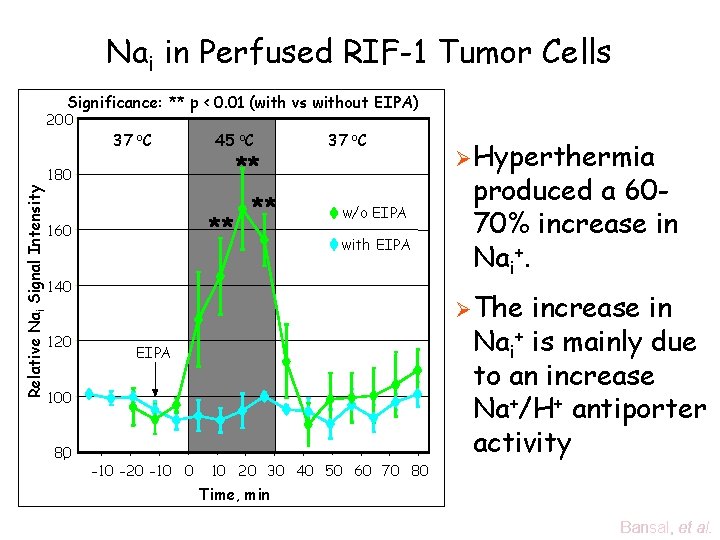
Nai in Perfused RIF-1 Tumor Cells Significance: ** p < 0. 01 (with vs without EIPA) Relative Nai Signal Intensity 200 37 o. C 45 o. C ** 180 ** 160 ** 37 o. C w/o EIPA with EIPA 140 120 produced a 6070% increase in Nai+. Ø The EIPA 100 80 Ø Hyperthermia -10 -20 -10 0 10 20 30 40 50 60 70 80 increase in Nai+ is mainly due to an increase Na+/H+ antiporter activity Time, min Bansal, et al.

Multiple-Quantum Filters MQFs depend only on the relaxation properties of 23 Na. Thus, they do not produce any known physiological perturbation to the biological system and cab be applied to humans. Disadvantages • • Low signal-to-noise ratio Some Nae+ contribution Bansal, et al.

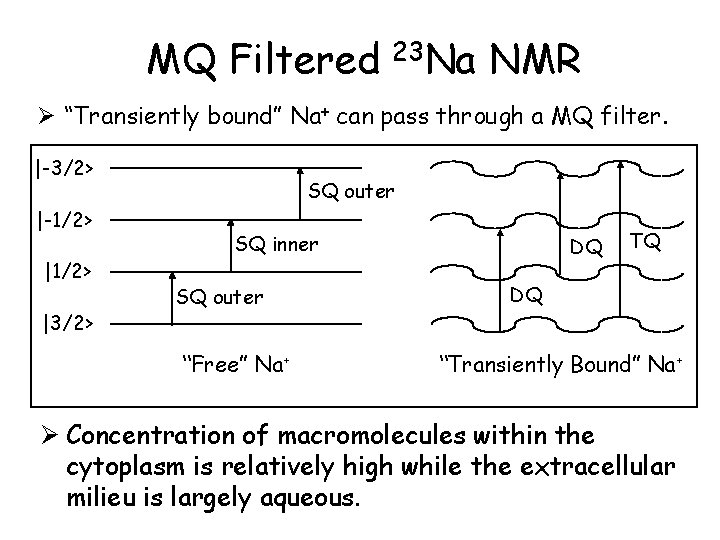
MQ Filtered 23 Na NMR Ø “Transiently bound” Na+ can pass through a MQ filter. |-3/2> |-1/2> |3/2> SQ outer SQ inner SQ outer “Free” Na+ DQ TQ DQ “Transiently Bound” Na+ Ø Concentration of macromolecules within the cytoplasm is relatively high while the extracellular milieu is largely aqueous.

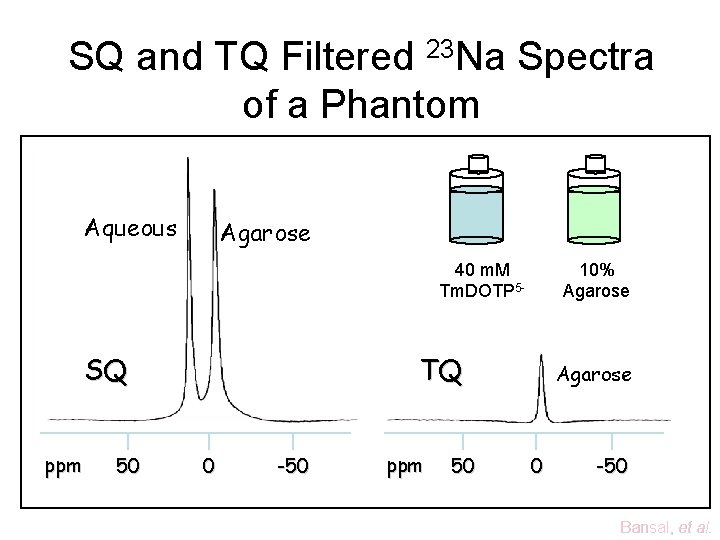
SQ and TQ Filtered 23 Na Spectra of a Phantom Aqueous Agarose 40 m. M Tm. DOTP 5 - SQ ppm 50 10% Agarose TQ 0 -50 ppm 50 Agarose 0 -50 Bansal, et al.

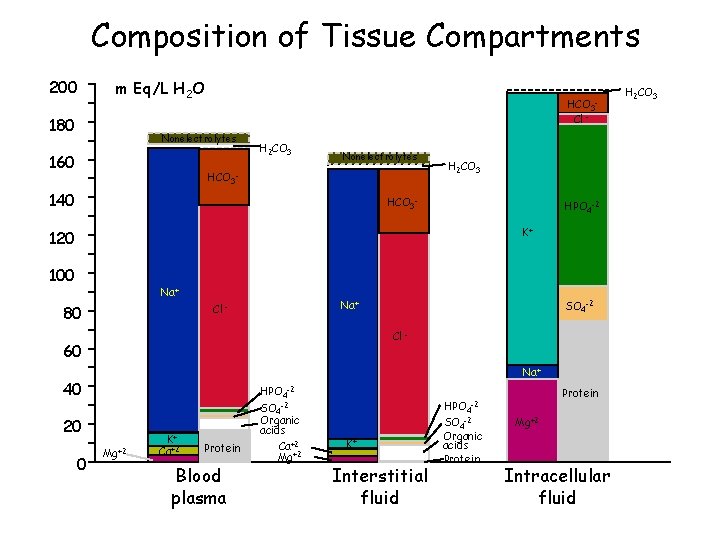
Composition of Tissue Compartments 200 m Eq/L H 2 O 180 HCO 3 Cl - Nonelectrolytes 160 H 2 CO 3 Nonelectrolytes HCO 3 - 140 H 2 CO 3 HCO 3 - HPO 4 -2 K+ 120 100 Na+ Cl - 80 Cl - 60 Na+ 40 20 0 SO 4 -2 Mg+2 K+ Ca+2 Protein Blood plasma HPO 4 -2 SO 4 -2 Organic acids Ca+2 Mg+2 K+ Interstitial fluid HPO 4 -2 SO 4 -2 Organic acids Protein Mg+2 Intracellular fluid H 2 CO 3

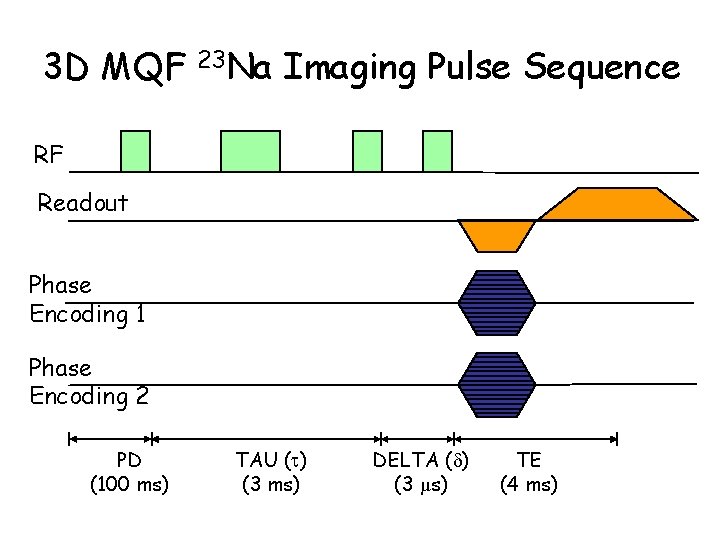
3 D MQF 23 Na Imaging Pulse Sequence RF Readout Phase Encoding 1 Phase Encoding 2 PD (100 ms) TAU ( ) (3 ms) DELTA ( ) (3 µs) TE (4 ms)

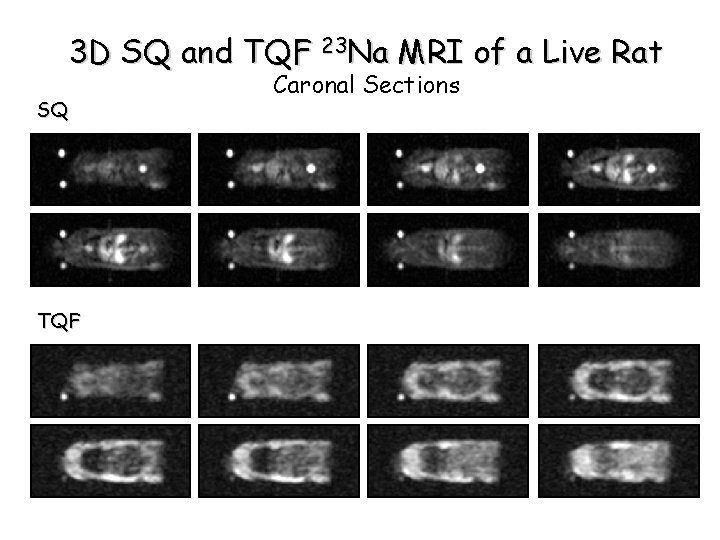
3 D SQ and TQF 23 Na MRI of a Live Rat SQ TQF Caronal Sections

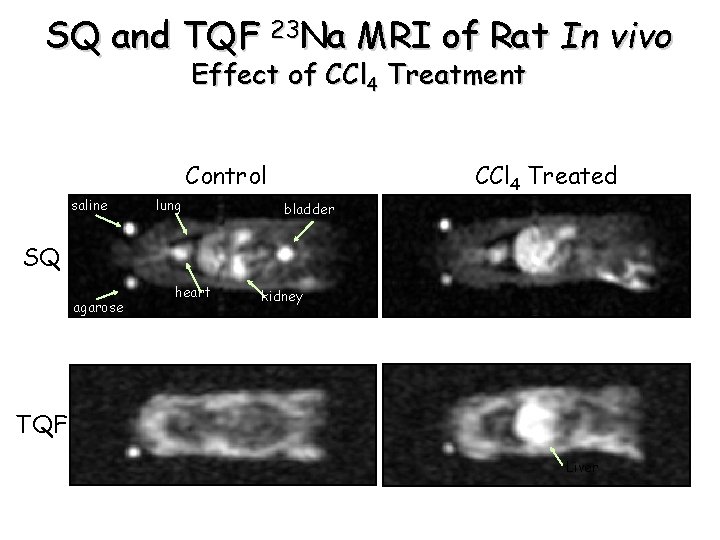
SQ and TQF 23 Na MRI of Rat In vivo Effect of CCl 4 Treatment Control saline lung CCl 4 Treated bladder SQ agarose heart kidney TQF Liver