RNA Switches Genetic Research Tools RAVINDER NAGPAL 1













- Slides: 40

RNA Switches Genetic Research Tools RAVINDER NAGPAL 1, MALVIKA MALIK 1, MONICA PUNIYA 2, AARTI BHARDWAJ 3, SHALINI JAIN 4 and HARIOM YADAV 4* 1 Dairy Microbiology, 2 Dairy Cattle Nutrition, 4 Animal Biochemistry, National Dairy Research Institute, Karnal 132001, Haryana, 3 Meerut Institute of Engineering and Technology, Meerut-250002, U. P. , India. Email: yadavhariom@gmail. com

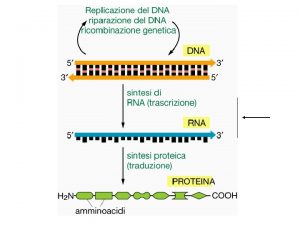
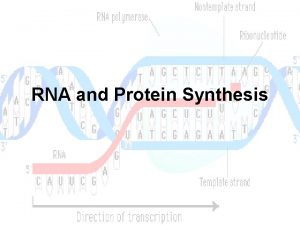
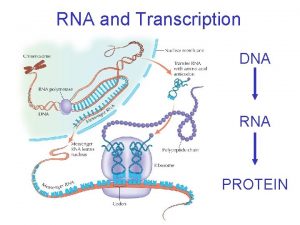
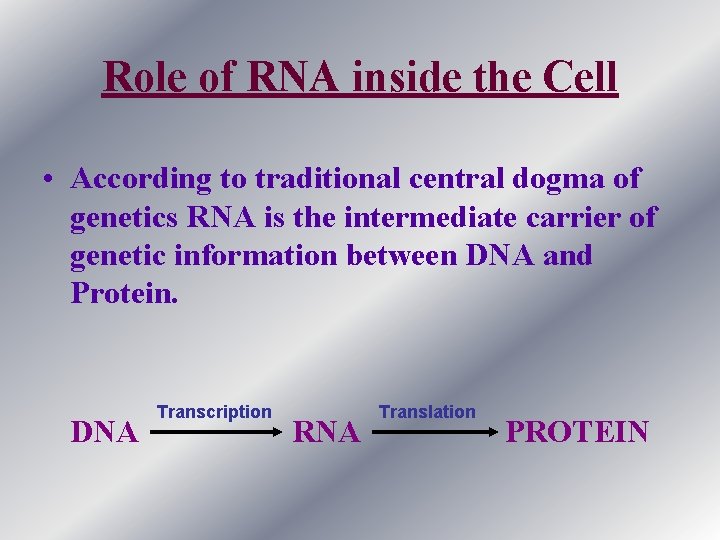
Role of RNA inside the Cell • According to traditional central dogma of genetics RNA is the intermediate carrier of genetic information between DNA and Protein. DNA Transcription RNA Translation PROTEIN

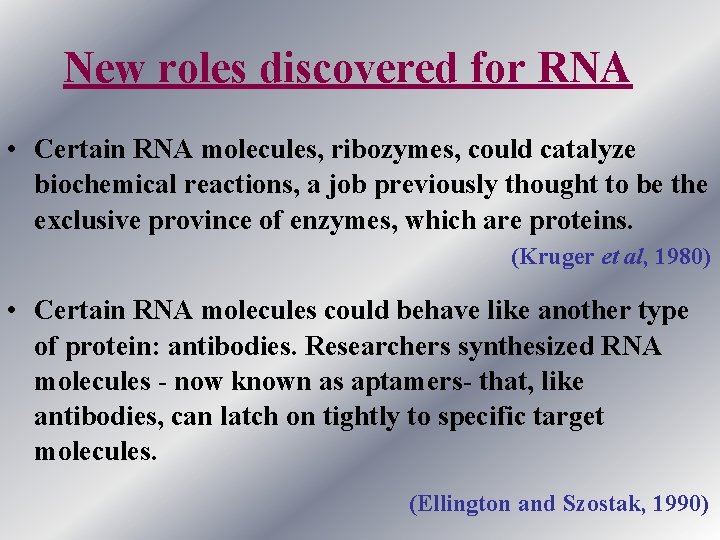
New roles discovered for RNA • Certain RNA molecules, ribozymes, could catalyze biochemical reactions, a job previously thought to be the exclusive province of enzymes, which are proteins. (Kruger et al, 1980) • Certain RNA molecules could behave like another type of protein: antibodies. Researchers synthesized RNA molecules - now known as aptamers- that, like antibodies, can latch on tightly to specific target molecules. (Ellington and Szostak, 1990)

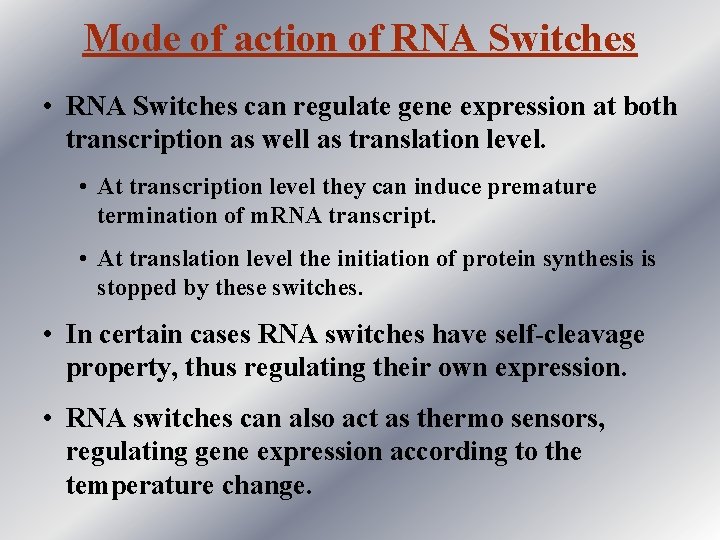
contd. . . • Certain RNA sequences can directly sense environmental factors and small molecule metabolites, this allow the associated m. RNA to regulate their own transcription or translation. (Breaker, 2002) • These self regulatory messages are called as RNA sensors and RNA switches.

RNA Switches • RNA switches are m. RNAs that sense the environment directly, shutting themselves down in response to particular chemical clues. • Breaker, Nudler, Yura and Cossart laboratories report that specific RNA sequences can act as environmental sensors of vitamin cofactors (including vitamins B 1, B 2 and B 12) and temperature, which allow them to directly regulate the transcription or translation of associated m. RNAs.

contd. . . • Distinct RNA molecules directly perform or mediate enzymatic processes such as RNA cleavage, splicing and translation. • Non-coding RNAs are involved in a tremendous variety of gene regulatory mechanisms that operate at both the DNA and m. RNA level. • Seven types of RNA switches have been found in bacteria so far. These include switches controlling the manufacture of the vitamins B 1 and B 2 and the nucleotide guanine.

Mode of action of RNA Switches • RNA Switches can regulate gene expression at both transcription as well as translation level. • At transcription level they can induce premature termination of m. RNA transcript. • At translation level the initiation of protein synthesis is stopped by these switches. • In certain cases RNA switches have self-cleavage property, thus regulating their own expression. • RNA switches can also act as thermo sensors, regulating gene expression according to the temperature change.

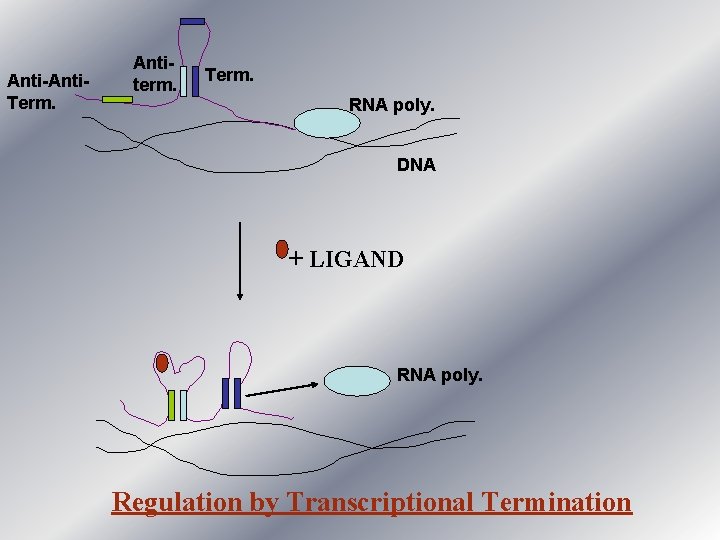
Anti-Anti. Term. Antiterm. Term. RNA poly. DNA + LIGAND RNA poly. Regulation by Transcriptional Termination

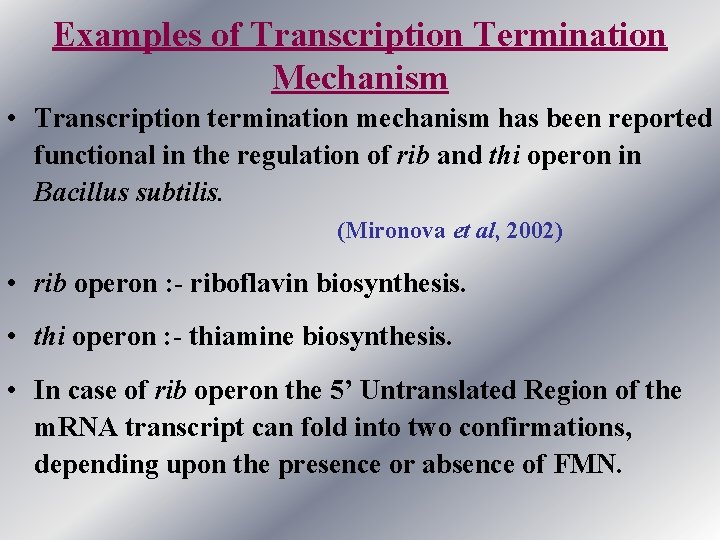
Examples of Transcription Termination Mechanism • Transcription termination mechanism has been reported functional in the regulation of rib and thi operon in Bacillus subtilis. (Mironova et al, 2002) • rib operon : - riboflavin biosynthesis. • thi operon : - thiamine biosynthesis. • In case of rib operon the 5’ Untranslated Region of the m. RNA transcript can fold into two confirmations, depending upon the presence or absence of FMN.

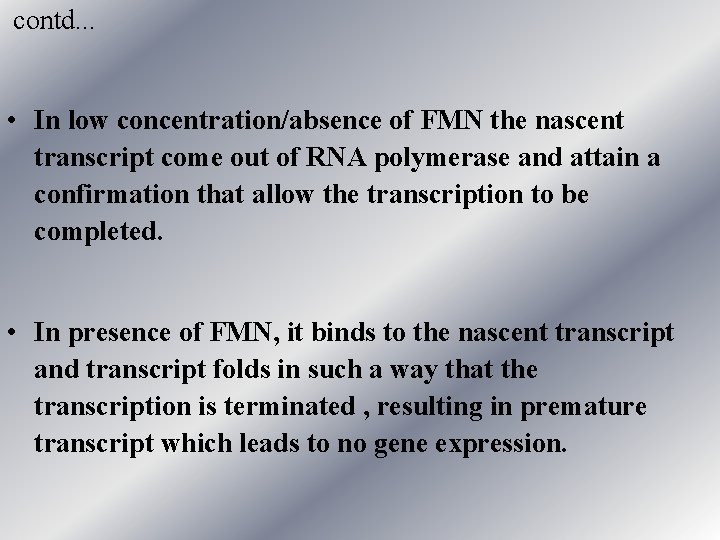
contd. . . • In low concentration/absence of FMN the nascent transcript come out of RNA polymerase and attain a confirmation that allow the transcription to be completed. • In presence of FMN, it binds to the nascent transcript and transcript folds in such a way that the transcription is terminated , resulting in premature transcript which leads to no gene expression.

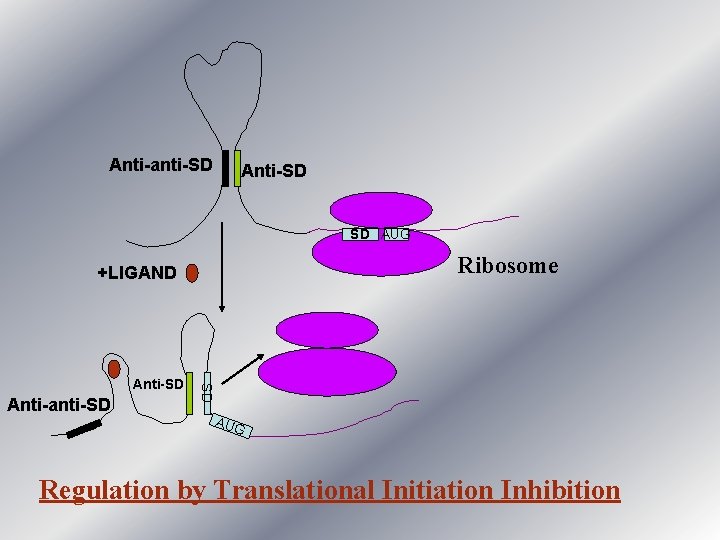
Anti-anti-SD Anti-SD SD AUG Ribosome +LIGAND Anti-anti-SD SD Anti-SD AUG Regulation by Translational Initiation Inhibition

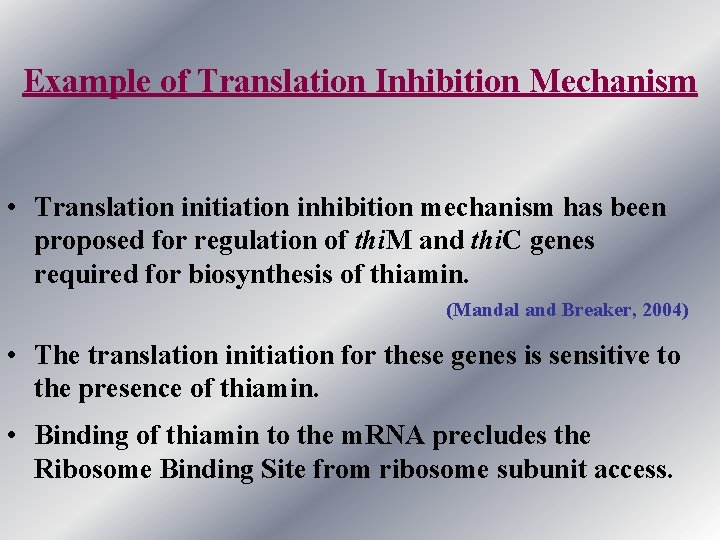
Example of Translation Inhibition Mechanism • Translation initiation inhibition mechanism has been proposed for regulation of thi. M and thi. C genes required for biosynthesis of thiamin. (Mandal and Breaker, 2004) • The translation initiation for these genes is sensitive to the presence of thiamin. • Binding of thiamin to the m. RNA precludes the Ribosome Binding Site from ribosome subunit access.

Observations: • Gram-positive bacteria are more likely to couple these elements to a terminator/anti-terminator system, that is at the level of transcription. • Gram-negative bacteria more typically link these elements to a SD-sequestration mechanism, that is at the level of translation.

contd. . . • This type of regulation is found to be economic for the bacterial system as: – In Gram positive bacteria the genes for one biosynthetic pathway are arranged in a cluster, therefore they are regulated at the transcription level. – In Gram negative bacteria the genes for one biosynthetic pathway are scattered along the whole genome and are therefore regulated at the translation level.

Self-cleavage of the RNA molecules • The regulation of glm. S gene(glutamine-fructose-6 phosphate amidotransferase)in B. subtilis is by associated activity of ribozyme and riboswitches. • In this case the 5’ UTR of glm. S binds with gluosamin-6 -phosphate, product of Glm. S activity and start acting as ribozyme, due to conformational change. • This results in the self cleavage of the m. RNA by an internal phosphoester transfer.

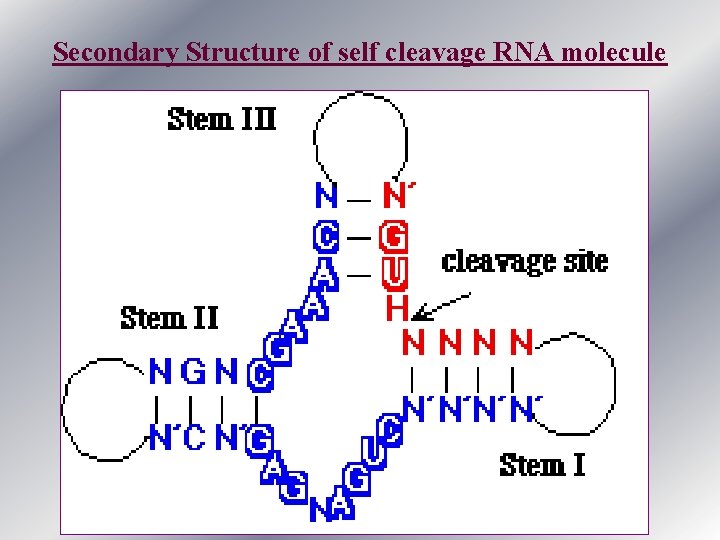
Secondary Structure of self cleavage RNA molecule

Activation of gene expression by RNA Thermo sensors • There are two well-studied examples of temperature-dependent regulation of gene expression and/or activity: – during the heat shock response – during pathogenic invasion • Almost all the known thermo sensors regulate the gene expression at the translational level.

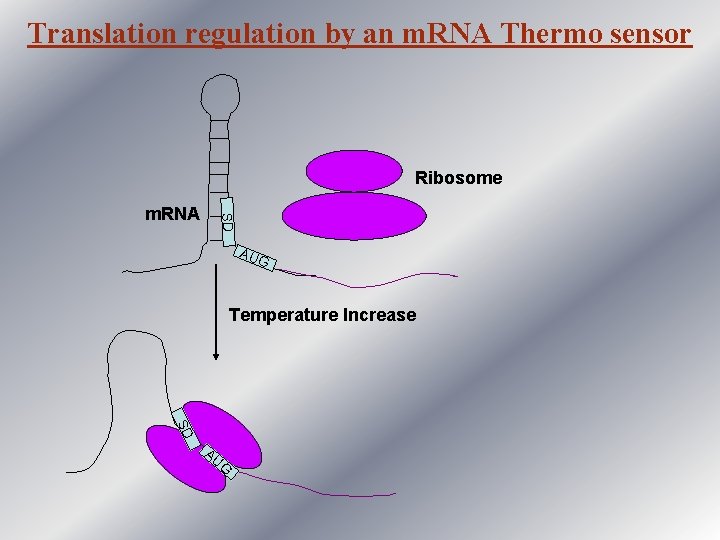
Translation regulation by an m. RNA Thermo sensor Ribosome SD m. RNA AU G Temperature Increase SD AU G

Examples of RNA Thermo sensors • Translation of Lcr. F, a general activator of virulencerelated gene expression in Yersinia pestis, was found to be thermally regulated. (Hoe and Goguen, 1993) • A new report from the Cossart lab (2002) now provides strong evidence for an RNA thermo sensor that regulates translation of Prf. A, a general activator of virulence genes in a pathogenic variety of Listeria, L. monocytogenes.

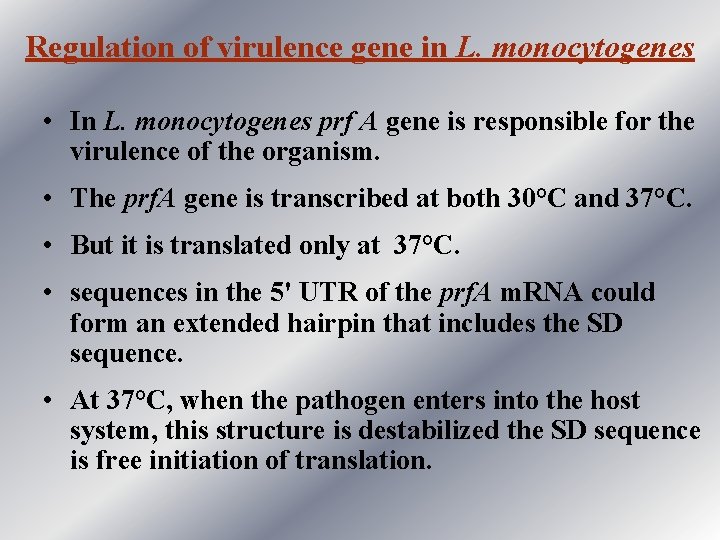
Regulation of virulence gene in L. monocytogenes • In L. monocytogenes prf A gene is responsible for the virulence of the organism. • The prf. A gene is transcribed at both 30°C and 37°C. • But it is translated only at 37°C. • sequences in the 5' UTR of the prf. A m. RNA could form an extended hairpin that includes the SD sequence. • At 37°C, when the pathogen enters into the host system, this structure is destabilized the SD sequence is free initiation of translation.

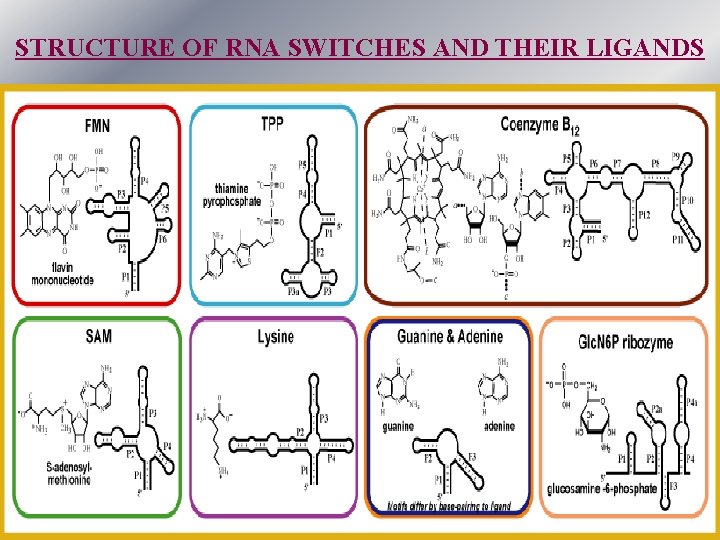
STRUCTURE OF RNA SWITCHES AND THEIR LIGANDS

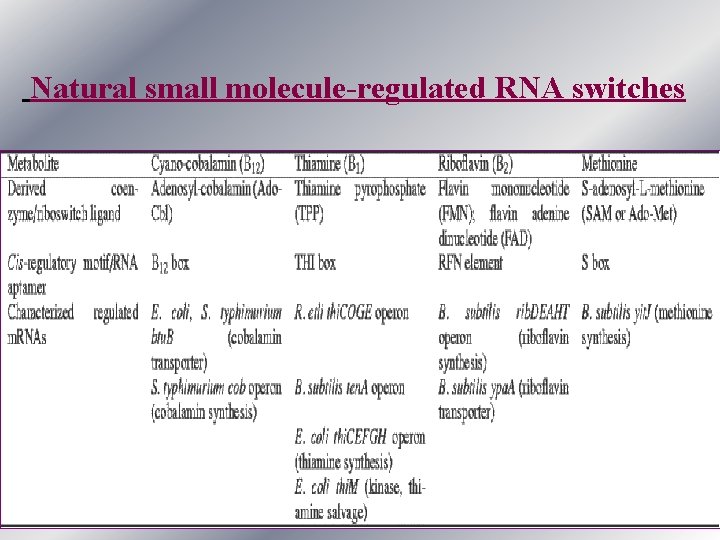
Natural small molecule-regulated RNA switches

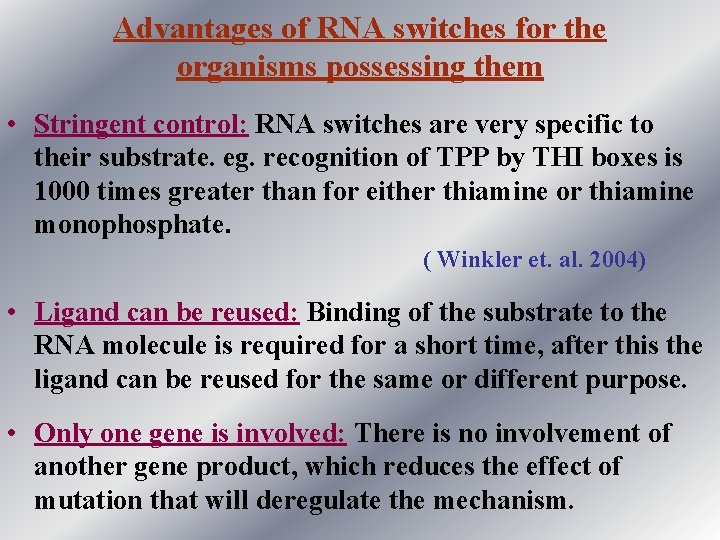
Advantages of RNA switches for the organisms possessing them • Stringent control: RNA switches are very specific to their substrate. eg. recognition of TPP by THI boxes is 1000 times greater than for either thiamine or thiamine monophosphate. ( Winkler et. al. 2004) • Ligand can be reused: Binding of the substrate to the RNA molecule is required for a short time, after this the ligand can be reused for the same or different purpose. • Only one gene is involved: There is no involvement of another gene product, which reduces the effect of mutation that will deregulate the mechanism.

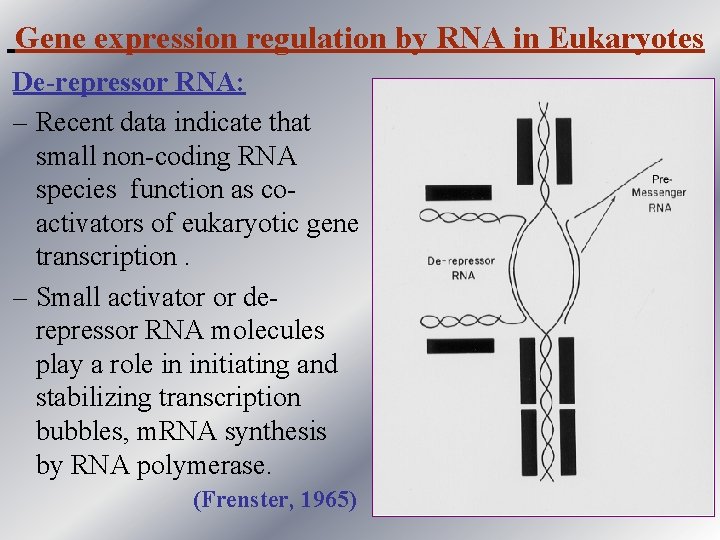
Gene expression regulation by RNA in Eukaryotes De-repressor RNA: – Recent data indicate that small non-coding RNA species function as coactivators of eukaryotic gene transcription. – Small activator or derepressor RNA molecules play a role in initiating and stabilizing transcription bubbles, m. RNA synthesis by RNA polymerase. (Frenster, 1965)

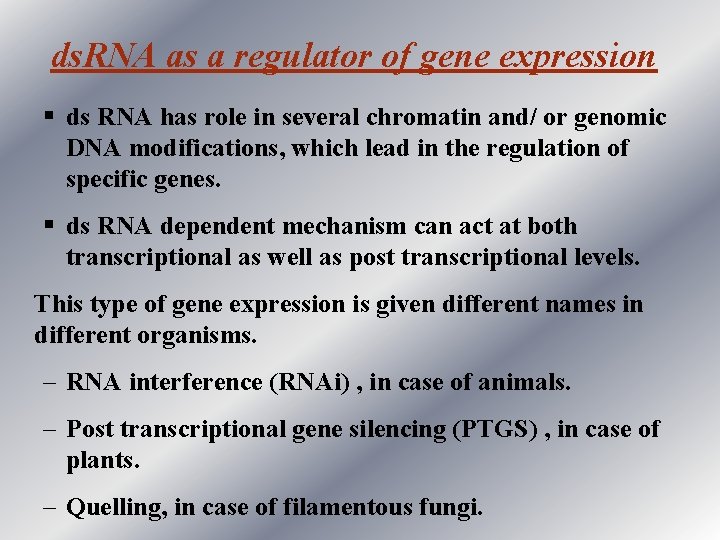
ds. RNA as a regulator of gene expression § ds RNA has role in several chromatin and/ or genomic DNA modifications, which lead in the regulation of specific genes. § ds RNA dependent mechanism can act at both transcriptional as well as post transcriptional levels. This type of gene expression is given different names in different organisms. – RNA interference (RNAi) , in case of animals. – Post transcriptional gene silencing (PTGS) , in case of plants. – Quelling, in case of filamentous fungi.

Other RNAs involved in regulation of gene expression – micro RNA (mi RNA)- a class of noncoding small RNA identified for their role in translational repression in some animals. – short hairpin RNA (sh RNA)- a special class of RNA that becomes double stranded by folding of the RNA strand over itself, has been shown to be responsible for the transcriptional regulation.

Applications of RNA switches in genetic research • RNA switches can be used to create small moleculeregulated transgenes, which may allow researchers to manipulate expression of any individual construct within a battery of simultaneously introduced experimental constructs. (Werstuck and Green, 1998) • We can imagine exploiting RNA thermoregulation to create heat-inducible transgenes. (Brand Perrimon, 1993)

contd. . . • Aptamers have been placed within 5' UTRs and shown to inhibit gene expression upon introduction of the appropriate ligand. (Werstuck and Green, 1998) • Self-cleaving RNAs can be placed under allosteric control by various small molecules, which can then be used to analyze the composition of chemical and biological mixtures. (Seetharaman et al, 2001) • RNA switches can be used in gene therapy, allowing the patient to take pills to switch introduced gene on or off.

contd. . . • It may be possible to design drugs using RNA switches’ cognate ligands that would constitutively repress associated gene activity. – Such compounds would efficiently inhibit bacterial growth by simultaneously repressing multiple components in a given biosynthetic/metabolic/transport pathway. – Such compounds might be likely to have relatively low toxicity, as they would be designed to target RNA, not protein.

RNA on a Chip • In 1995, Breaker successfully engineered RNA-based molecular switches. – A molecular switch is a molecule that turns on or off by another molecule or compound. • With dozens of these switches on hand, Breaker created an array of biosensors that use RNA to measure or detect compounds.

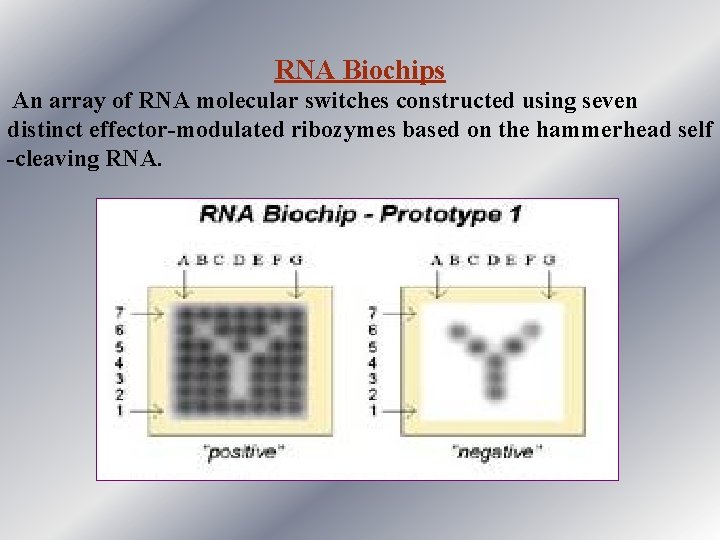
RNA Biochips An array of RNA molecular switches constructed using seven distinct effector-modulated ribozymes based on the hammerhead self -cleaving RNA.

Array for RNA • Breaker placed the RNA switches on a gold-coated silicon surface and arranged them in clusters. • Each switch was designed to bind only to a specific molecule its “target” and then to release a signal that identifies the target molecule. (In the prototype, the switches released a radioactive signal. ) • This array of RNA switches was tested on a variety of complex mixtures. In one experiment different strains of E. coli found in bacterial cultures were successfully identified.

• The array's ability to simultaneously identify a potentially large number of compounds, combined with the precise exclusivity of each switch, adds up to a recipe for a powerful and wide-ranging laboratory on a dime-sized slice of silicon.

RNA Super chip • Future RNA chips, capable of revealing the molecular composition of complex mixtures-like blood serum and industrial waste-far more comprehensively than current biochips can be synthesized. • Advanced versions of RNA biochip could be used for many different targets like drugs, toxins and metabolites, as well as proteins and nucleic acids.

Benefit of using RNA biochip • Benefit of RNA switches is their ability to withstand the sometimes unpredictable and harsh environment outside the lab as compared to a protein biochip. • Breaker's RNA switches have been engineered to refold back to their original form after heating, this snap-back character will give RNA biochips a considerable advantage for use in more exotic test environments.

Obstacles • Manufacturing costs. • The chemical stability of the switches. RNA is vulnerable to certain chemicals often found in test situations that can disintegrate a switch. • The finer points of molecular recognition. • This technology is so new that it is unclear just how many different compounds it will prove possible to recognize.

Future Prospects • RNA switches must be engineered to release a fluorescent, rather than a radioactive signal. • Use of an RNA chip in diverse fields like chemical engineering, environmental science, and even biological and chemical warfare defense. • Use of RNA chip in the detection and identification of pathogens.


Conclusion • Apart from being the carrier of genetic information , RNA can also act as the regulator of expression this information. These regulators are called as RNA switches. • RNA switches have been discovered to be involved in the regulation of gene expression in prokaryotes. • RNA switches can be used in genetic research to study the effect of environmental changes on particular gene expression.

• RNA switches can be used to in gene therapy and as drugs. • Researchers are trying to synthesize artificial RNA switches and we can hope that in future we will be able to use RNA switches not only in genetic research but also in antimicrobial therapy, as biosensors for different chemicals and in detection of pathogenic microorganisms.
 Radhika nagpal
Radhika nagpal Man kit leung
Man kit leung Dr rajeev nagpal
Dr rajeev nagpal Nagpal surgery
Nagpal surgery Gene flow vs genetic drift
Gene flow vs genetic drift Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm A gene pool consists of
A gene pool consists of Gene flow vs genetic drift
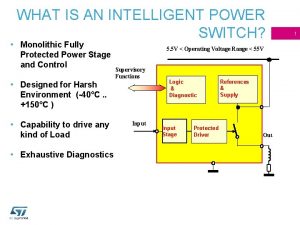
Gene flow vs genetic drift Intelligent power switches
Intelligent power switches Three generations of packet switches
Three generations of packet switches Kundan switches models
Kundan switches models Which type of reaction
Which type of reaction Bridges vs switches
Bridges vs switches Cisco rv120w price
Cisco rv120w price Cisco 100 series switches
Cisco 100 series switches Bridges vs switches
Bridges vs switches Dominant off circuit
Dominant off circuit Mercury switches in cars
Mercury switches in cars Radiall rf microwave switches
Radiall rf microwave switches Two technicians are discussing schematic symbols
Two technicians are discussing schematic symbols High performance switches
High performance switches Slotted optical switches
Slotted optical switches Unica solutions
Unica solutions Series resonant inverter with bidirectional switches
Series resonant inverter with bidirectional switches High performance switches and routers
High performance switches and routers A switch combines crossbar switches in several stages
A switch combines crossbar switches in several stages Uma multiprocessors using crossbar switches
Uma multiprocessors using crossbar switches A switch combines crossbar switches in several stages
A switch combines crossbar switches in several stages Zenith automatic transfer switches
Zenith automatic transfer switches Netgear gsm/fsm fully managed switches
Netgear gsm/fsm fully managed switches Refurbished netgear gsm/fsm fully managed switches
Refurbished netgear gsm/fsm fully managed switches Limit switches
Limit switches We should not touch electric switches with wet hands. why
We should not touch electric switches with wet hands. why High performance core router
High performance core router Banofge
Banofge Compliance objectives definition
Compliance objectives definition Sewing tools measuring tools
Sewing tools measuring tools Legal analysis methods
Legal analysis methods Tools for secondary research
Tools for secondary research Nucleotide in rna
Nucleotide in rna