Roles of RNA m RNA messenger r RNA












- Slides: 35

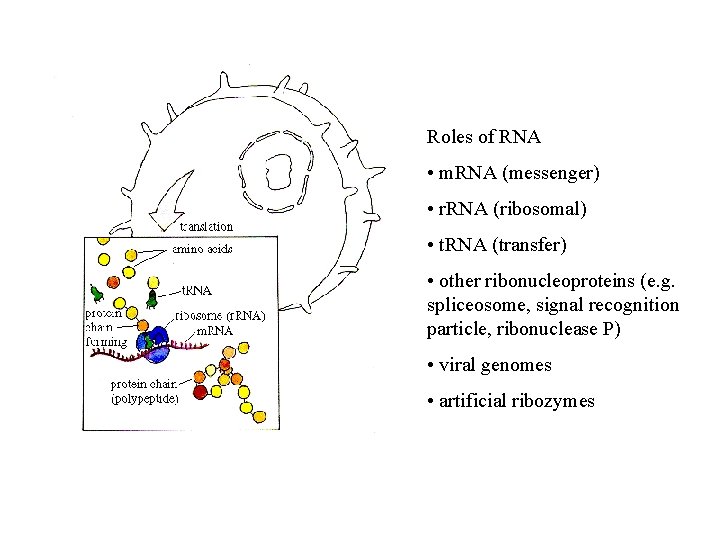
Roles of RNA • m. RNA (messenger) • r. RNA (ribosomal) • t. RNA (transfer) • other ribonucleoproteins (e. g. spliceosome, signal recognition particle, ribonuclease P) • viral genomes • artificial ribozymes

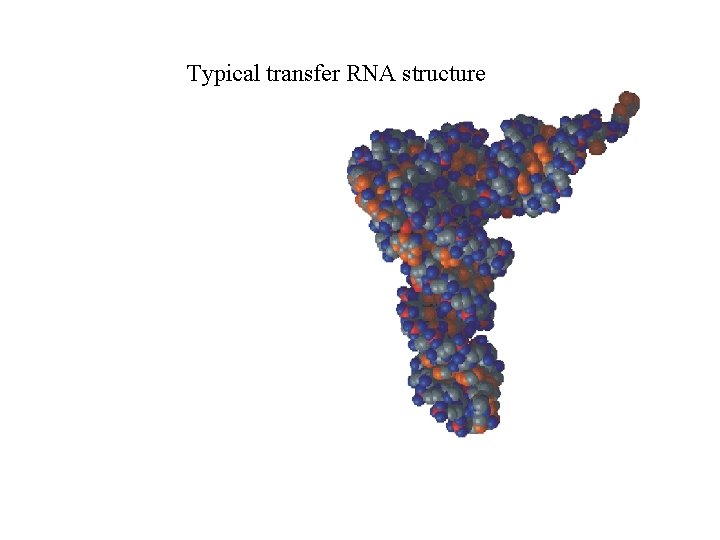
Typical transfer RNA structure



Thermodynamics parameters are measured on real molecules. Helix formation = hydrogen bonds + stacking G C U A C U Multi-branched loop Bulges Internal loops } DG = -1. 2 kcal/mol U loop DG = + 4. 5 kcal/mol C Entropic penalty for loop formation. } DG = -2. 1 kcal/mol Sum up contributions of helices and loops over the whole structure. Hairpin loop

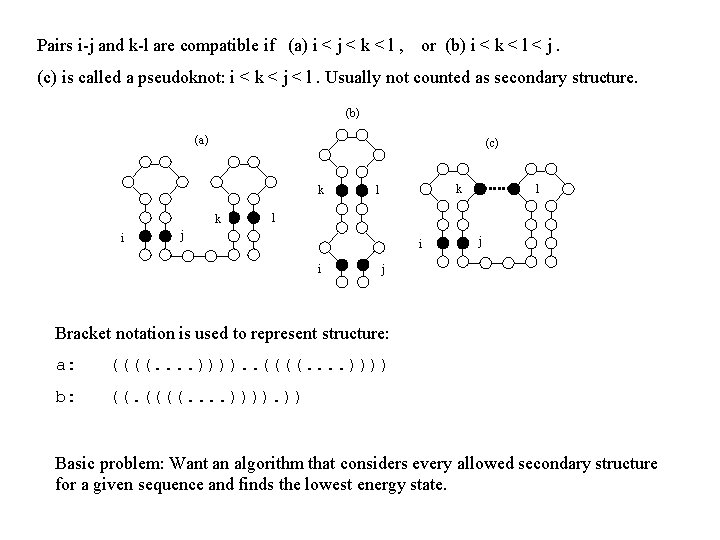
Pairs i-j and k-l are compatible if (a) i < j < k < l , or (b) i < k < l < j. (c) is called a pseudoknot: i < k < j < l. Usually not counted as secondary structure. (b) (a) (c) k k i k l l l j i i j j Bracket notation is used to represent structure: a: ((((. . )))). . ((((. . )))) b: ((. ((((. . )))). )) Basic problem: Want an algorithm that considers every allowed secondary structure for a given sequence and finds the lowest energy state.

Simplest case: find structure which maximizes number of base pairs. Let = -1 if bases can pair and + if not. Ignore loop contributions. E(i, j) = energy of min energy structure for chain segment from i to j. We want E(1, N). or = i j-1 j i k j Algorithms that work by recursion relations like this are called dynamic programming. The algorithm is O(N 3) although the number of structures increases exponentially with N. Also need to do backtracking to work out the minimum energy structure: Set B(i, j) = k if j is paired with k, or 0 if unpaired.

Partition Function Algorithm (for simplest energy rules) or = i j-1 j i k where Real Energy Rules : Need to consider many special cases. What type of loop are you closing? Algorithm is more complex but still is O(N 3). j

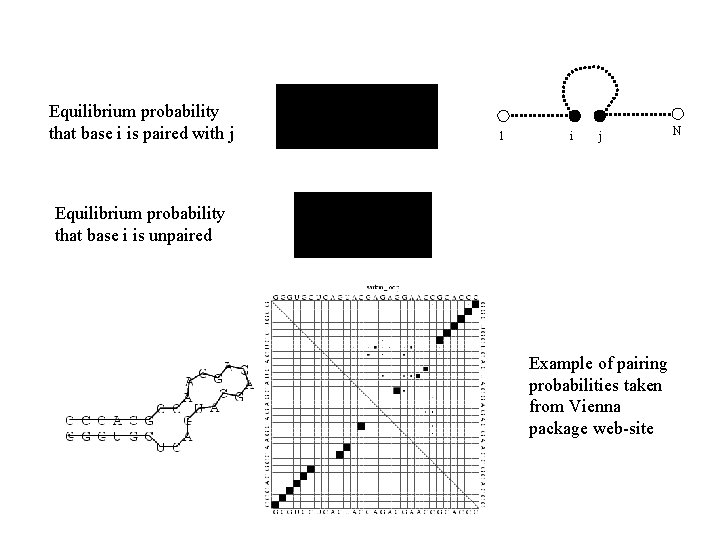
Equilibrium probability that base i is paired with j 1 i j Equilibrium probability that base i is unpaired Example of pairing probabilities taken from Vienna package web-site N

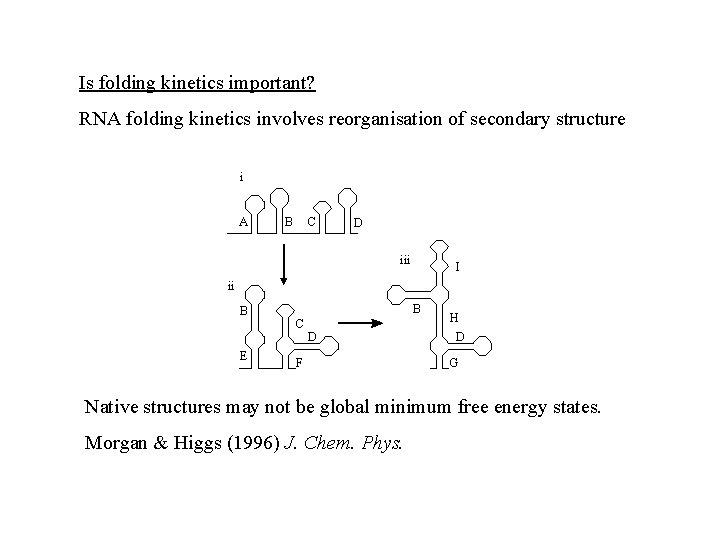
Is folding kinetics important? RNA folding kinetics involves reorganisation of secondary structure i A C B D iii I ii B E B C D F H D G Native structures may not be global minimum free energy states. Morgan & Higgs (1996) J. Chem. Phys.

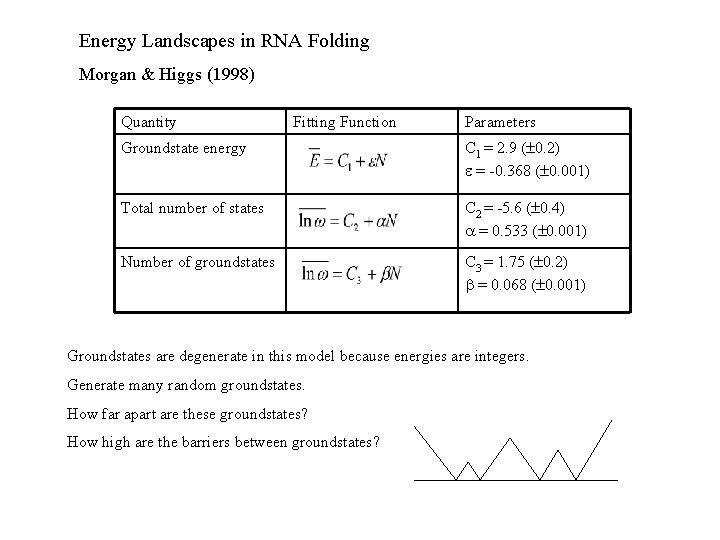
Energy Landscapes in RNA Folding Morgan & Higgs (1998) Quantity Fitting Function Parameters Groundstate energy C 1 = 2. 9 ( 0. 2) = -0. 368 ( 0. 001) Total number of states C 2 = -5. 6 ( 0. 4) = 0. 533 ( 0. 001) Number of groundstates C 3 = 1. 75 ( 0. 2) = 0. 068 ( 0. 001) Groundstates are degenerate in this model because energies are integers. Generate many random groundstates. How far apart are these groundstates? How high are the barriers between groundstates?

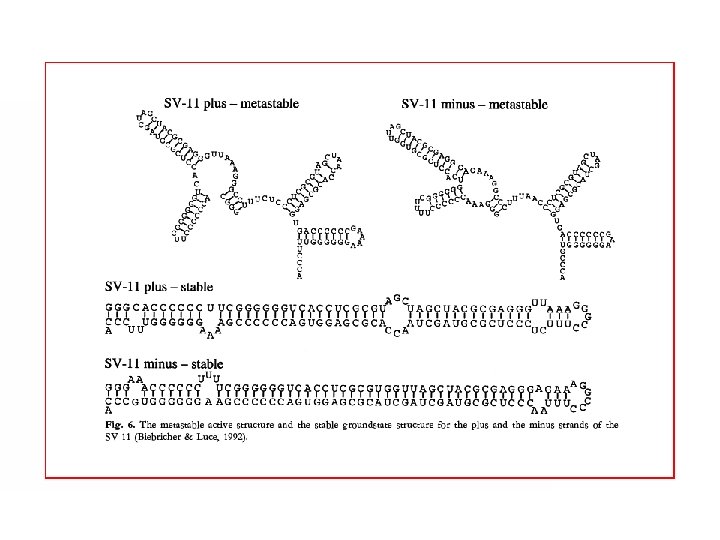
We found Frozen pairs (present in every groundstate) This figure shows the frozen pairs only. The molecule is divided into independent unfrozen loops. Define Neff as the length of the longest loop. Two groundstates for the same sequence

Minimum Free Energy Prediction Deterministic. Always gets MFE structure for a given set of energy rules. If MFE structure is not the same as biological structure, this could be because (i) energy rules are inaccurate or insufficient (ii) kinetics is important and molecule is trapped in metastable state. Monte Carlo simulations of folding kinetics. Store a current structure. Estimate rates of removal of existing helices and rates of addition of other compatible helices. Choose one helix to be added or removed with probability proportional to its rate. Repeat this many times. Can simulate structure formation from an unfolded state.

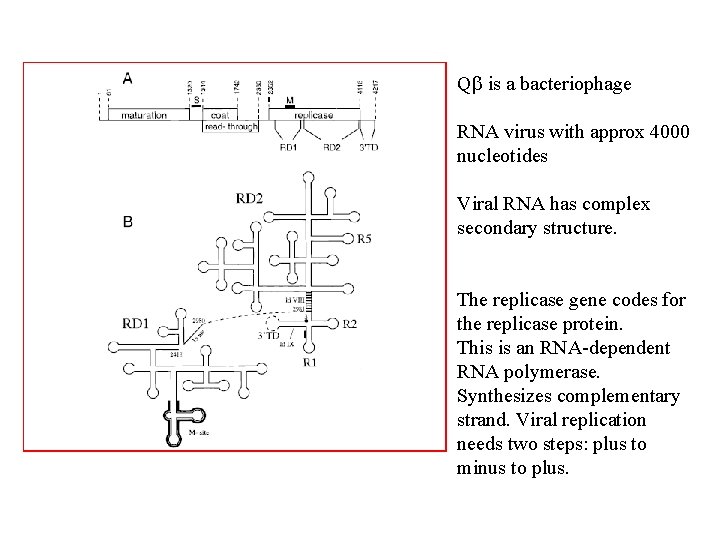
Q is a bacteriophage RNA virus with approx 4000 nucleotides Viral RNA has complex secondary structure. The replicase gene codes for the replicase protein. This is an RNA-dependent RNA polymerase. Synthesizes complementary strand. Viral replication needs two steps: plus to minus to plus.

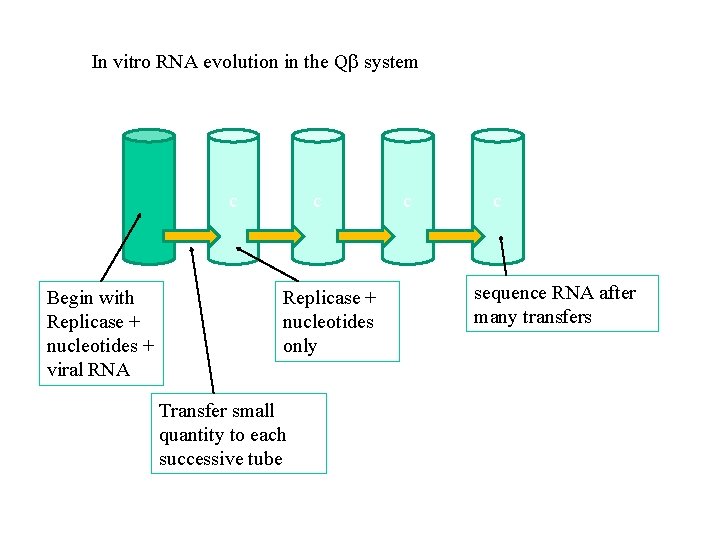
In vitro RNA evolution in the Q system c Begin with Replicase + nucleotides + viral RNA c Replicase + nucleotides only Transfer small quantity to each successive tube c c sequence RNA after many transfers

Barrier heights between alternative groundstates Observation: Mean barrier height between groundstates scales as <h> ~ Neff 0. 5 Neff ~ 0. 3 N Therefore barriers become significant for large enough sequences.


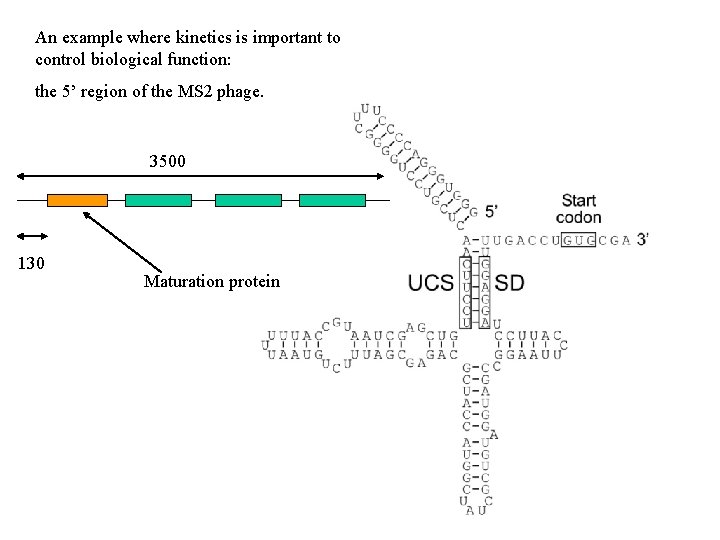
An example where kinetics is important to control biological function: the 5’ region of the MS 2 phage. 3500 130 Maturation protein

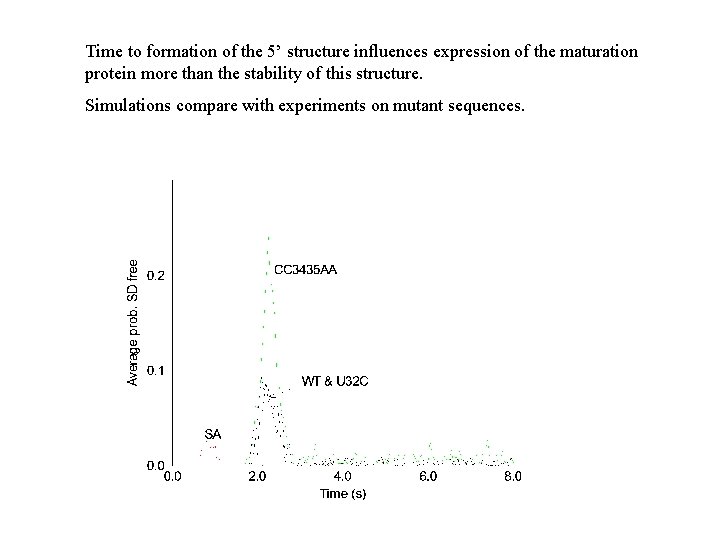
Time to formation of the 5’ structure influences expression of the maturation protein more than the stability of this structure. Simulations compare with experiments on mutant sequences.

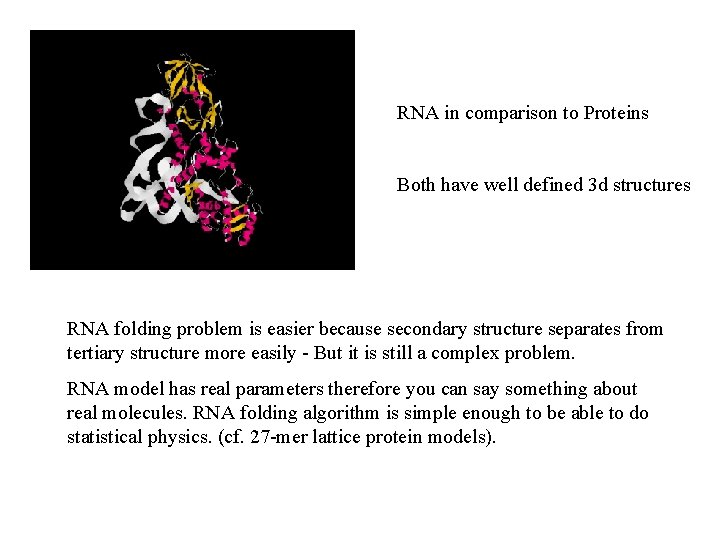
RNA in comparison to Proteins Both have well defined 3 d structures RNA folding problem is easier because secondary structure separates from tertiary structure more easily - But it is still a complex problem. RNA model has real parameters therefore you can say something about real molecules. RNA folding algorithm is simple enough to be able to do statistical physics. (cf. 27 -mer lattice protein models).

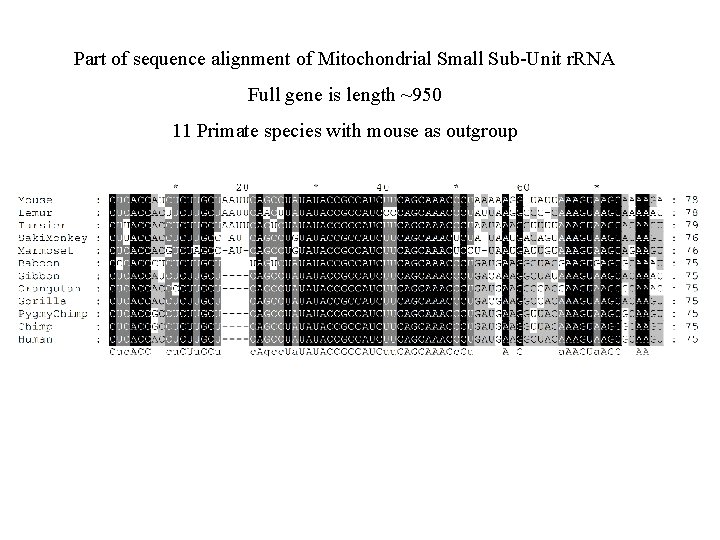
Part of sequence alignment of Mitochondrial Small Sub-Unit r. RNA Full gene is length ~950 11 Primate species with mouse as outgroup

Murphy et al. Nature (2001) uses 15 nuclear plus 3 mitochondrial proteins

Afrotheria / Laurasiatheria Striking examples of convergent evolution

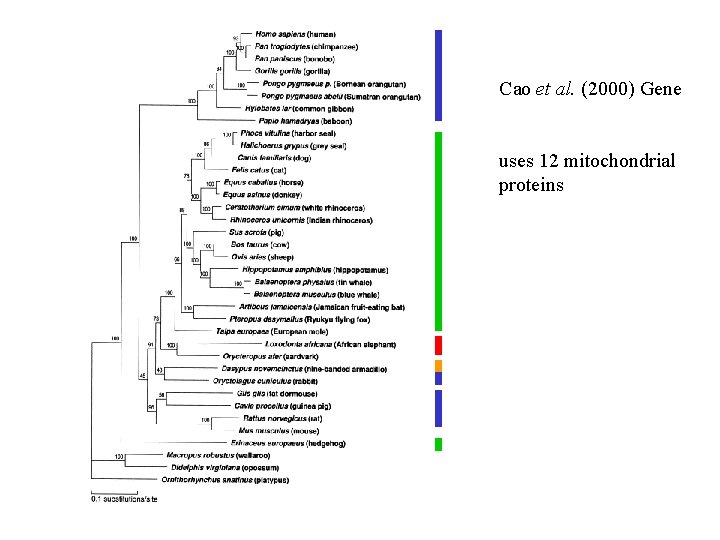
Cao et al. (2000) Gene uses 12 mitochondrial proteins

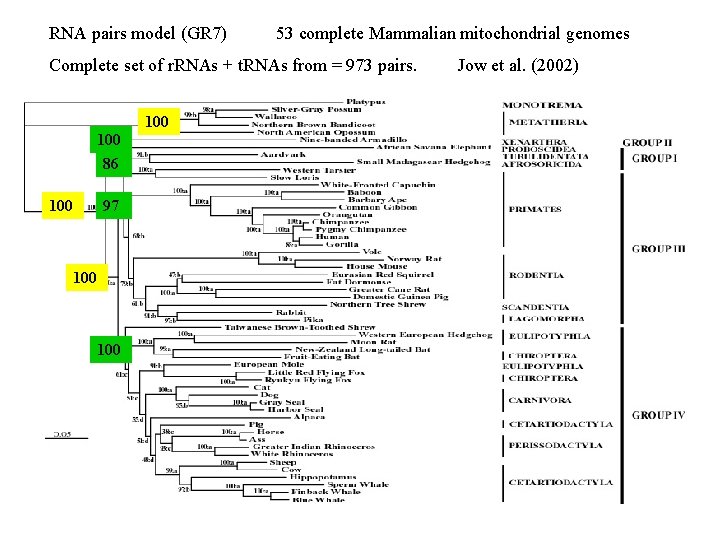
RNA pairs model (GR 7) 53 complete Mammalian mitochondrial genomes Complete set of r. RNAs + t. RNAs from = 973 pairs. 100 86 100 97 100 Jow et al. (2002)

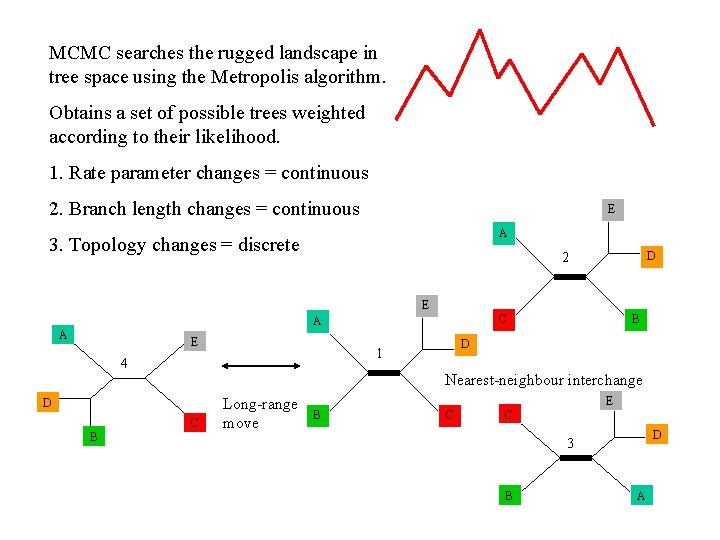
MCMC searches the rugged landscape in tree space using the Metropolis algorithm. Obtains a set of possible trees weighted according to their likelihood. 1. Rate parameter changes = continuous 2. Branch length changes = continuous E A 3. Topology changes = discrete D 2 E C A A E Nearest-neighbour interchange D C B D 1 4 B Long-range move E B C C D 3 B A

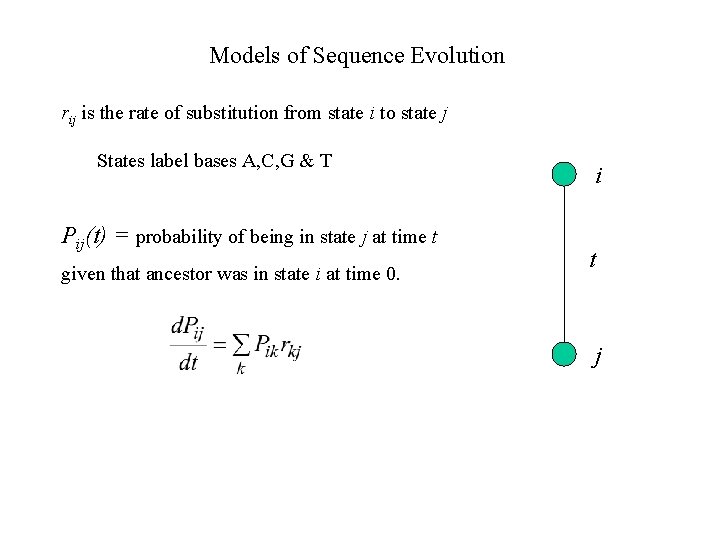
Models of Sequence Evolution rij is the rate of substitution from state i to state j States label bases A, C, G & T Pij(t) = probability of being in state j at time t given that ancestor was in state i at time 0. i t j

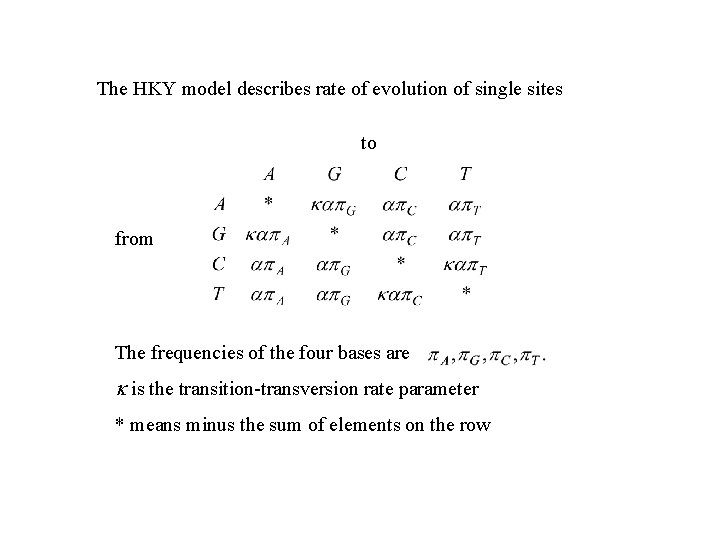
The HKY model describes rate of evolution of single sites to from The frequencies of the four bases are k is the transition-transversion rate parameter * means minus the sum of elements on the row

Compensatory Substitutions Two sides of the acceptor stem from a t. RNA are shown. Due to structure conservation alignment is possible in widely different species. Bacillus subtilis Escherichia coli Saccharomyces cerevisiae Drosophila melanogaster Homo sapiens 1234567 ((((((( 7654321 ))))))) GGCUCGG GCCCGGA GCGGAUU GCCGAAA CCGAGCC UCCGGGC AAUUCGC UUUCGGC

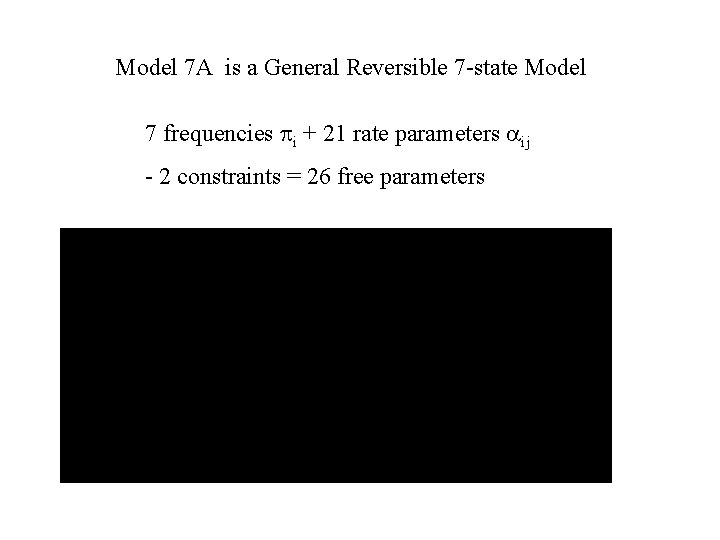
Model 7 A is a General Reversible 7 -state Model 7 frequencies pi + 21 rate parameters ij - 2 constraints = 26 free parameters

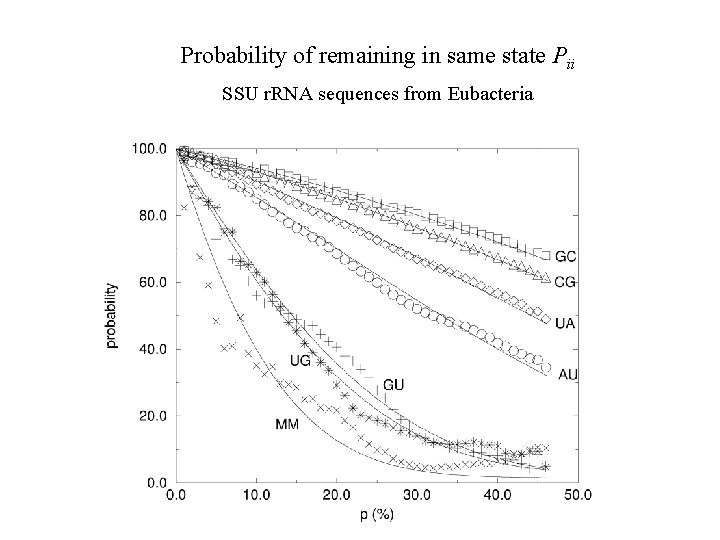
Probability of remaining in same state Pii SSU r. RNA sequences from Eubacteria

Probability Pij of changes from CG to other pairs SSU r. RNA from Eubacteria

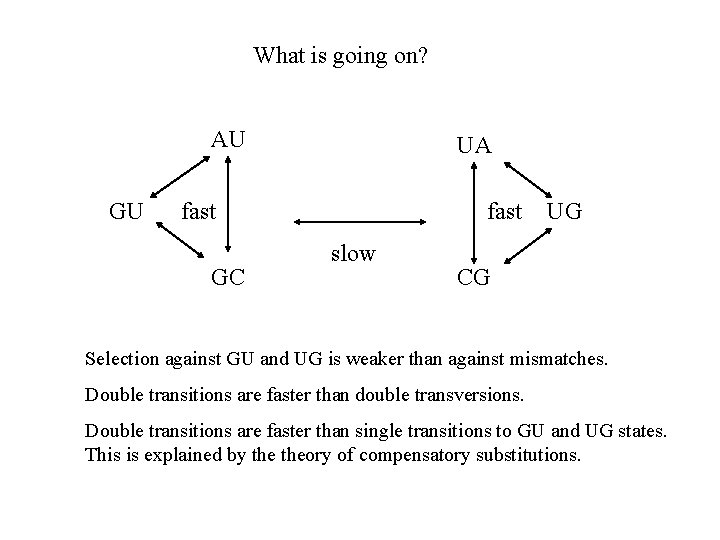
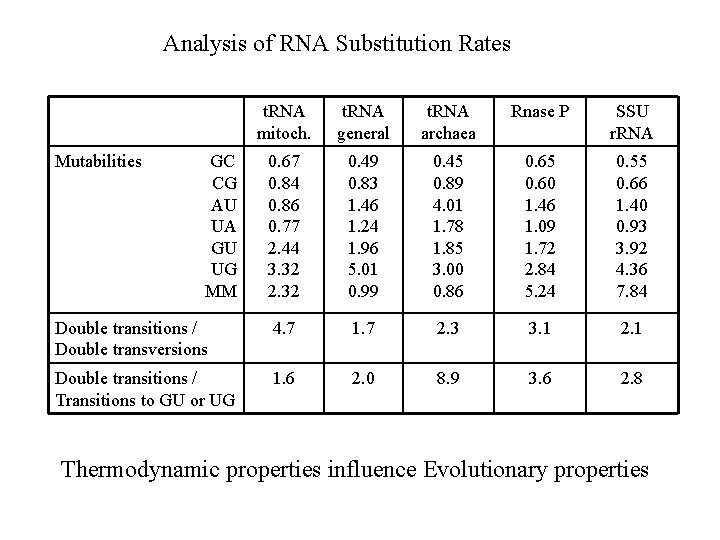
What is going on? AU GU UA fast GC fast slow UG CG Selection against GU and UG is weaker than against mismatches. Double transitions are faster than double transversions. Double transitions are faster than single transitions to GU and UG states. This is explained by theory of compensatory substitutions.

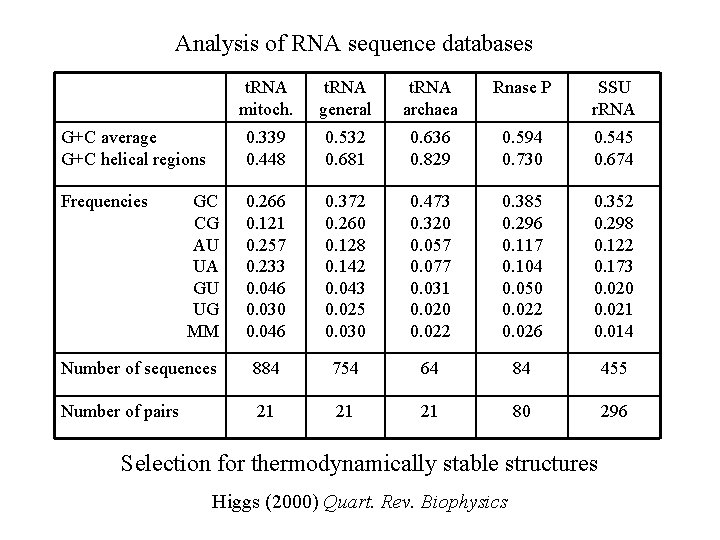
Analysis of RNA sequence databases t. RNA mitoch. t. RNA general t. RNA archaea Rnase P SSU r. RNA G+C average G+C helical regions 0. 339 0. 448 0. 532 0. 681 0. 636 0. 829 0. 594 0. 730 0. 545 0. 674 Frequencies 0. 266 0. 121 0. 257 0. 233 0. 046 0. 030 0. 046 0. 372 0. 260 0. 128 0. 142 0. 043 0. 025 0. 030 0. 473 0. 320 0. 057 0. 077 0. 031 0. 020 0. 022 0. 385 0. 296 0. 117 0. 104 0. 050 0. 022 0. 026 0. 352 0. 298 0. 122 0. 173 0. 020 0. 021 0. 014 Number of sequences 884 754 64 84 455 Number of pairs 21 21 21 80 296 GC CG AU UA GU UG MM Selection for thermodynamically stable structures Higgs (2000) Quart. Rev. Biophysics

Analysis of RNA Substitution Rates t. RNA mitoch. t. RNA general t. RNA archaea Rnase P SSU r. RNA 0. 67 0. 84 0. 86 0. 77 2. 44 3. 32 2. 32 0. 49 0. 83 1. 46 1. 24 1. 96 5. 01 0. 99 0. 45 0. 89 4. 01 1. 78 1. 85 3. 00 0. 86 0. 65 0. 60 1. 46 1. 09 1. 72 2. 84 5. 24 0. 55 0. 66 1. 40 0. 93 3. 92 4. 36 7. 84 Double transitions / Double transversions 4. 7 1. 7 2. 3 3. 1 2. 1 Double transitions / Transitions to GU or UG 1. 6 2. 0 8. 9 3. 6 2. 8 Mutabilities GC CG AU UA GU UG MM Thermodynamic properties influence Evolutionary properties
 Rna protein synthesis
Rna protein synthesis Transversion
Transversion Transcription
Transcription Codon chart
Codon chart Messenger rna sequence
Messenger rna sequence Messenger
Messenger Msn messenger 7
Msn messenger 7 Messenger web
Messenger web Messenger
Messenger Open source lan messenger
Open source lan messenger Messenger
Messenger Messenger of god planet
Messenger of god planet Indeed in the messenger of allah
Indeed in the messenger of allah 2 corinthians 12 10
2 corinthians 12 10 The messenger ed character analysis
The messenger ed character analysis Rsview
Rsview Dynamic messenger
Dynamic messenger Pidgin facebook messenger
Pidgin facebook messenger Calculator messenger
Calculator messenger Oedipus rex parados
Oedipus rex parados The messenger trainer
The messenger trainer Gh
Gh I am the messenger symbols
I am the messenger symbols Messenger brak ankiety
Messenger brak ankiety Greek messenger
Greek messenger Second messenger hypothesis
Second messenger hypothesis Who does creon advise oedipus to send for
Who does creon advise oedipus to send for What are the two main parts of a lipid?
What are the two main parts of a lipid? Yahoo messenger
Yahoo messenger Messenger
Messenger Web msn messenger live
Web msn messenger live Secondary messenger
Secondary messenger Msn messenger chat rooms
Msn messenger chat rooms Real life calculator
Real life calculator I miss you messenger
I miss you messenger Messenger
Messenger