Gene to Protein Gene Expression Figure 17 UN



- Slides: 39

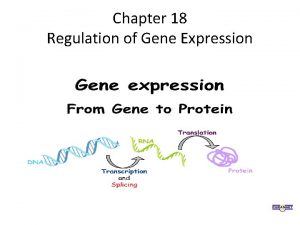
Gene to Protein Gene Expression

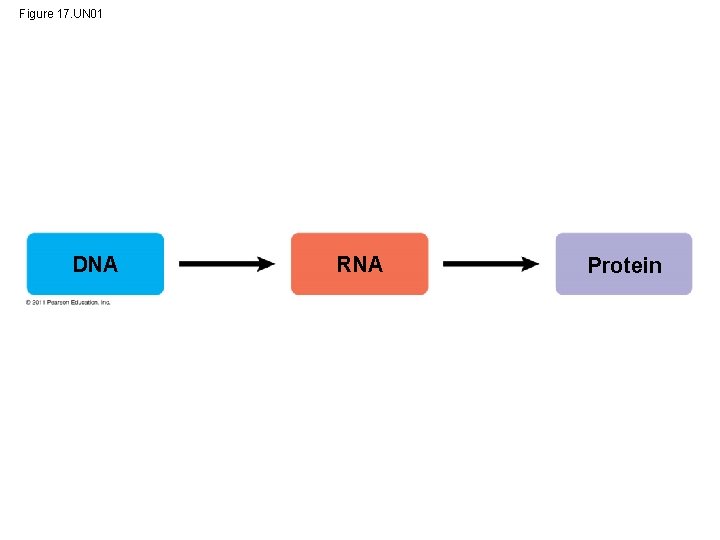
Figure 17. UN 01 DNA RNA Protein

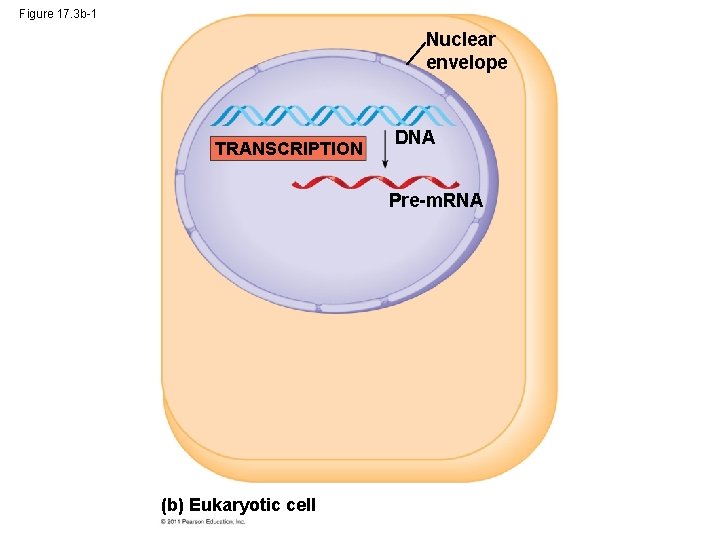
Figure 17. 3 b-1 Nuclear envelope TRANSCRIPTION DNA Pre-m. RNA (b) Eukaryotic cell

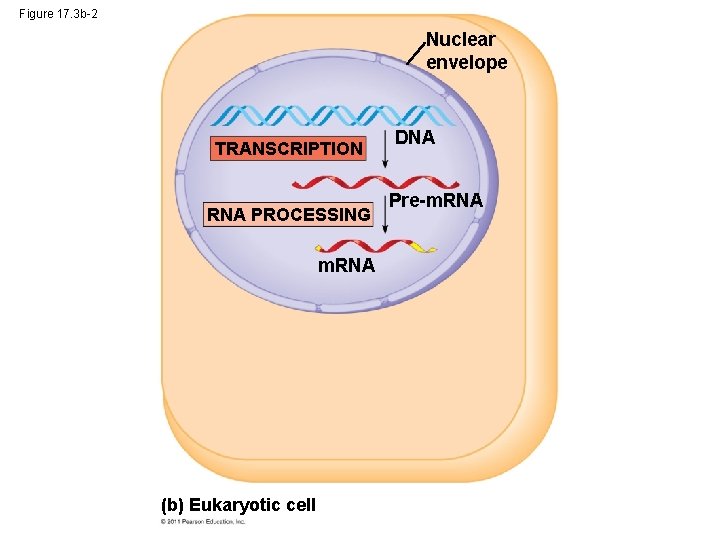
Figure 17. 3 b-2 Nuclear envelope TRANSCRIPTION RNA PROCESSING m. RNA (b) Eukaryotic cell DNA Pre-m. RNA

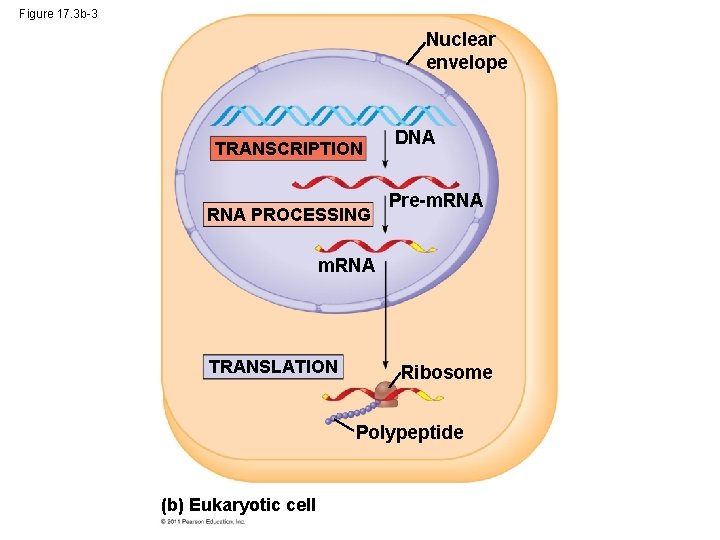
Figure 17. 3 b-3 Nuclear envelope TRANSCRIPTION RNA PROCESSING DNA Pre-m. RNA TRANSLATION Ribosome Polypeptide (b) Eukaryotic cell

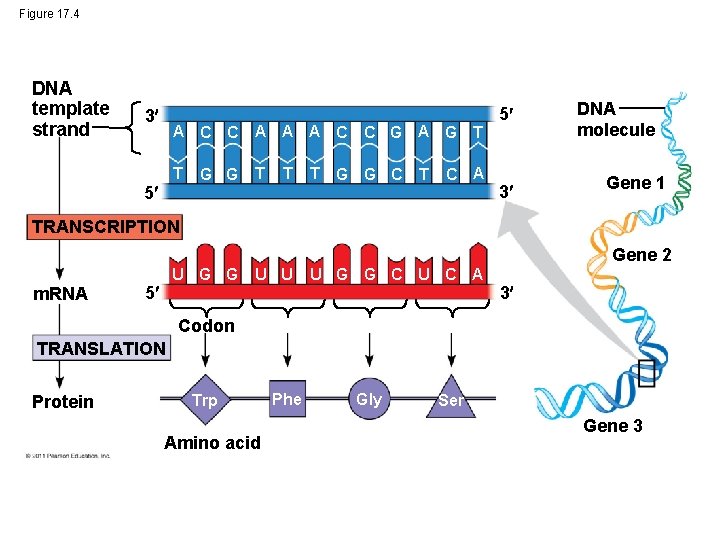
Figure 17. 4 DNA template strand 3 A C C A A A C T T 5 G G T T C G A G G G C T T C A 5 3 DNA molecule Gene 1 TRANSCRIPTION Gene 2 m. RNA U G G 5 U U U G G C U C A 3 Codon TRANSLATION Protein Trp Amino acid Phe Gly Ser Gene 3

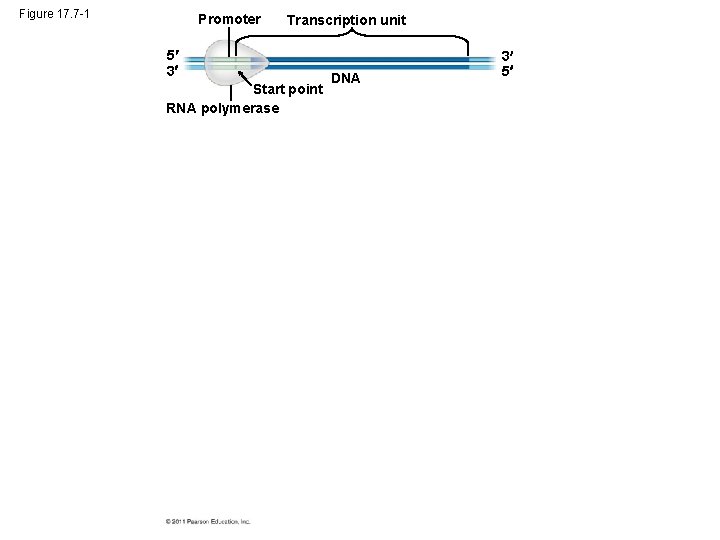
Figure 17. 7 -1 Promoter Transcription unit 5 3 Start point RNA polymerase DNA 3 5

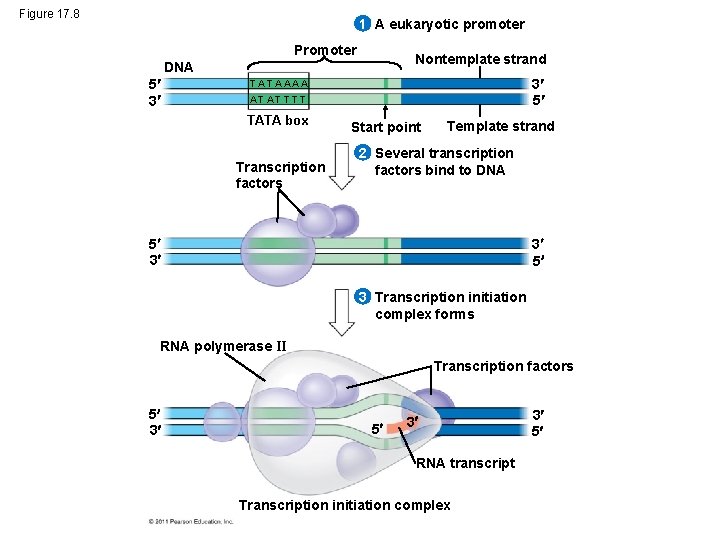
Figure 17. 8 1 A eukaryotic promoter Promoter Nontemplate strand DNA 5 3 3 5 T A A AA A T AT T TATA box Transcription factors Start point Template strand 2 Several transcription factors bind to DNA 5 3 3 5 3 Transcription initiation complex forms RNA polymerase II Transcription factors 5 3 5 3 RNA transcript Transcription initiation complex 3 5

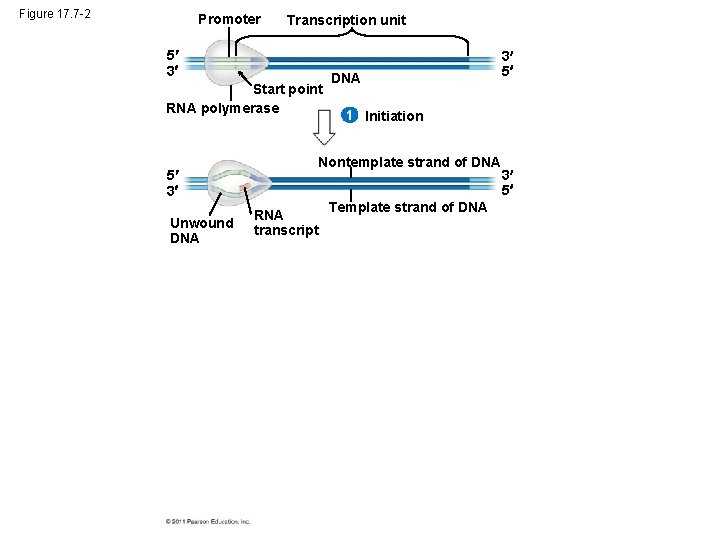
Figure 17. 7 -2 Promoter Transcription unit 5 3 Start point RNA polymerase 5 3 Unwound DNA 1 Initiation Nontemplate strand of DNA RNA transcript 3 5 Template strand of DNA 3 5

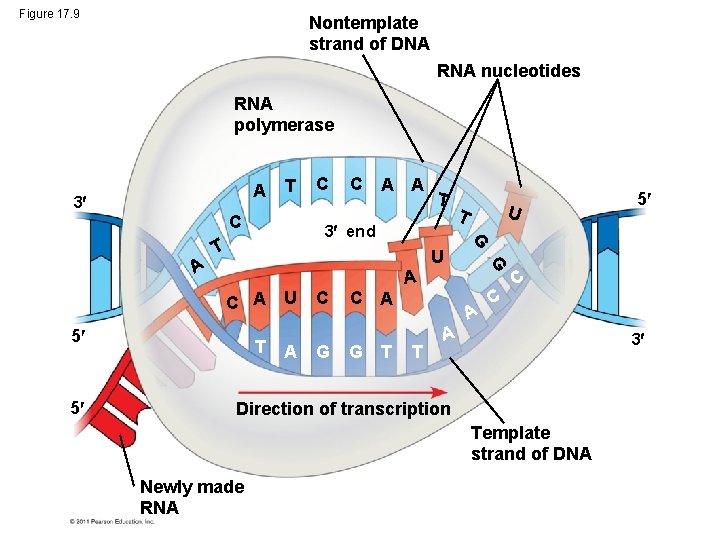
Figure 17. 9 Nontemplate strand of DNA RNA nucleotides RNA polymerase A 3 T C A A T 3 end T 5 T U A C G C A G T U T G U C A 5 C A A T C C A 3 Direction of transcription Template strand of DNA Newly made RNA 5 G A C

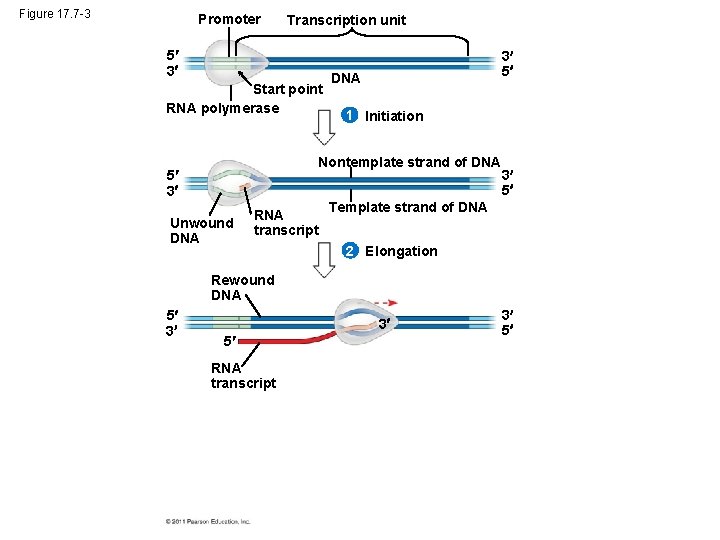
Figure 17. 7 -3 Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation Nontemplate strand of DNA 5 3 Unwound DNA RNA transcript 3 5 Template strand of DNA 2 Elongation Rewound DNA 5 3 3 5 RNA transcript 3 5

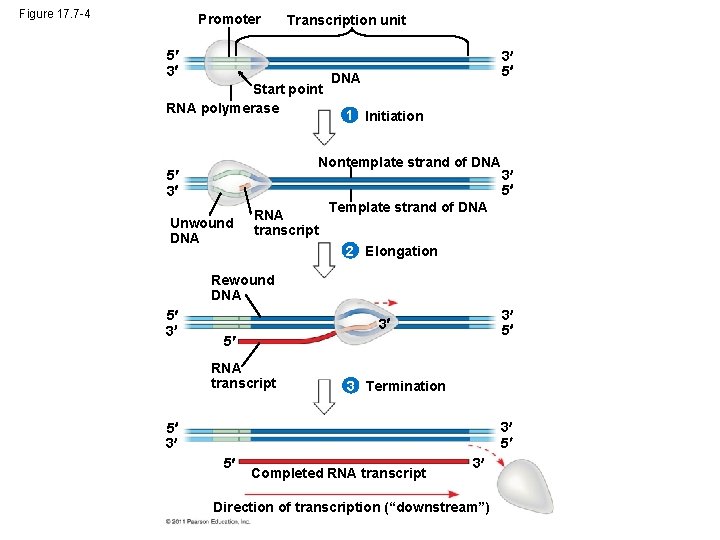
Figure 17. 7 -4 Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation Nontemplate strand of DNA 5 3 Unwound DNA RNA transcript 3 5 Template strand of DNA 2 Elongation Rewound DNA 5 3 3 5 RNA transcript 3 Termination 3 5 5 3 5 Completed RNA transcript 3 Direction of transcription (“downstream”)

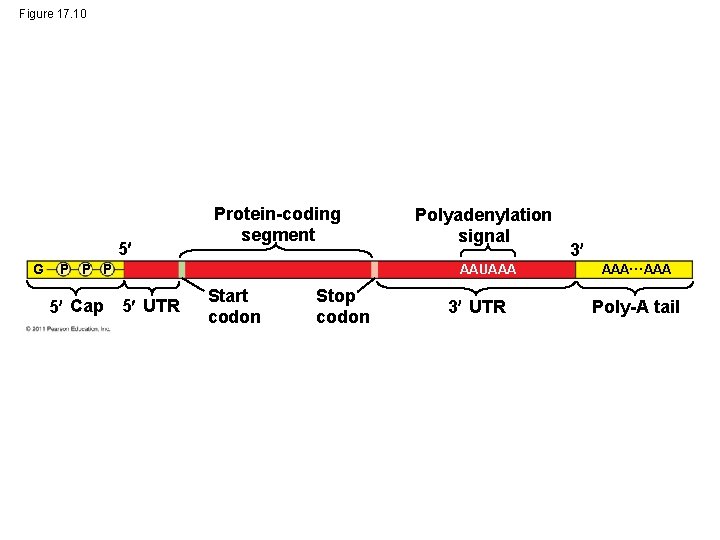
Figure 17. 10 5 G Protein-coding segment P P P 5 Cap 5 UTR Polyadenylation signal AAUAAA Start codon Stop codon 3 UTR 3 AAA… AAA Poly-A tail

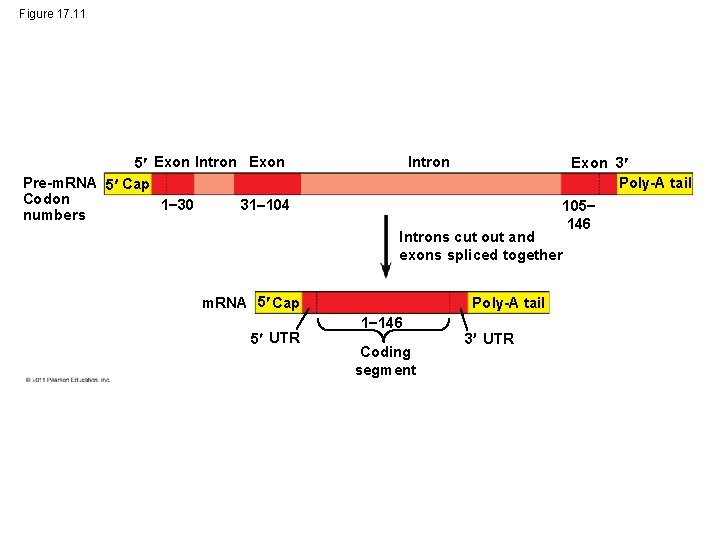
Figure 17. 11 5 Exon Intron Exon Pre-m. RNA 5 Cap Codon 1 30 31 104 numbers Intron Exon 3 Poly-A tail 105 146 Introns cut out and exons spliced together m. RNA 5 Cap 5 UTR Poly-A tail 1 146 Coding segment 3 UTR

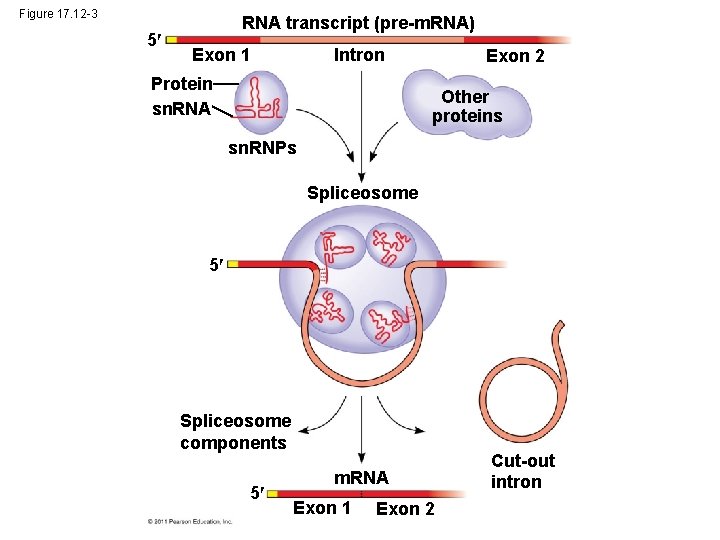
Figure 17. 12 -3 5 RNA transcript (pre-m. RNA) Exon 1 Intron Protein sn. RNA Exon 2 Other proteins sn. RNPs Spliceosome 5 Spliceosome components 5 m. RNA Exon 1 Exon 2 Cut-out intron

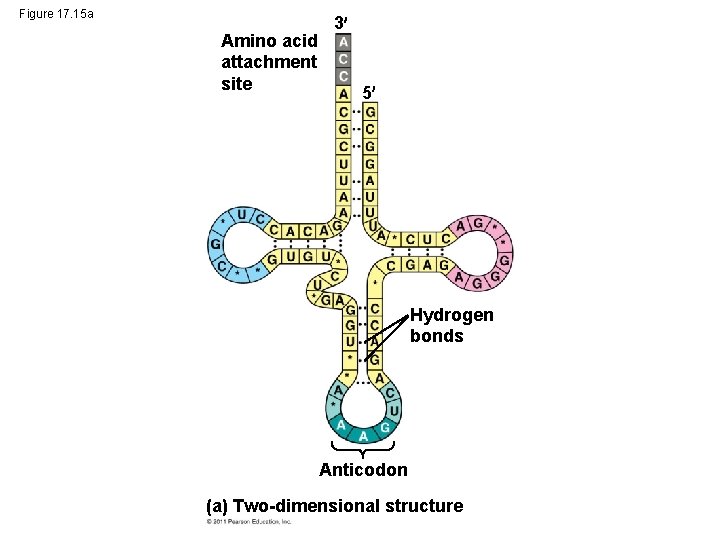
Figure 17. 15 a Amino acid attachment site 3 5 Hydrogen bonds Anticodon (a) Two-dimensional structure

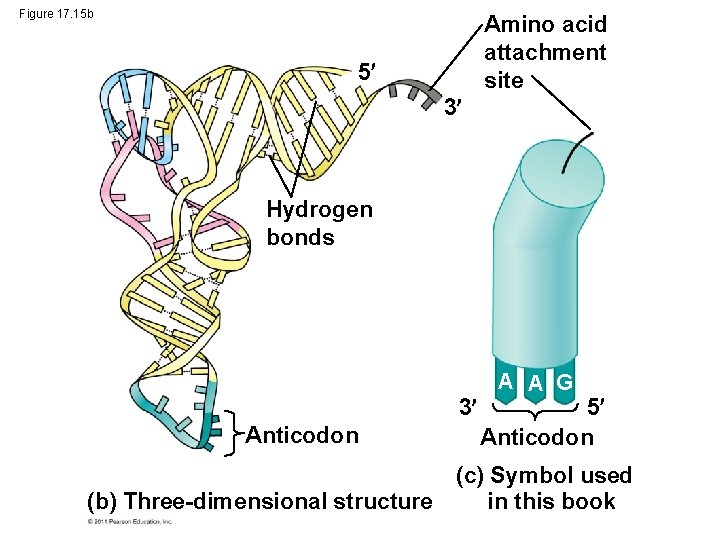
Figure 17. 15 b Amino acid attachment site 5 3 Hydrogen bonds 3 Anticodon A A G 5 Anticodon (c) Symbol used (b) Three-dimensional structure in this book

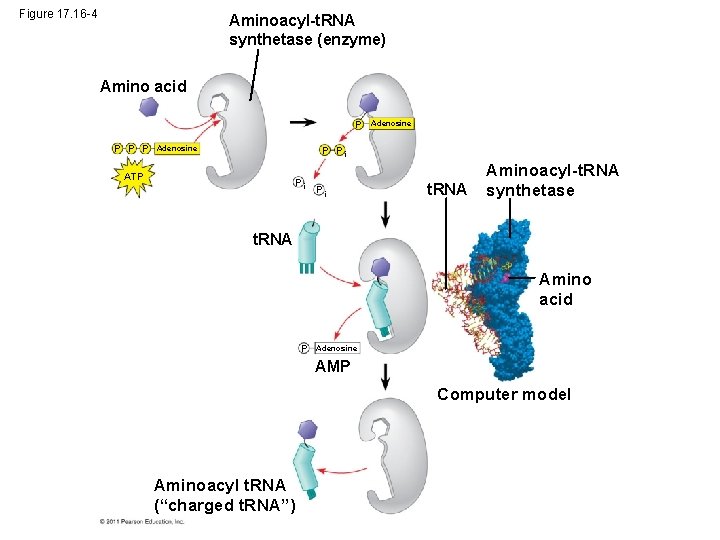
Figure 17. 16 -4 Aminoacyl-t. RNA synthetase (enzyme) Amino acid P P Adenosine P Pi ATP Pi Pi t. RNA Aminoacyl-t. RNA synthetase t. RNA Amino acid P Adenosine AMP Computer model Aminoacyl t. RNA (“charged t. RNA”)

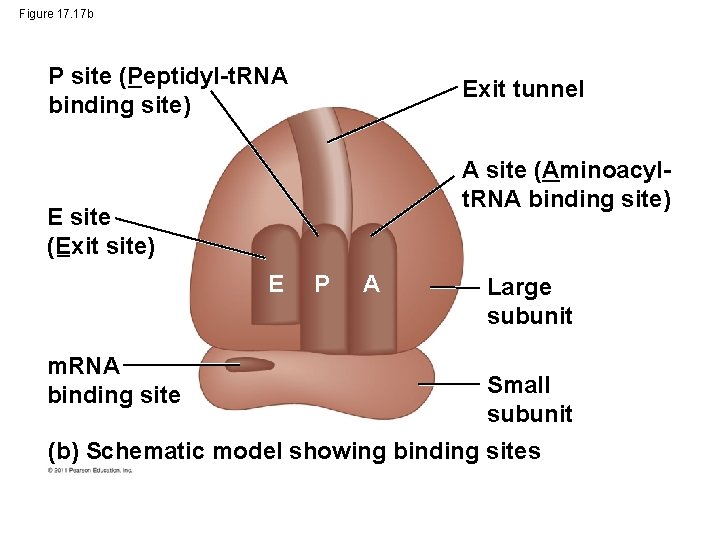
Figure 17. 17 b P site (Peptidyl-t. RNA binding site) Exit tunnel A site (Aminoacylt. RNA binding site) E site (Exit site) E m. RNA binding site P A Large subunit Small subunit (b) Schematic model showing binding sites

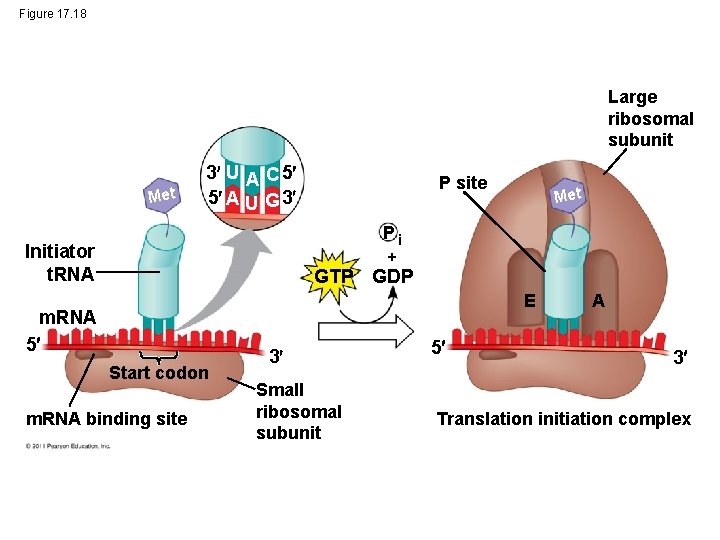
Figure 17. 18 Large ribosomal subunit Met 3 U A C 5 5 A U G 3 P site Met Pi Initiator t. RNA GTP GDP E m. RNA 5 Start codon m. RNA binding site 3 Small ribosomal subunit 5 A 3 Translation initiation complex

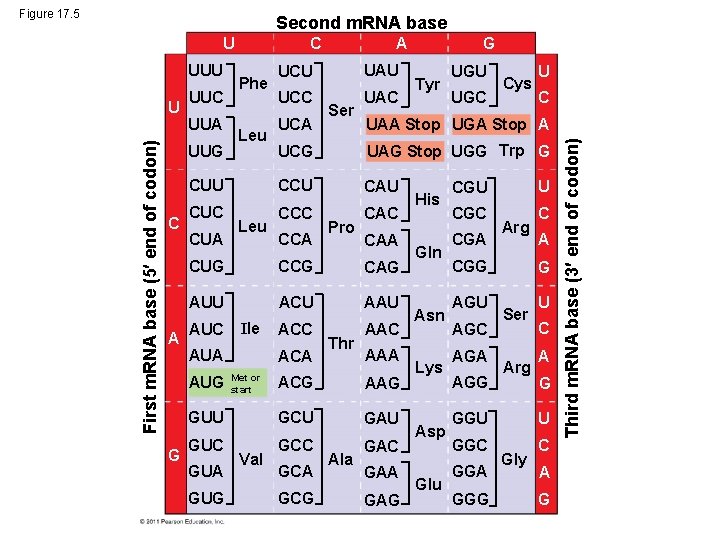
Figure 17. 5 Second m. RNA base UUU First m. RNA base (5 end of codon) U UUC UUA A Leu UCU UAU UCC UAC UCA Ser Tyr UGU UGC Cys U C UAA Stop UGA Stop A UAG Stop UGG Trp G CUU CCU CAU CUC CCC CAC CUA Leu CCA Pro CAA CUG CCG CAG AUU ACU AAU ACC AAC AUC Ile AUA AUG G Phe G UCG UUG C ACA Met or start Thr AAA ACG AAG GUU GCU GAU GUC GCC GAC GUA GUG Val GCA GCG Ala GAA GAG His Gln Asn Lys Asp Glu CGU U CGC C CGA Arg CGG AGU AGC AGA AGG A G Ser Arg U C A G GGU U GGC C GGA GGG Gly A G Third m. RNA base (3 end of codon) U

Figure 17. 6 (a) Tobacco plant expressing a firefly gene (b) Pig expressing a jellyfish gene

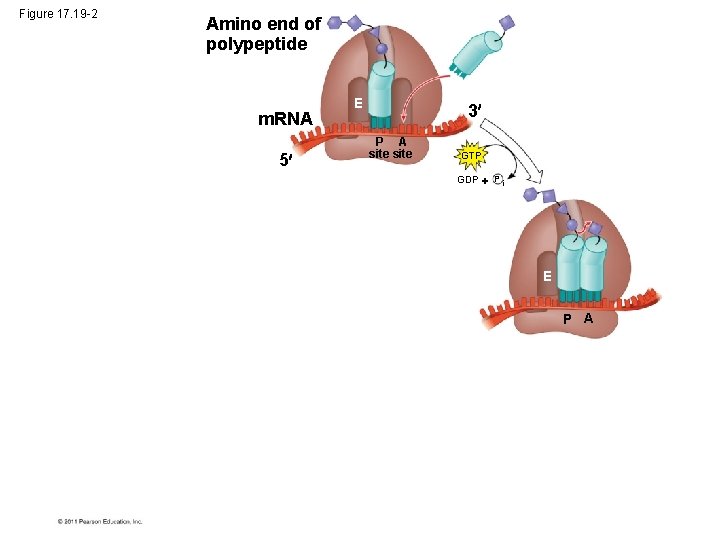
Figure 17. 19 -2 Amino end of polypeptide m. RNA 5 E 3 P A site GTP GDP P i E P A

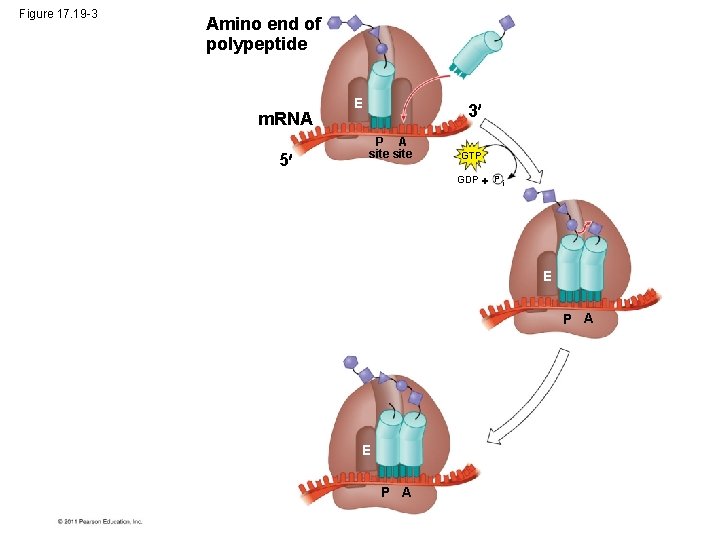
Figure 17. 19 -3 Amino end of polypeptide m. RNA 5 E 3 P A site GTP GDP P i E P A

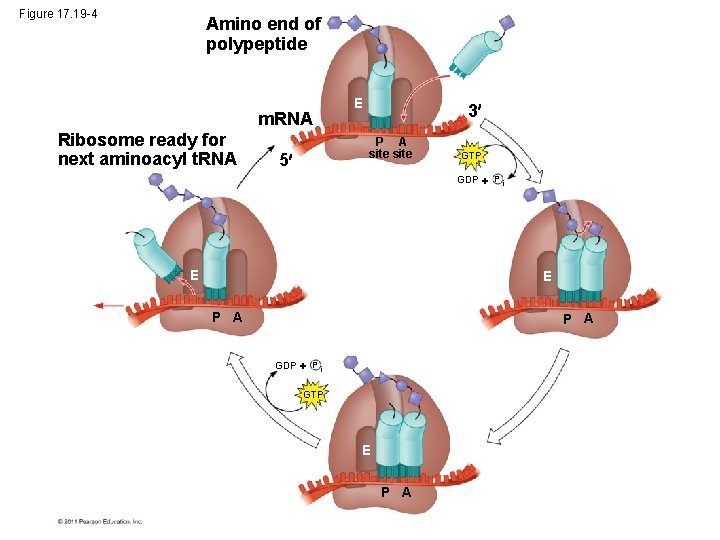
Figure 17. 19 -4 Amino end of polypeptide m. RNA Ribosome ready for next aminoacyl t. RNA E 3 P A site 5 GTP GDP P i E E P A GDP P i GTP E P A

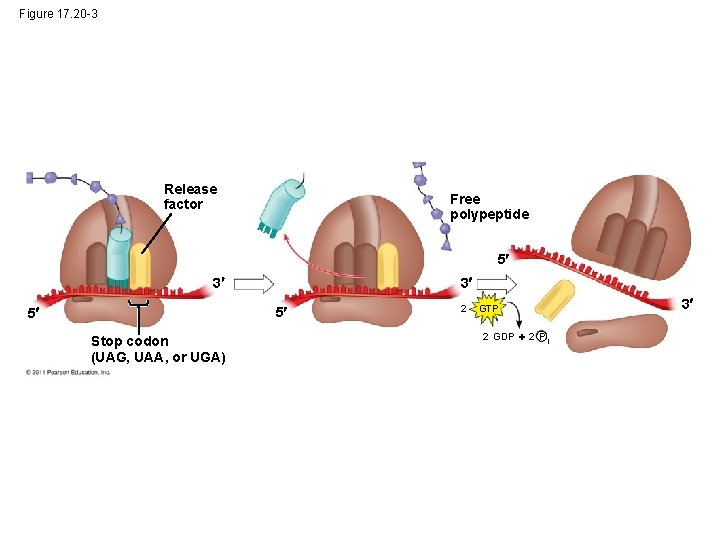
Figure 17. 20 -3 Release factor Free polypeptide 5 3 3 5 5 Stop codon (UAG, UAA, or UGA) 2 GTP 2 GDP 2 P i 3

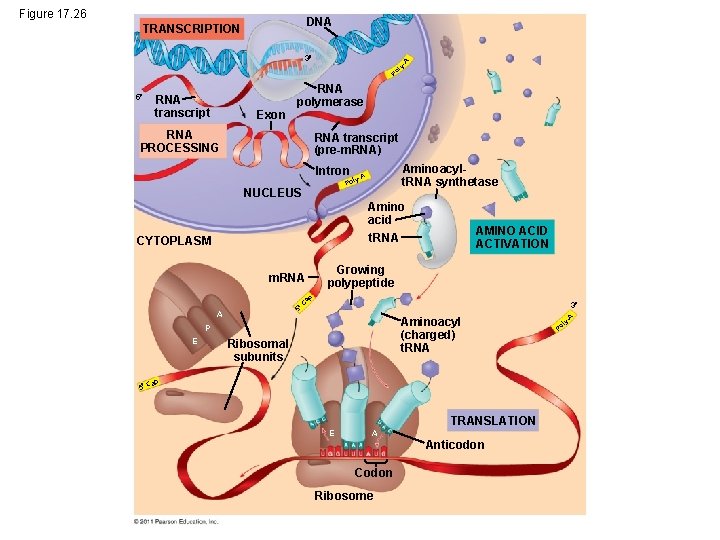
Figure 17. 26 DNA TRANSCRIPTION Po ly - A 3 5 RNA transcript Exon RNA polymerase RNA PROCESSING RNA transcript (pre-m. RNA) Intron Aminoacylt. RNA synthetase y-A Pol NUCLEUS Amino acid AMINO ACID ACTIVATION t. RNA CYTOPLASM Growing polypeptide ap m. RNA 3 5 C A Aminoacyl (charged) t. RNA P E Ribosomal subunits p a 5 C TRANSLATION E A Anticodon Codon Ribosome Po l A y-

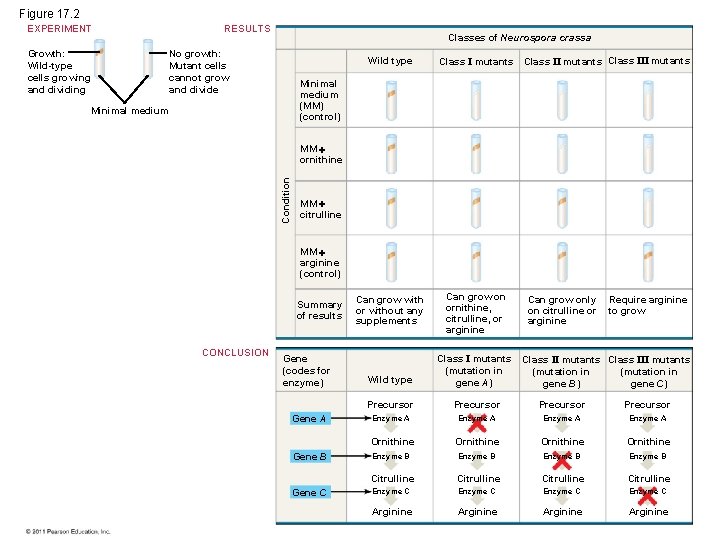
Figure 17. 2 RESULTS EXPERIMENT Growth: Wild-type cells growing and dividing Classes of Neurospora crassa No growth: Mutant cells cannot grow and divide Wild type Class I mutants Class III mutants Minimal medium (MM) (control) Minimal medium Condition MM ornithine MM citrulline MM arginine (control) Summary of results CONCLUSION Gene (codes for enzyme) Gene A Gene B Gene C Can grow with or without any supplements Can grow on ornithine, citrulline, or arginine Can grow only on citrulline or arginine Require arginine to grow Wild type Class I mutants (mutation in gene A) Precursor Enzyme A Ornithine Enzyme B Citrulline Enzyme C Arginine Class II mutants Class III mutants (mutation in gene B) gene C)

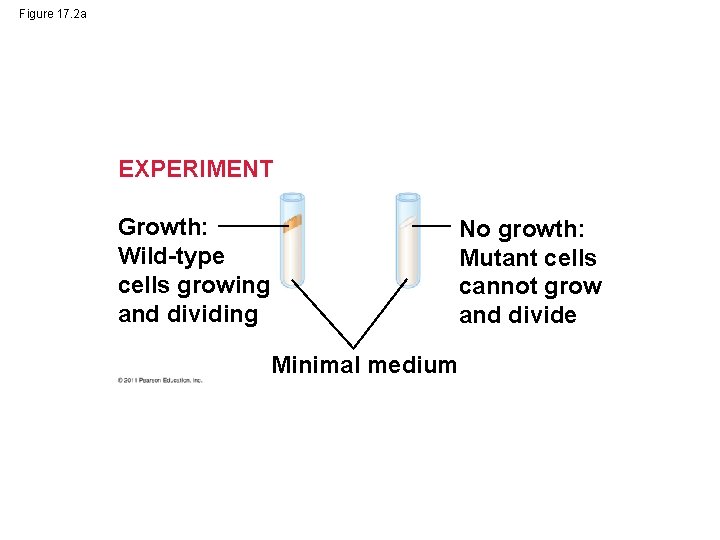
Figure 17. 2 a EXPERIMENT Growth: Wild-type cells growing and dividing No growth: Mutant cells cannot grow and divide Minimal medium

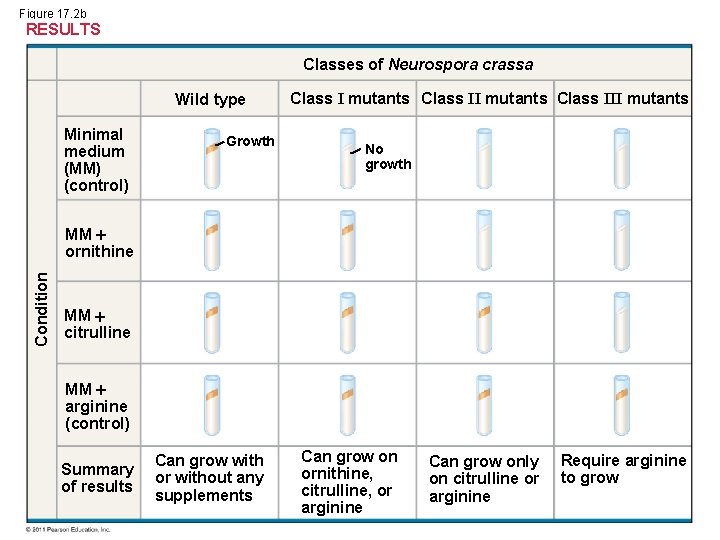
Figure 17. 2 b RESULTS Classes of Neurospora crassa Wild type Minimal medium (MM) (control) Growth Class I mutants Class III mutants No growth Condition MM ornithine MM citrulline MM arginine (control) Summary of results Can grow with or without any supplements Can grow on ornithine, citrulline, or arginine Can grow only on citrulline or arginine Require arginine to grow

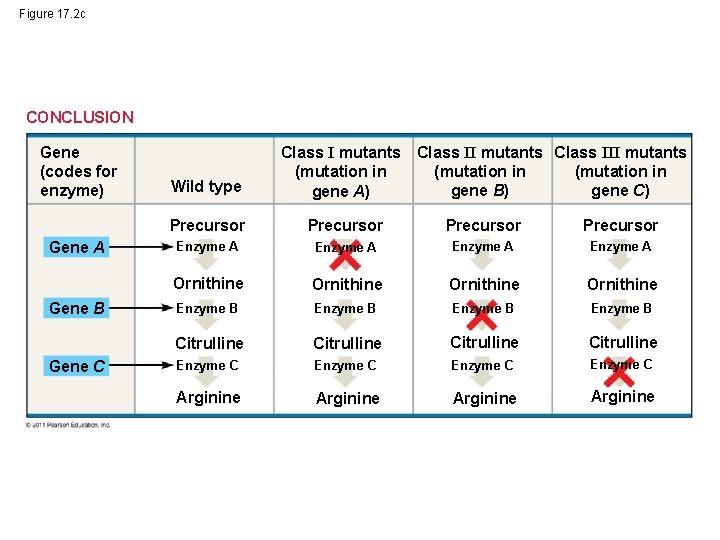
Figure 17. 2 c CONCLUSION Gene (codes for enzyme) Gene A Gene B Gene C Class II mutants Class III mutants (mutation in gene B) gene C) Wild type Class I mutants (mutation in gene A) Precursor Enzyme A Ornithine Enzyme B Citrulline Enzyme C Arginine

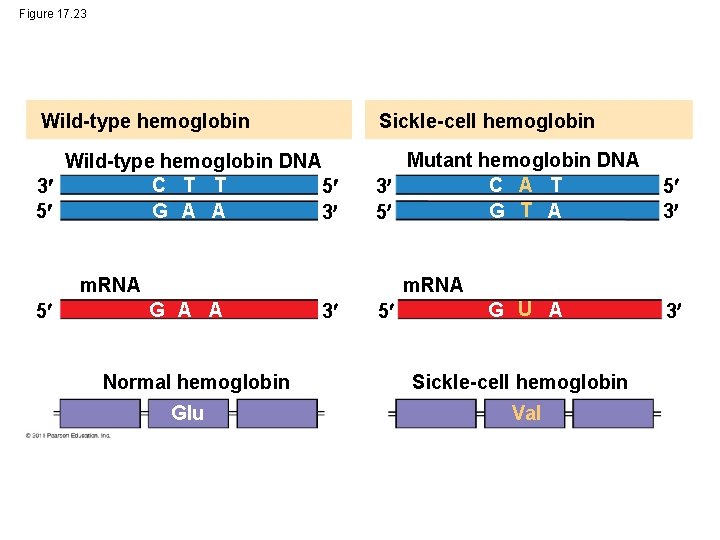
Figure 17. 23 Wild-type hemoglobin Sickle-cell hemoglobin Wild-type hemoglobin DNA C T T 3 5 G A A 5 3 Mutant hemoglobin DNA C A T 3 G T A 5 m. RNA 5 5 3 m. RNA G A A Normal hemoglobin Glu 3 5 G U A Sickle-cell hemoglobin Val 3

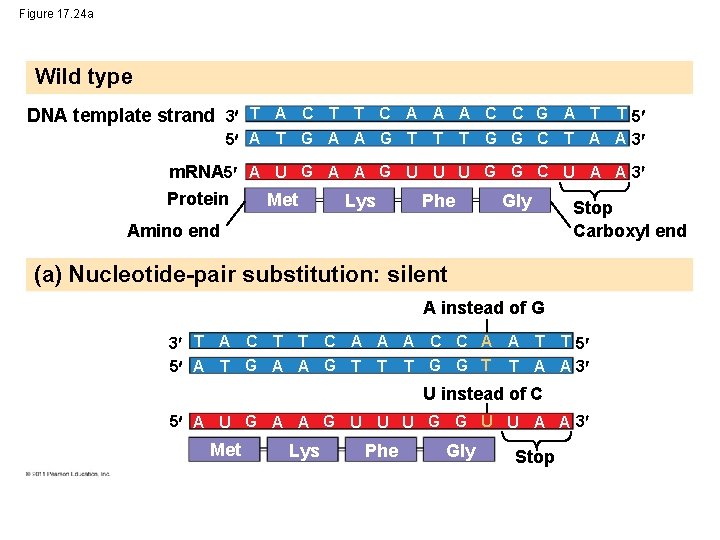
Figure 17. 24 a Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T G G C T A A 3 T m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Lys Phe Gly Stop Amino end Carboxyl end (a) Nucleotide-pair substitution: silent A instead of G 3 T A C T T C A A A C C A A T T 5 5 A T G A A G T T T G G T T A A 3 U instead of C 5 A U G A A G U U U G G U U A A 3 Met Lys Phe Gly Stop

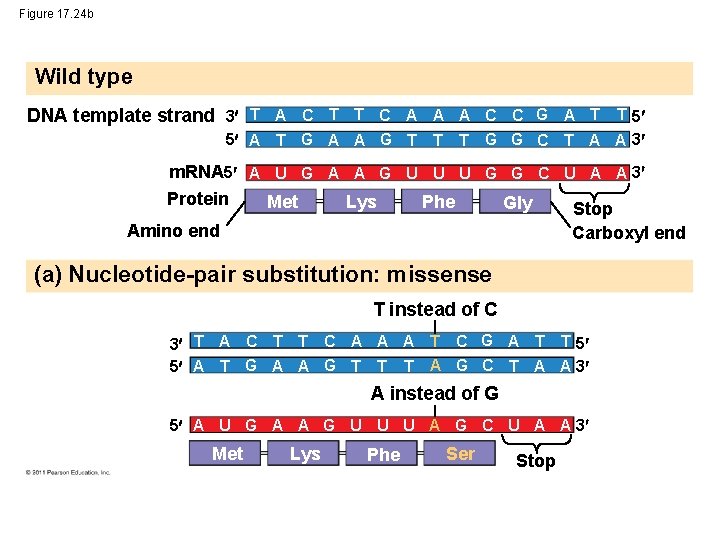
Figure 17. 24 b Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T T G G C T A A 3 m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Lys Phe Gly Stop Amino end Carboxyl end (a) Nucleotide-pair substitution: missense T instead of C 3 T A C T T C A A A T C G A T T 5 5 A T G A A G T T T A G C T A A 3 A instead of G 5 A U G A A G U U U A G C U A A 3 Met Lys Phe Ser Stop

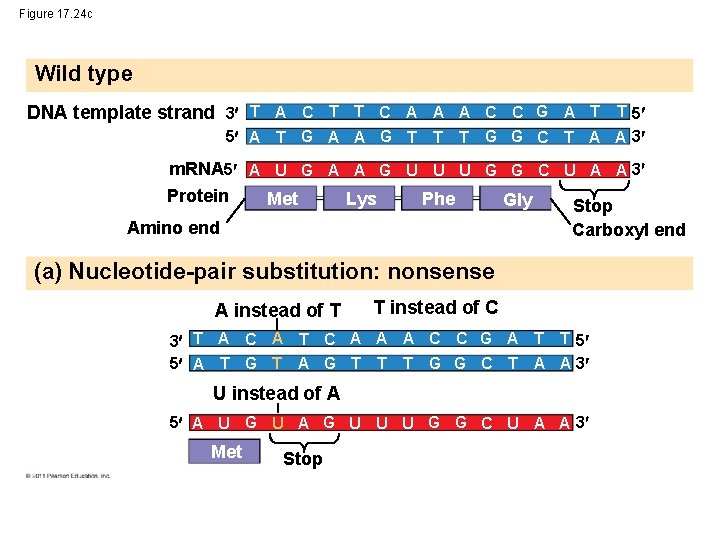
Figure 17. 24 c Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T T G G C T A A 3 m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Lys Phe Gly Stop Amino end Carboxyl end (a) Nucleotide-pair substitution: nonsense A instead of T T instead of C 3 T A C A T C A A A C C G A T T 5 5 A T G T A G T T T G G C T A A 3 U instead of A 5 A U G U A G U U U G G C U A A 3 Met Stop

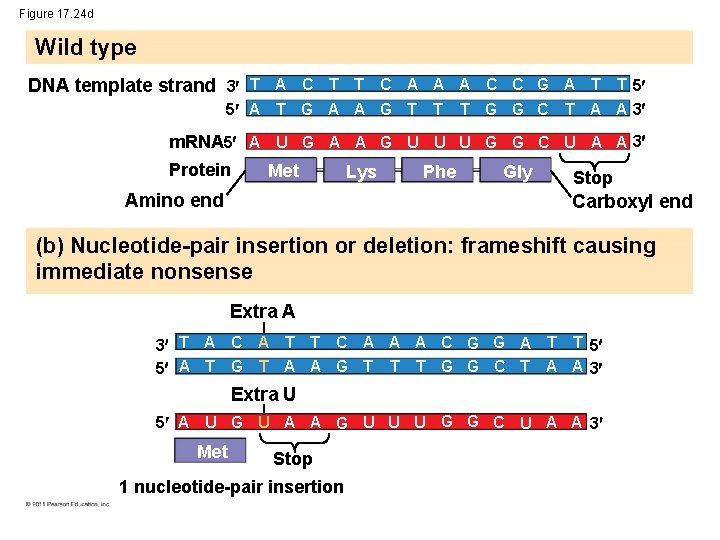
Figure 17. 24 d Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T T G G C T A A 3 m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Amino end Lys Phe Gly Stop Carboxyl end (b) Nucleotide-pair insertion or deletion: frameshift causing immediate nonsense Extra A 3 T A C A T T C A A A C G G A T T 5 5 A T G T A A G T T T G G C T A A 3 Extra U 5 A U G U A A G U U U G G C U A A 3 Met Stop 1 nucleotide-pair insertion

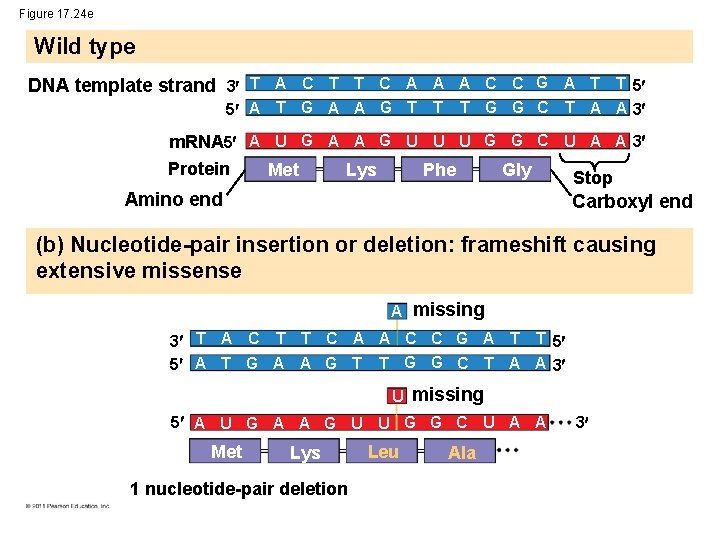
Figure 17. 24 e Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T T G G C T A A 3 m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Gly Lys Phe Stop Amino end Carboxyl end (b) Nucleotide-pair insertion or deletion: frameshift causing extensive missense A missing 3 T A C T T C A A C C G A T T 5 5 A T G A A G T T G G C T A A 3 U missing 5 A U G A A G U U G G C U A A Met Lys 1 nucleotide-pair deletion Leu Ala 3


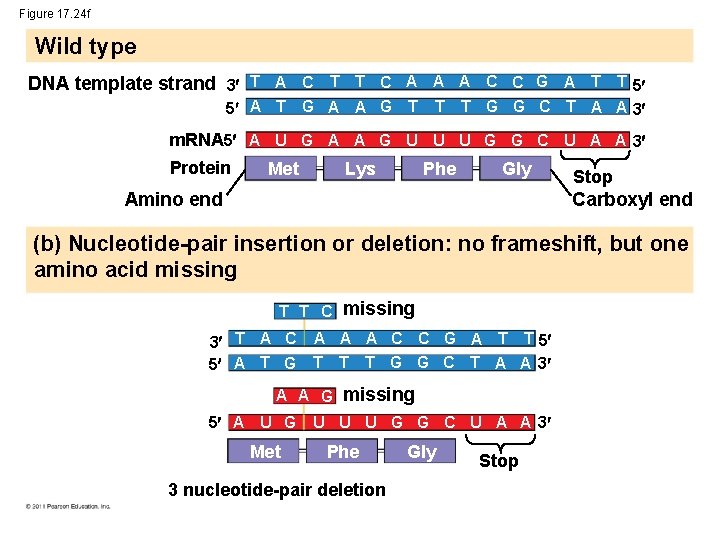
Figure 17. 24 f Wild type DNA template strand 3 T A C T T C A A A C C G A T T 5 5 A T G A A G T T T G G C T A A 3 m. RNA 5 A U G A A G U U U G G C U A A 3 Protein Met Gly Lys Phe Stop Carboxyl end Amino end (b) Nucleotide-pair insertion or deletion: no frameshift, but one amino acid missing T T C missing 3 T A C A A A C C G A T T 5 5 A T G T T T G G C T A A 3 A A G missing 5 A U G Met U U U G G C U A A 3 Phe 3 nucleotide-pair deletion Gly Stop
 Chapter 17: from gene to protein
Chapter 17: from gene to protein Carrier vs channel proteins
Carrier vs channel proteins Protein-protein docking
Protein-protein docking Chapter 17 from gene to protein
Chapter 17 from gene to protein Chapter 17 from gene to protein
Chapter 17 from gene to protein Gene by gene test results
Gene by gene test results Regulation of gene expression in bacteria
Regulation of gene expression in bacteria Gene expression
Gene expression טרנסלציה
טרנסלציה Cells must control gene expression so that __________.
Cells must control gene expression so that __________. Fret rt pcr
Fret rt pcr Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Gene expression
Gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Lyonization of gene expression
Lyonization of gene expression Prokaryotic gene expression
Prokaryotic gene expression Gene expression omnibus tutorial
Gene expression omnibus tutorial Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Gene expression
Gene expression Negative vs positive control operon
Negative vs positive control operon Genetic effects on gene expression across human tissues
Genetic effects on gene expression across human tissues Dr kevin ahern
Dr kevin ahern Gene expression
Gene expression Regulation of gene expression
Regulation of gene expression Repressible operon
Repressible operon Chapter 18 regulation of gene expression
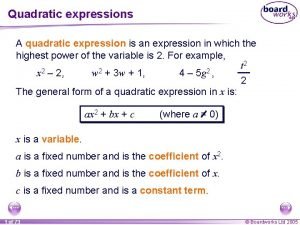
Chapter 18 regulation of gene expression Quadratic formula
Quadratic formula In figure 1 what facial expression did mommy lenny show
In figure 1 what facial expression did mommy lenny show Understand rigid transformations
Understand rigid transformations An operation that maps an original figure onto a new figure
An operation that maps an original figure onto a new figure Is a trapezoid a plane figure
Is a trapezoid a plane figure 4 figure and 6 figure grid references
4 figure and 6 figure grid references Biological function of protein ppt
Biological function of protein ppt Protein losing enteropathy diagnosis
Protein losing enteropathy diagnosis Void volume
Void volume Protein analiz yöntemleri
Protein analiz yöntemleri Is myoglobin a globular protein
Is myoglobin a globular protein Kids growth powder
Kids growth powder Transcortin protein
Transcortin protein Transcortin protein
Transcortin protein