Les polypeptides Diversit et complexit fonctionelles Gnes Gnome












































- Slides: 74

Les polypeptides

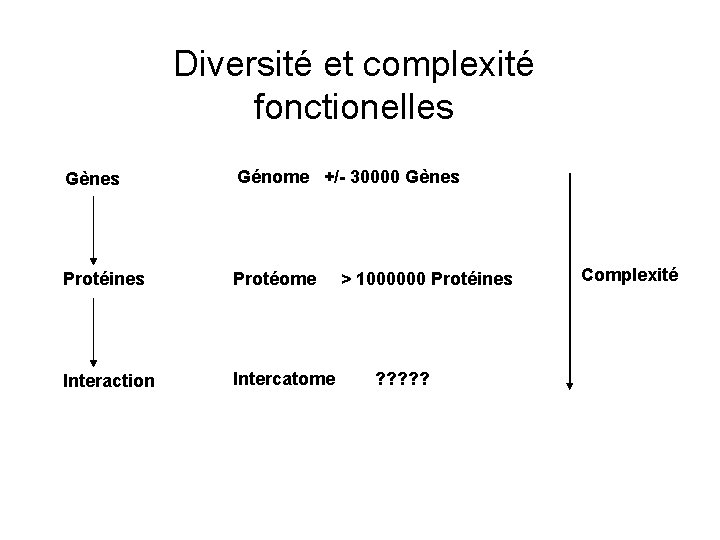
Diversité et complexité fonctionelles Gènes Génome +/- 30000 Gènes Protéines Protéome Interaction Intercatome > 1000000 Protéines ? ? ? Complexité

In 2003, Human genome sequence was deciphered! • • Genome is the complete set of genes of a living thing. In 2003, the human genome sequencing was completed. The human genome contains about 3 billion base pairs. The number of genes is estimated to be between 20, 000 to 25, 000. • The difference between the genome of human and that of chimpanzee is only 1. 23%! 3 billion base pair => 6 G letters & 1 letter => 1 byte The whole genome can be recorded in just 10 CD-ROMs!

Our life is maintained by molecular network systems Molecular network system in a cell (From Ex. PASy Biochemical Pathways; http: //www. expasy. org/cgi-bin/show_thumbnails. pl? 2)

Diversité structurale Protéines simples Protéines conjuguées Goupe Prosthétique

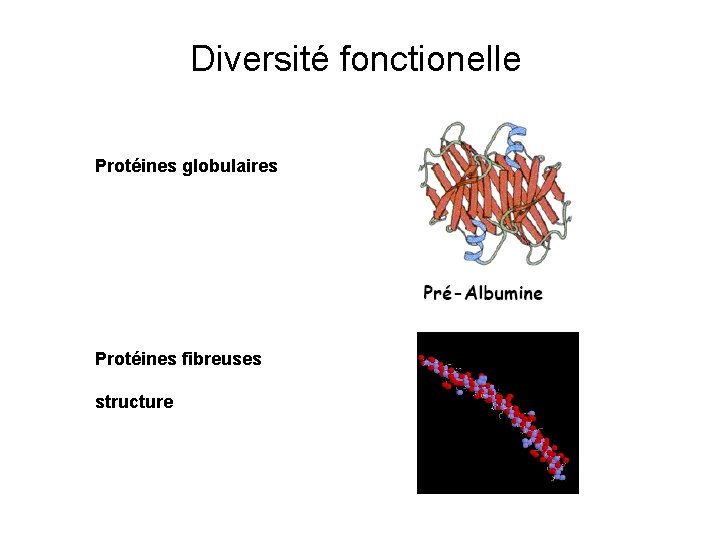
Diversité fonctionelle Protéines globulaires Protéines fibreuses structure

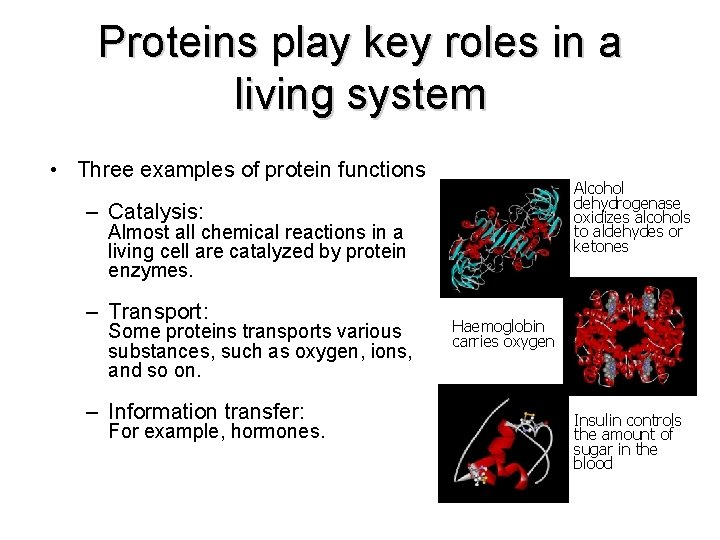
Proteins play key roles in a living system • Three examples of protein functions Alcohol dehydrogenase oxidizes alcohols to aldehydes or ketones – Catalysis: Almost all chemical reactions in a living cell are catalyzed by protein enzymes. – Transport: Some proteins transports various substances, such as oxygen, ions, and so on. – Information transfer: For example, hormones. Haemoglobin carries oxygen Insulin controls the amount of sugar in the blood

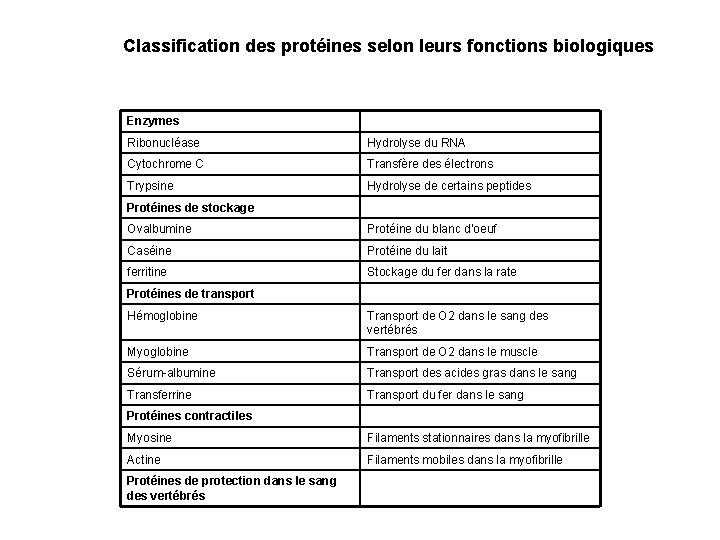
Classification des protéines selon leurs fonctions biologiques Enzymes Ribonucléase Hydrolyse du RNA Cytochrome C Transfère des électrons Trypsine Hydrolyse de certains peptides Protéines de stockage Ovalbumine Protéine du blanc d’oeuf Caséine Protéine du lait ferritine Stockage du fer dans la rate Protéines de transport Hémoglobine Transport de O 2 dans le sang des vertébrés Myoglobine Transport de O 2 dans le muscle Sérum-albumine Transport des acides gras dans le sang Transferrine Transport du fer dans le sang Protéines contractiles Myosine Filaments stationnaires dans la myofibrille Actine Filaments mobiles dans la myofibrille Protéines de protection dans le sang des vertébrés

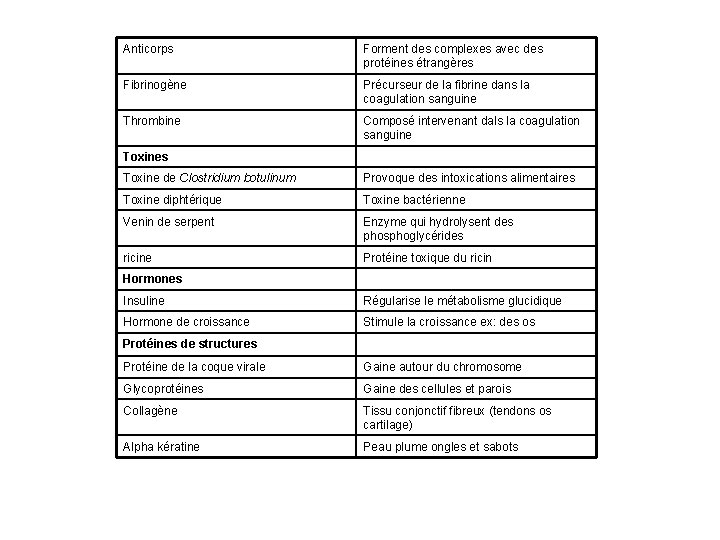
Anticorps Forment des complexes avec des protéines étrangères Fibrinogène Précurseur de la fibrine dans la coagulation sanguine Thrombine Composé intervenant dals la coagulation sanguine Toxines Toxine de Clostridium botulinum Provoque des intoxications alimentaires Toxine diphtérique Toxine bactérienne Venin de serpent Enzyme qui hydrolysent des phosphoglycérides ricine Protéine toxique du ricin Hormones Insuline Régularise le métabolisme glucidique Hormone de croissance Stimule la croissance ex: des os Protéines de structures Protéine de la coque virale Gaine autour du chromosome Glycoprotéines Gaine des cellules et parois Collagène Tissu conjonctif fibreux (tendons os cartilage) Alpha kératine Peau plume ongles et sabots

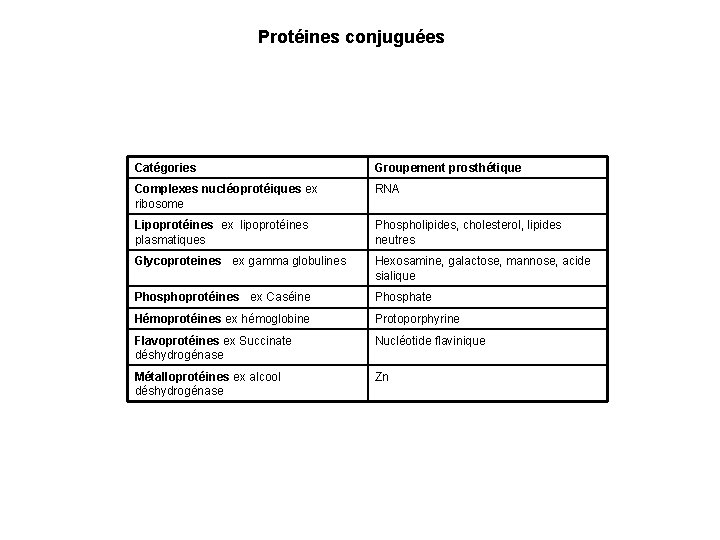
Protéines conjuguées Catégories Groupement prosthétique Complexes nucléoprotéiques ex ribosome RNA Lipoprotéines ex lipoprotéines plasmatiques Phospholipides, cholesterol, lipides neutres Glycoproteines ex gamma globulines Hexosamine, galactose, mannose, acide sialique Phosphoprotéines ex Caséine Phosphate Hémoprotéines ex hémoglobine Protoporphyrine Flavoprotéines ex Succinate déshydrogénase Nucléotide flavinique Métalloprotéines ex alcool déshydrogénase Zn

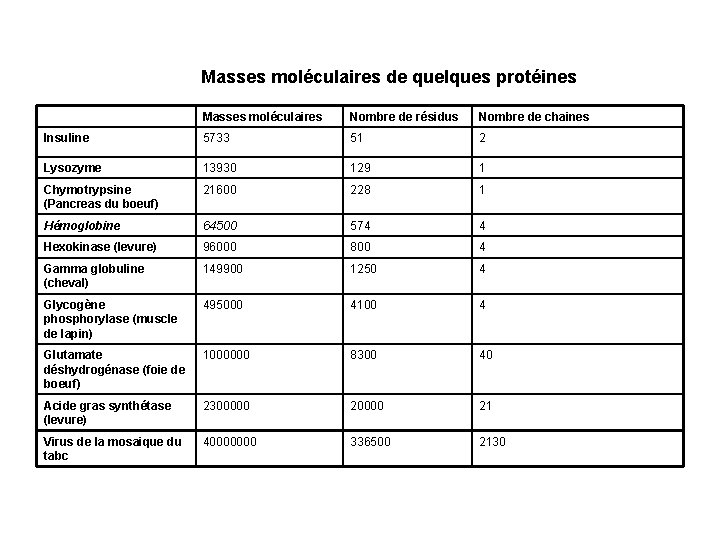
Masses moléculaires de quelques protéines Masses moléculaires Nombre de résidus Nombre de chaines Insuline 5733 51 2 Lysozyme 13930 129 1 Chymotrypsine (Pancreas du boeuf) 21600 228 1 Hémoglobine 64500 574 4 Hexokinase (levure) 96000 800 4 Gamma globuline (cheval) 149900 1250 4 Glycogène phosphorylase (muscle de lapin) 495000 4100 4 Glutamate déshydrogénase (foie de boeuf) 1000000 8300 40 Acide gras synthétase (levure) 2300000 21 Virus de la mosaique du tabc 40000000 336500 2130

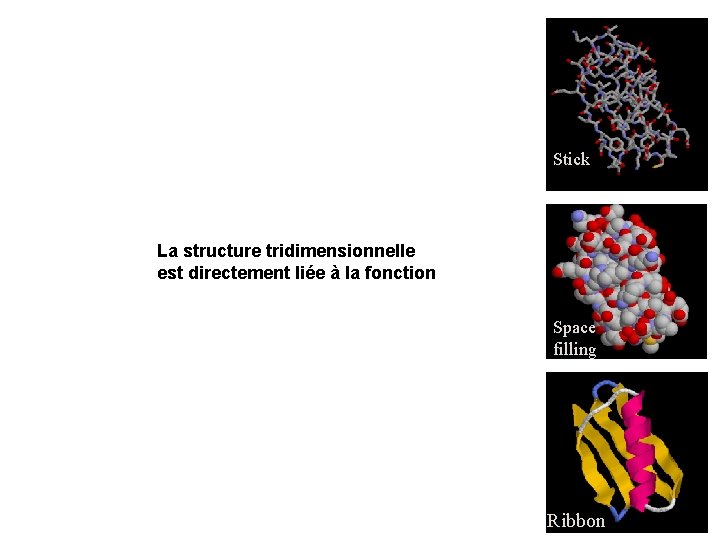
Stick La structure tridimensionnelle est directement liée à la fonction Space filling Ribbon



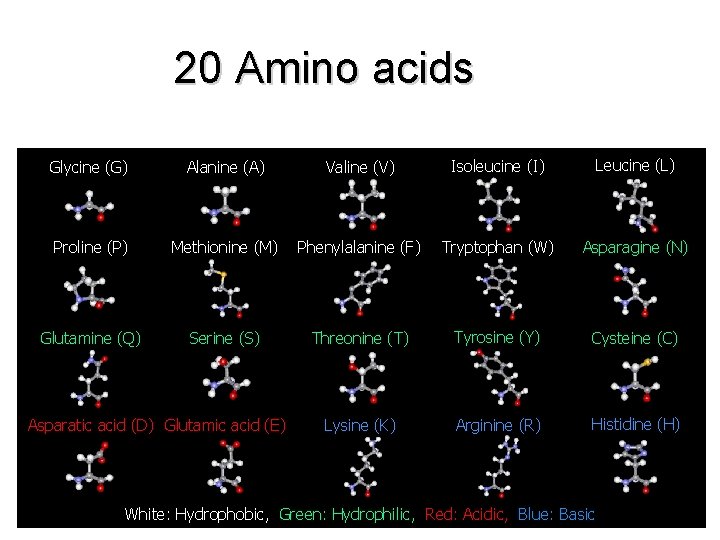
• There are 20 “standard” amino acids that make up all the proteins in all organisms • There are more potential combinations of these 20 amino acids than visible stars in the sky

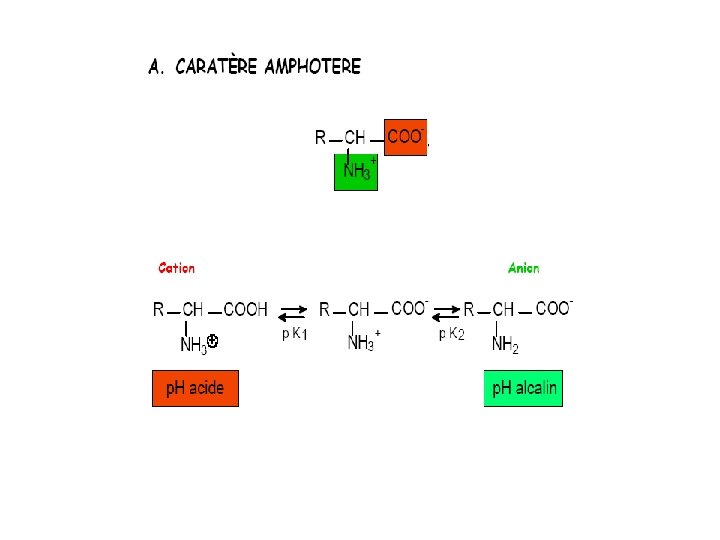
Acide aminé • The a-carbon has two functional groups: – a primary amine (-NH 2) – a primary carboxylic acid (-COOH) • The 20 biochemical amino acids are all distinguished by their side chain group ‘R’

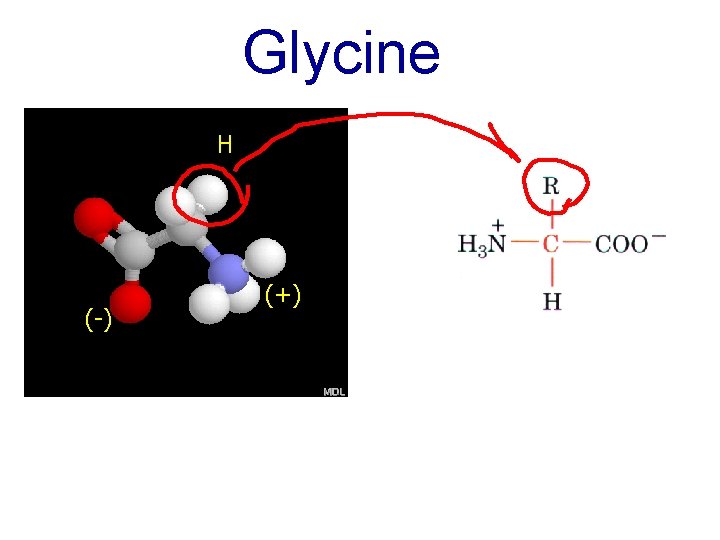
Glycine H (-) (+)

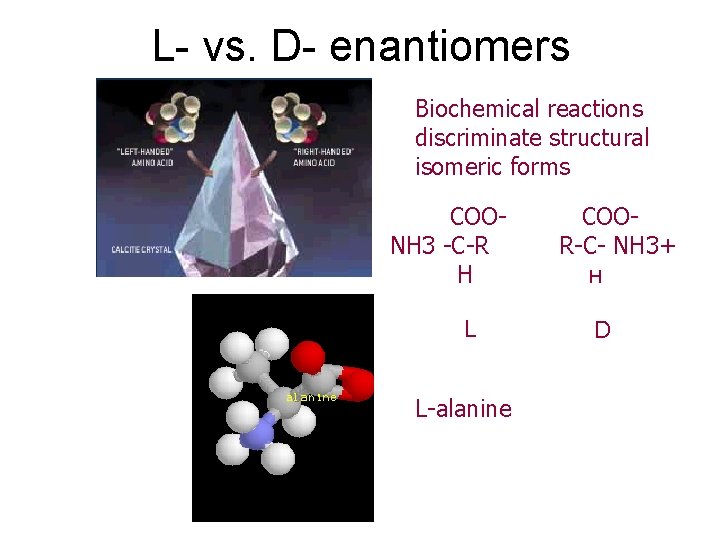
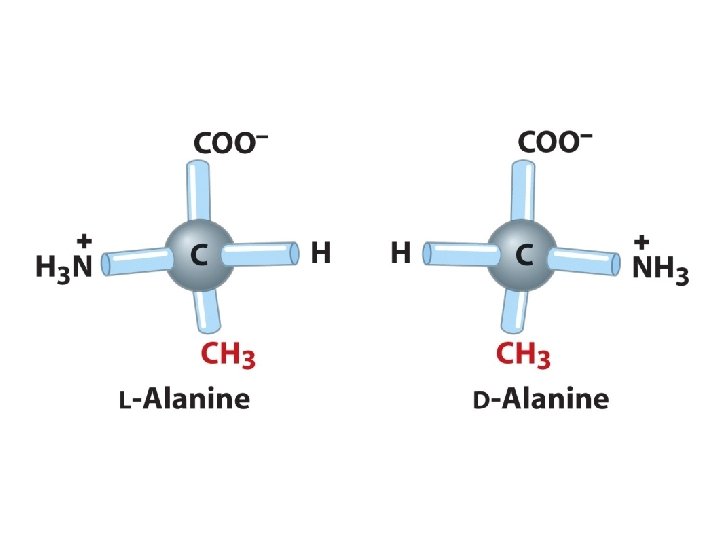
L- vs. D- enantiomers Biochemical reactions discriminate structural isomeric forms COONH 3 -C-R H L L-alanine COOR-C- NH 3+ H D



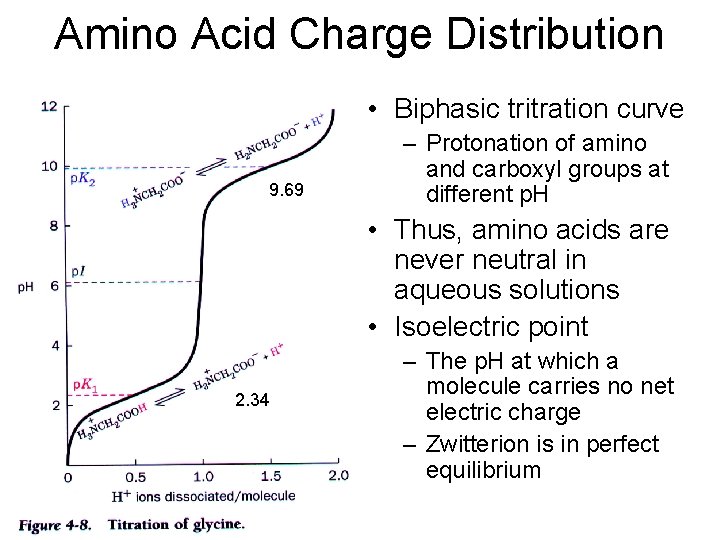
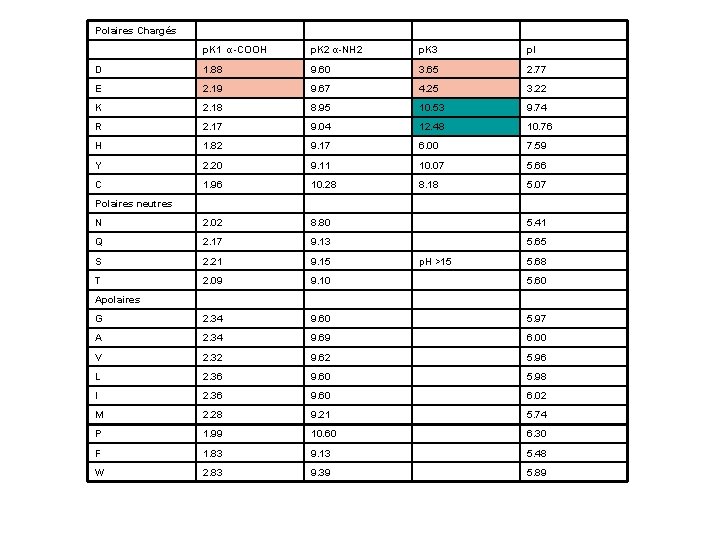
Amino Acid Charge Distribution • Biphasic tritration curve 9. 69 – Protonation of amino and carboxyl groups at different p. H • Thus, amino acids are never neutral in aqueous solutions • Isoelectric point 2. 34 – The p. H at which a molecule carries no net electric charge – Zwitterion is in perfect equilibrium

a-amino acids: at p. H 7 • At physiological p. H, AA are dipolar: – The amino group is protonated (-NH 3+) – The carboxylic acid is deprotonated (-COO-) • Ions with a structural polarity are: zwitterions – Very soluble in polar solvents (cytoplasm)

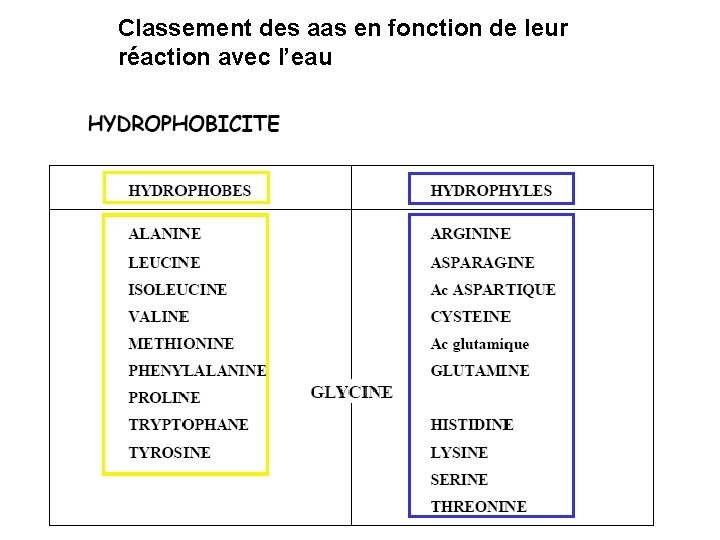
Classement des aas en fonction de leur réaction avec l’eau








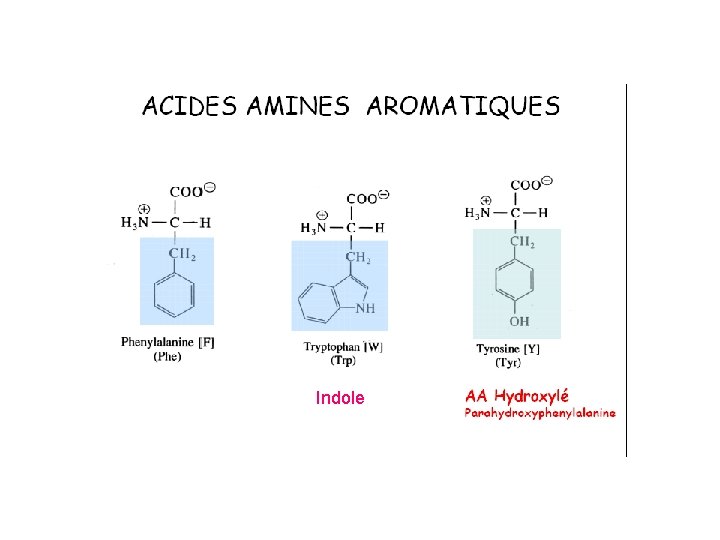
Indole



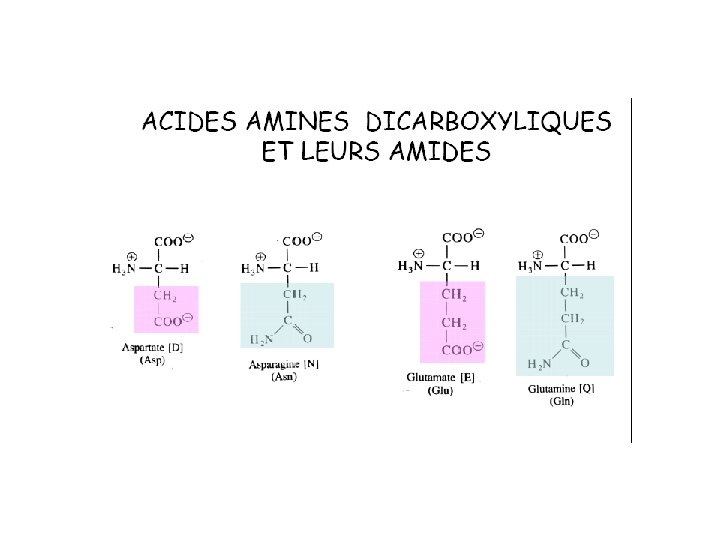
20 Amino acids Glycine (G) Alanine (A) Valine (V) Isoleucine (I) Leucine (L) Proline (P) Methionine (M) Phenylalanine (F) Tryptophan (W) Asparagine (N) Glutamine (Q) Serine (S) Threonine (T) Tyrosine (Y) Cysteine (C) Lysine (K) Arginine (R) Histidine (H) Asparatic acid (D) Glutamic acid (E) White: Hydrophobic, Green: Hydrophilic, Red: Acidic, Blue: Basic

Polaires Chargés p. K 1 α-COOH p. K 2 α-NH 2 p. K 3 p. I D 1. 88 9. 60 3. 65 2. 77 E 2. 19 9. 67 4. 25 3. 22 K 2. 18 8. 95 10. 53 9. 74 R 2. 17 9. 04 12. 48 10. 76 H 1. 82 9. 17 6. 00 7. 59 Y 2. 20 9. 11 10. 07 5. 66 C 1. 96 10. 28 8. 18 5. 07 N 2. 02 8. 80 5. 41 Q 2. 17 9. 13 5. 65 S 2. 21 9. 15 T 2. 09 9. 10 5. 60 G 2. 34 9. 60 5. 97 A 2. 34 9. 69 6. 00 V 2. 32 9. 62 5. 96 L 2. 36 9. 60 5. 98 I 2. 36 9. 60 6. 02 M 2. 28 9. 21 5. 74 P 1. 99 10. 60 6. 30 F 1. 83 9. 13 5. 48 W 2. 83 9. 39 5. 89 Polaires neutres p. H >15 5. 68 Apolaires

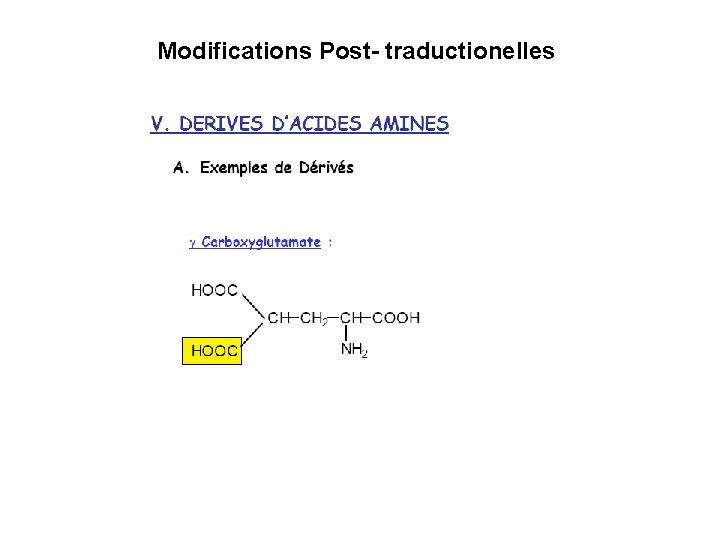
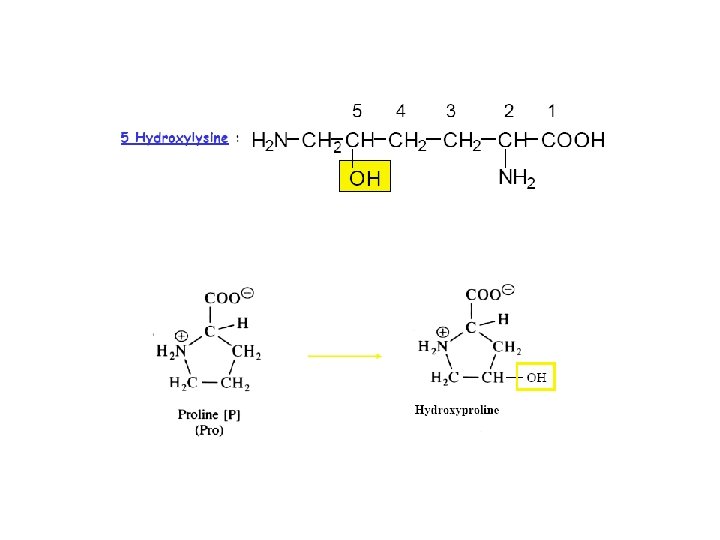
Modifications Post- traductionelles








Les peptides

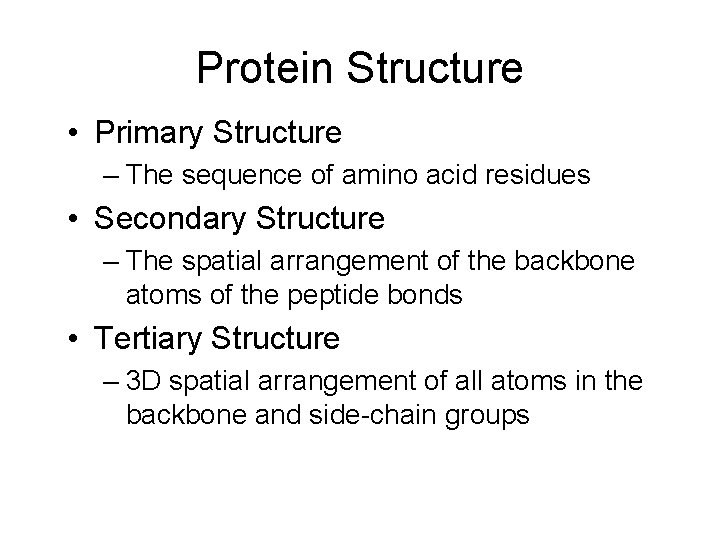
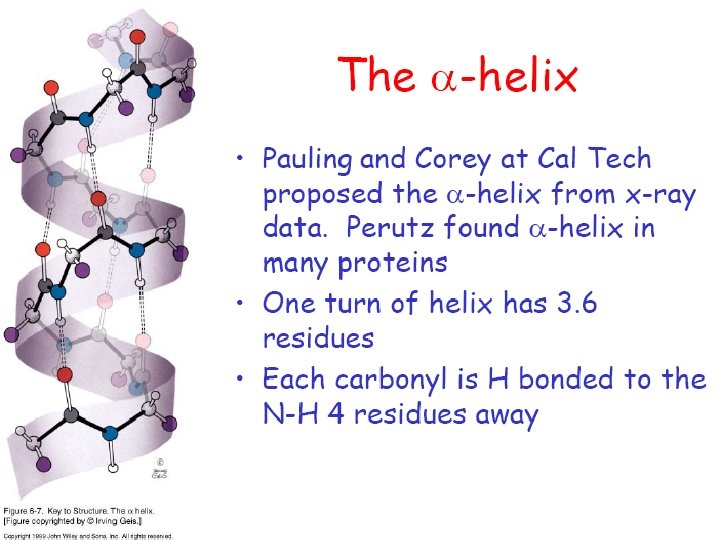
Protein Structure • Primary Structure – The sequence of amino acid residues • Secondary Structure – The spatial arrangement of the backbone atoms of the peptide bonds • Tertiary Structure – 3 D spatial arrangement of all atoms in the backbone and side-chain groups


N aas >>>> N-1 lien peptidiques


Peptide Resonance • Resonance interactions give the peptide bond ~40% of the structural character of a double-bond • Configuration Trans <> Cis • Lien peptidique polaire


Structure primaire

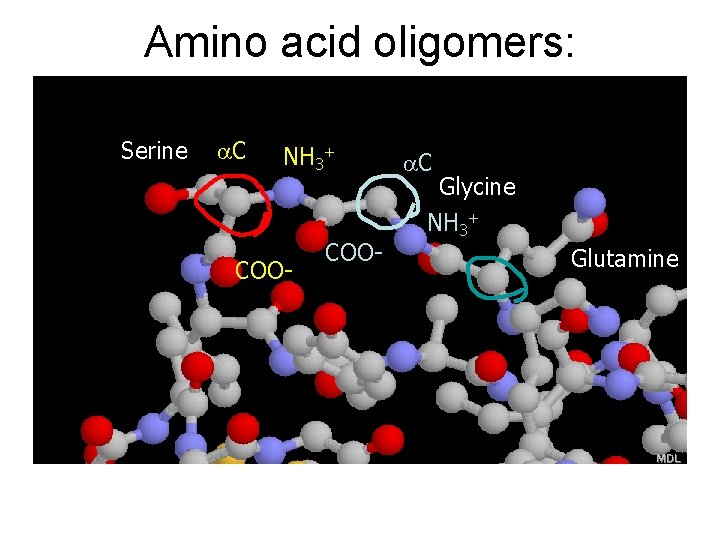
Amino acid oligomers: polypeptides Serine a. C NH 3+ COO- a. C Glycine NH 3+ Glutamine

• • Rotational flexibility only possible between different planar units Ca – N: – f bond angle Ca – C: – y bond angle These angle are constrained by – Amide hydrogens – Carbonyl oxygen – Side chain groups

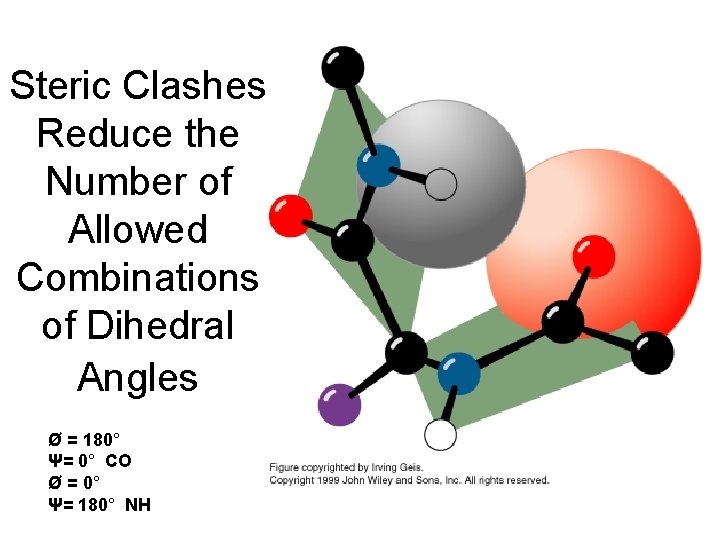
Steric Clashes Reduce the Number of Allowed Combinations of Dihedral Angles Ø = 180° Ψ= 0° CO Ø = 0° Ψ= 180° NH





Experimental data




Solution Proteique • Conservation -800 C > -200 C > -40 C • lyophilisation • Eviter congélation/décongélation (perte d’activité et précipitation) • Stable hautement concentrée (1 -10 mg/ml) • Manipulation -40 C > température ambiante • Solution tamponnée (50 -100 m. M) • Antibactérien (Na. N 3 ou stérile) • Antiproteases • Agent stabilisant (antioxidant…)

Gel Filtration Chromatography

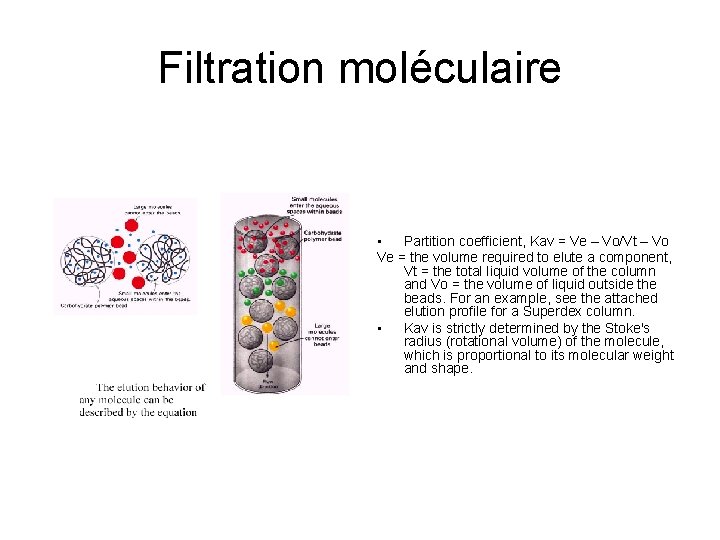
Filtration moléculaire • Partition coefficient, Kav = Ve – Vo/Vt – Vo Ve = the volume required to elute a component, Vt = the total liquid volume of the column and Vo = the volume of liquid outside the beads. For an example, see the attached elution profile for a Superdex column. • Kav is strictly determined by the Stoke's radius (rotational volume) of the molecule, which is proportional to its molecular weight and shape.

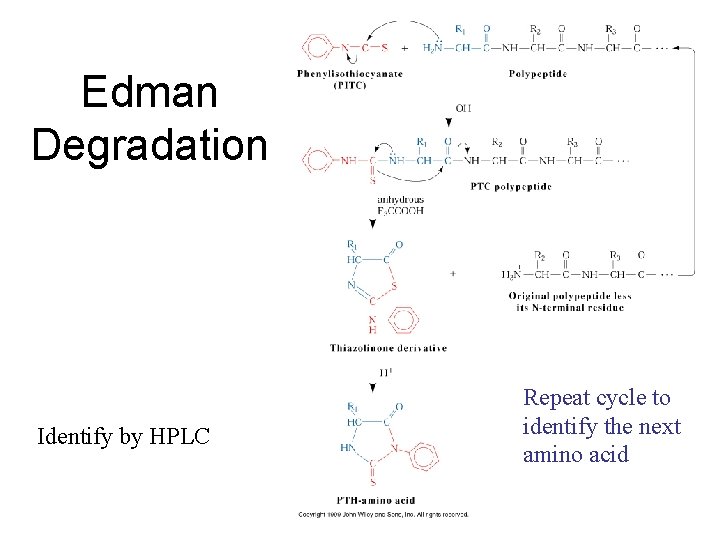
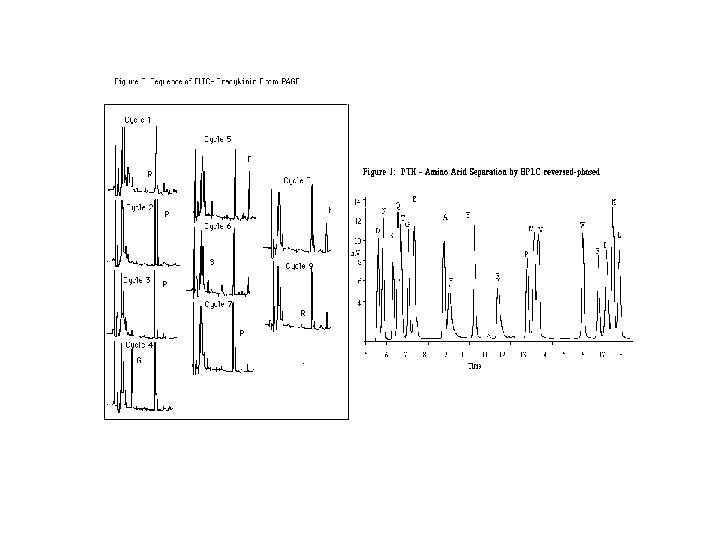
Edman reaction Edman Degradation chemistry is a stepwise process that has been automated (instruments perform the reactions without human intervention). PITC labeling of the unprotonated, terminal amine is followed by cleavage in anhydrous trifluoroacetic acid in the gas phase. The free, derivitized aa is moved away from the rest of the peptide, where it is dissolved in aqueous acid. The structure rearranges to a form that absorbs UV light at 254 nm. The derivitization cyclizes the aa, so that each PTH-aa is more hydrophobic (less polar) than the corresponding parent aa. In order to differentiate between the 20 hydrophobic PTH-aa, a reversed-phase HPLC column is used. The most common application is to determine the NH 2 -terminal sequence of proteins that have been separated by SDS-PAGE or 2 D electrophoresis.

Protein identification strategies Edman Degradation Electro-Transfert - Blocked N-terminus Slow Loss of material

Edman Degradation Identify by HPLC Repeat cycle to identify the next amino acid

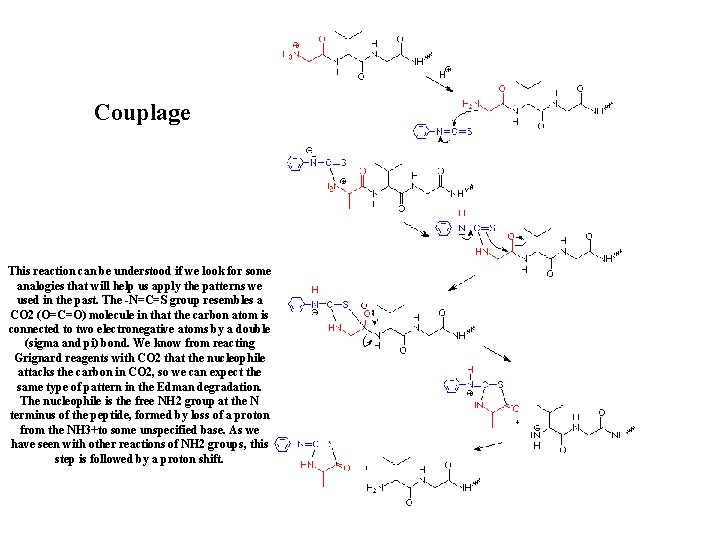
Couplage This reaction can be understood if we look for some analogies that will help us apply the patterns we used in the past. The -N=C=S group resembles a CO 2 (O=C=O) molecule in that the carbon atom is connected to two electronegative atoms by a double (sigma and pi) bond. We know from reacting Grignard reagents with CO 2 that the nucleophile attacks the carbon in CO 2, so we can expect the same type of pattern in the Edman degradation. The nucleophile is the free NH 2 group at the N terminus of the peptide, formed by loss of a proton from the NH 3+to some unspecified base. As we have seen with other reactions of NH 2 groups, this step is followed by a proton shift.

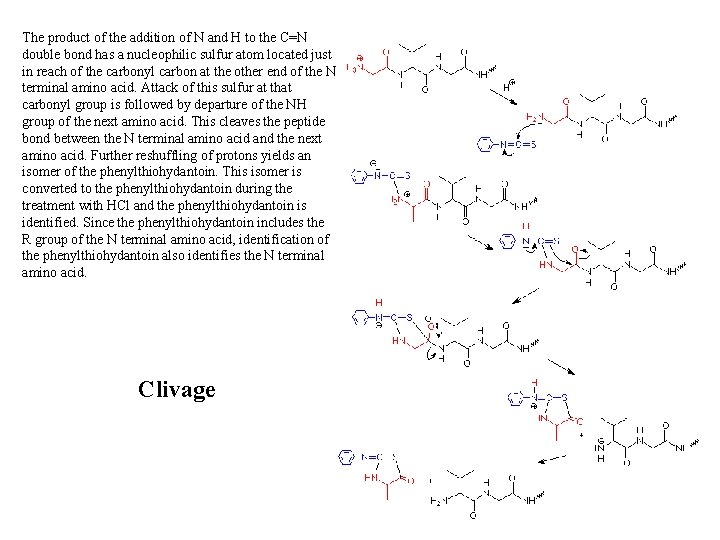
The product of the addition of N and H to the C=N double bond has a nucleophilic sulfur atom located just in reach of the carbonyl carbon at the other end of the N terminal amino acid. Attack of this sulfur at that carbonyl group is followed by departure of the NH group of the next amino acid. This cleaves the peptide bond between the N terminal amino acid and the next amino acid. Further reshuffling of protons yields an isomer of the phenylthiohydantoin. This isomer is converted to the phenylthiohydantoin during the treatment with HCl and the phenylthiohydantoin is identified. Since the phenylthiohydantoin includes the R group of the N terminal amino acid, identification of the phenylthiohydantoin also identifies the N terminal amino acid. Clivage




 Pyuria leukocytes urine
Pyuria leukocytes urine Complexité d'un algorithme
Complexité d'un algorithme Complexit
Complexit Alessi kettle david jones
Alessi kettle david jones Gnome outline
Gnome outline Gnome outline
Gnome outline Kde architecture
Kde architecture Johari window là gì
Johari window là gì Gnome code
Gnome code Les parts de les plantes
Les parts de les plantes Remplacer les mots soulignes par les pronoms convenables
Remplacer les mots soulignes par les pronoms convenables Les mots qu'on ne dit pas sont les fleurs du silence
Les mots qu'on ne dit pas sont les fleurs du silence Grand corps malade les voyages en train
Grand corps malade les voyages en train Les 10 volcans les plus dangereux du monde
Les 10 volcans les plus dangereux du monde Non non non ne failliront jamais lyrics
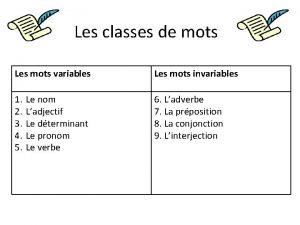
Non non non ne failliront jamais lyrics Nom variable et invariable
Nom variable et invariable Des organisateurs textuels
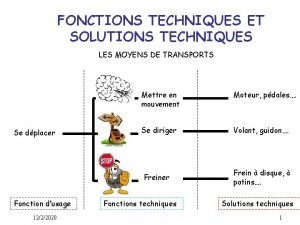
Des organisateurs textuels Fonctions techniques et solutions techniques
Fonctions techniques et solutions techniques Allez vous en sur les places et sur les parvis
Allez vous en sur les places et sur les parvis Le paratexte de la ficelle
Le paratexte de la ficelle Horloge stratégique
Horloge stratégique Les mot variable
Les mot variable Calorie dragibus
Calorie dragibus Parts de la flor
Parts de la flor Les voitures les plus rapides du monde
Les voitures les plus rapides du monde Les actionneurs et les préactionneurs
Les actionneurs et les préactionneurs Les constellations les plus connues
Les constellations les plus connues Qu'est-ce que tu aimes manger
Qu'est-ce que tu aimes manger Trouvez les réponses. écrivez-les en chiffres (numbers).
Trouvez les réponses. écrivez-les en chiffres (numbers). Exemple texte expressif
Exemple texte expressif Les lettres en français
Les lettres en français Les trois obstacles et les quatre démons
Les trois obstacles et les quatre démons Nahrungsnetz
Nahrungsnetz Les avantages du commerce international
Les avantages du commerce international Défense homme à homme handball
Défense homme à homme handball Les principes de l'absurde
Les principes de l'absurde Je vais aller en vacances
Je vais aller en vacances Entrave au dialogue
Entrave au dialogue Les pices
Les pices Backwards accent a
Backwards accent a Introduction sur les capteurs
Introduction sur les capteurs La fonction impressive
La fonction impressive Les indicateurs de l'imparfait
Les indicateurs de l'imparfait En croyant a des fleurs
En croyant a des fleurs Les compagnons de la chanson gondolier
Les compagnons de la chanson gondolier Pourquoi les plongeurs plongent en arrière
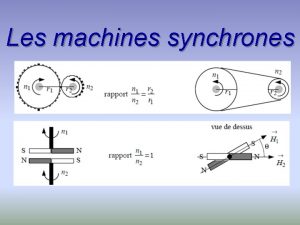
Pourquoi les plongeurs plongent en arrière Les machines synchrones
Les machines synchrones Les trois étapes de la repentance
Les trois étapes de la repentance Que mange les coccinelles à part des pucerons
Que mange les coccinelles à part des pucerons Style de gestion de classe
Style de gestion de classe Les propietats textuals
Les propietats textuals Les gens ne changent pas
Les gens ne changent pas Les amants quadro
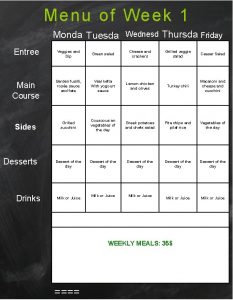
Les amants quadro Entree main and dessert
Entree main and dessert Tableau des pronom
Tableau des pronom Ecoute active def
Ecoute active def Les capes de la terra
Les capes de la terra Les mesure de tendance centrale
Les mesure de tendance centrale Les pomes
Les pomes Amara management
Amara management Slidetodoc.com
Slidetodoc.com Les cytones
Les cytones Adjectifs irréguliers
Adjectifs irréguliers Les sources d'énergie non renouvelable
Les sources d'énergie non renouvelable Coin science maternelle
Coin science maternelle Annette messager les pensionnaires
Annette messager les pensionnaires Regarder des seins
Regarder des seins Outils pour les cfds
Outils pour les cfds Les 10 maillons de la chaîne documentaire
Les 10 maillons de la chaîne documentaire J'aime les fruits
J'aime les fruits Les biotechnologies rouges
Les biotechnologies rouges Rangement lspcc
Rangement lspcc Les types de compte rendu
Les types de compte rendu Ecris en lettres les nombres suivants
Ecris en lettres les nombres suivants Les ongles de vos reves
Les ongles de vos reves