DNA Replication Repair DNA Replication Repair Dr Ketki

































- Slides: 88

DNA Replication & Repair

DNA Replication & Repair Dr. Ketki K Assistant Professor Dept of Biochemistry

The flow of information from DNA to RNA to protein is termed the …………. with the exception of some viruses that have RNA as the repository of their genetic information

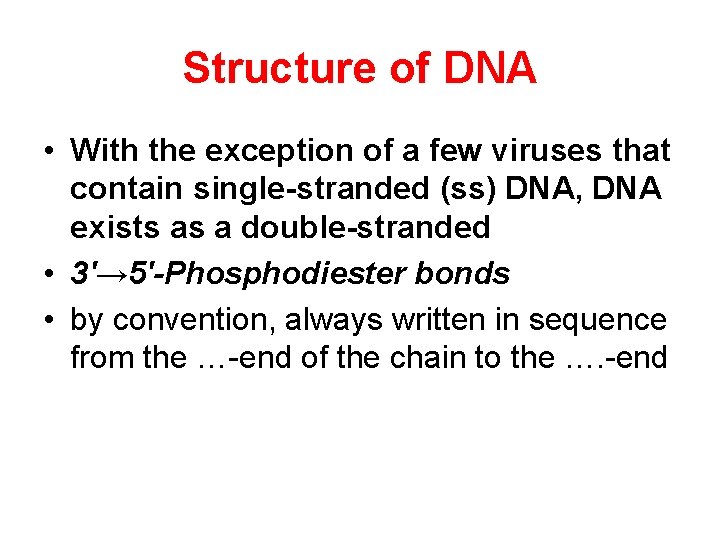
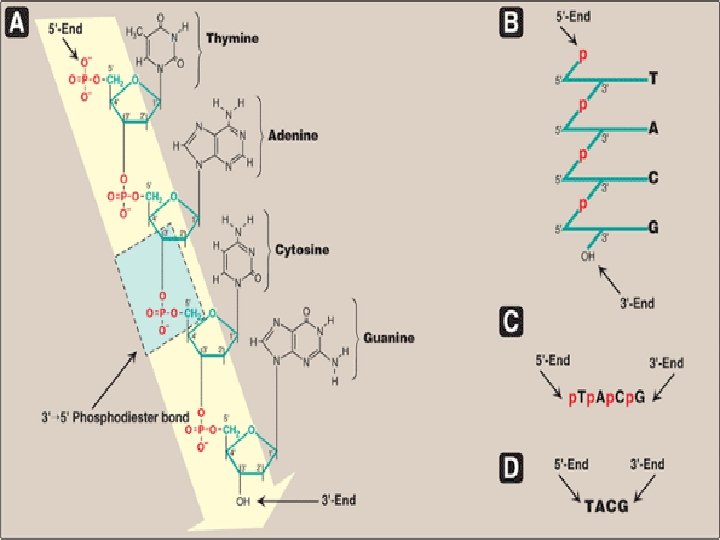
Structure of DNA • With the exception of a few viruses that contain single-stranded (ss) DNA, DNA exists as a double-stranded • 3′→ 5′-Phosphodiester bonds • by convention, always written in sequence from the …-end of the chain to the …. -end


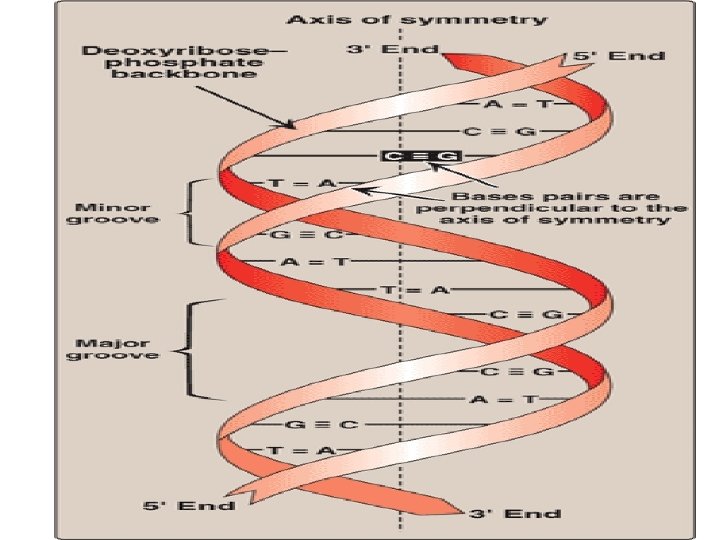
Double helix • Chains are paired in an …………. fashion • Hydrophobic bases are stacked………. . • hydrophilic deoxyribose–phosphate backbone of each chain is on the………. • Base pairing : ………… • Chargaff Rule : ………. .


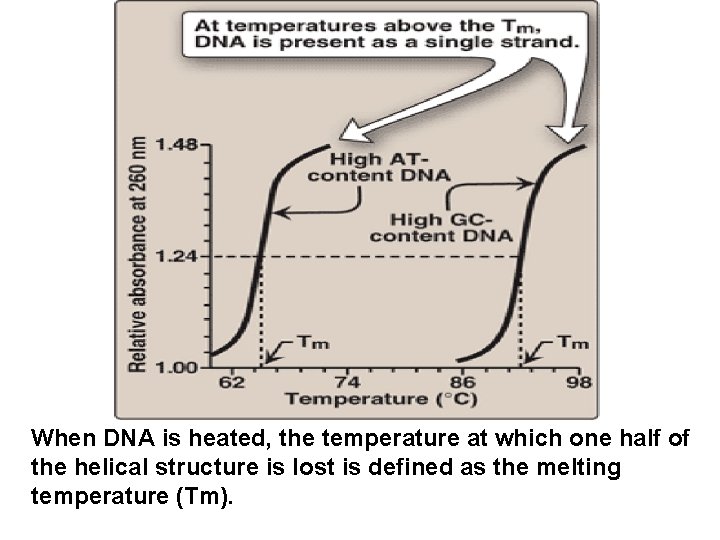
When DNA is heated, the temperature at which one half of the helical structure is lost is defined as the melting temperature (Tm).


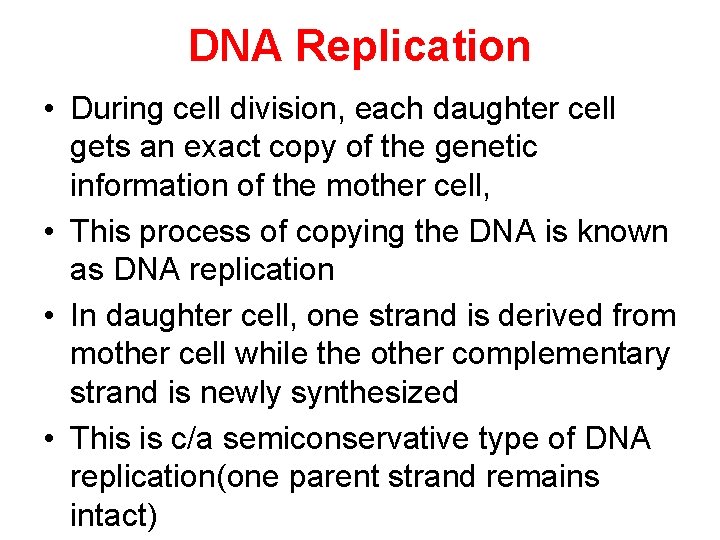
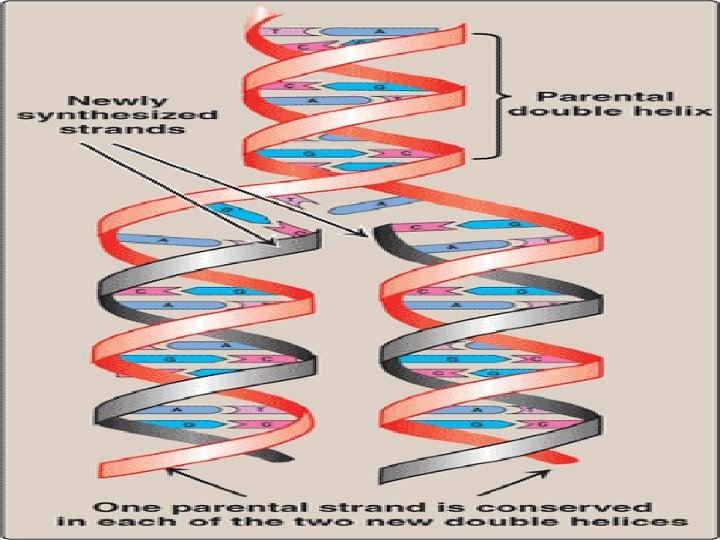
DNA Replication • During cell division, each daughter cell gets an exact copy of the genetic information of the mother cell, • This process of copying the DNA is known as DNA replication • In daughter cell, one strand is derived from mother cell while the other complementary strand is newly synthesized • This is c/a semiconservative type of DNA replication(one parent strand remains intact)


• For easy understanding, replication is divided into 1) Initiation 2) Elongation 3) Termination

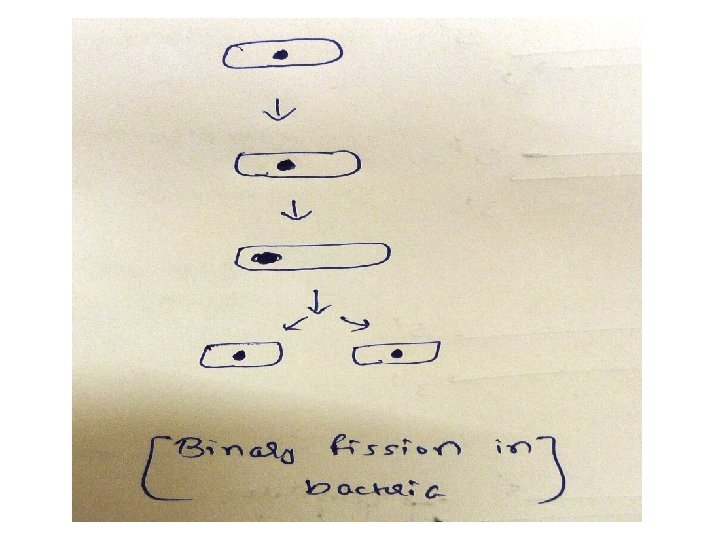
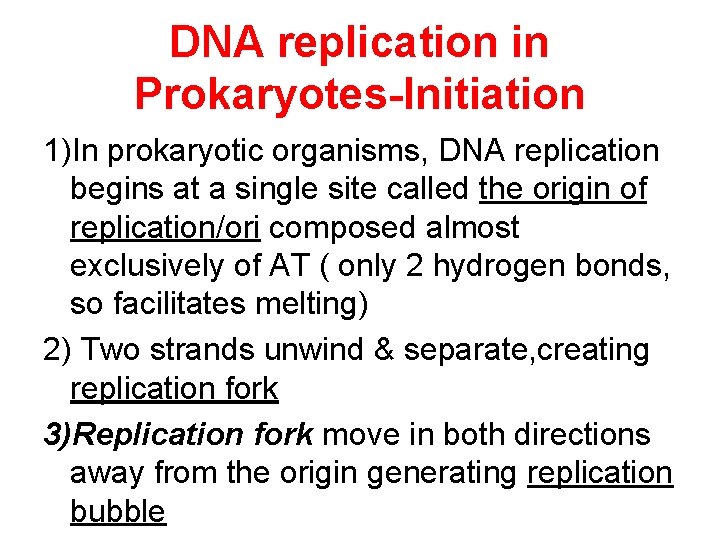
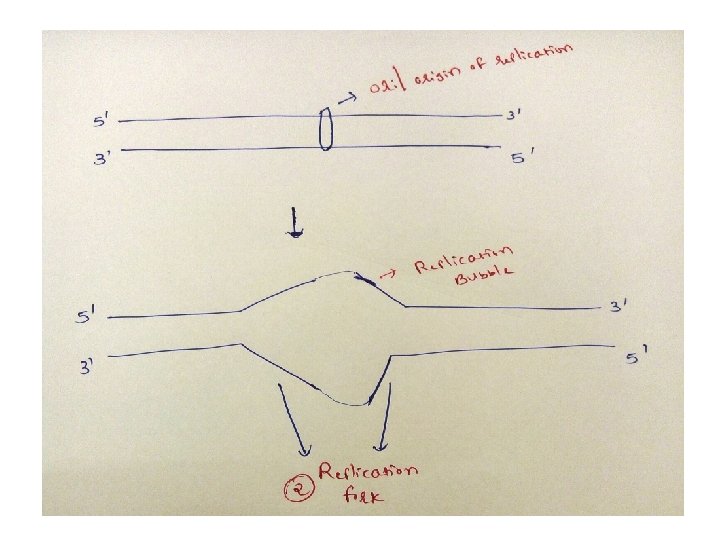
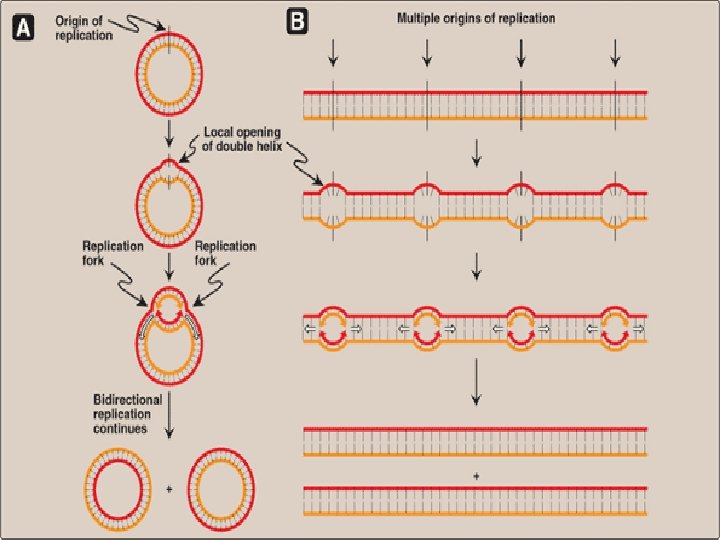
DNA replication in Prokaryotes-Initiation 1)In prokaryotic organisms, DNA replication begins at a single site called the origin of replication/ori composed almost exclusively of AT ( only 2 hydrogen bonds, so facilitates melting) 2) Two strands unwind & separate, creating replication fork 3)Replication fork move in both directions away from the origin generating replication bubble



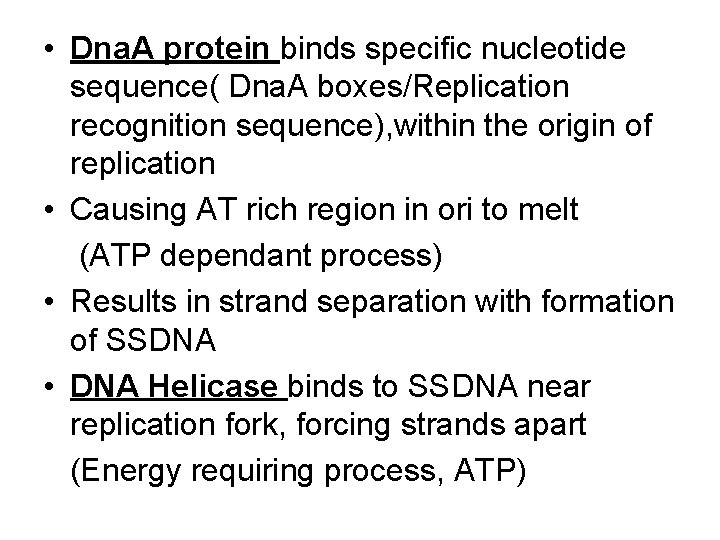
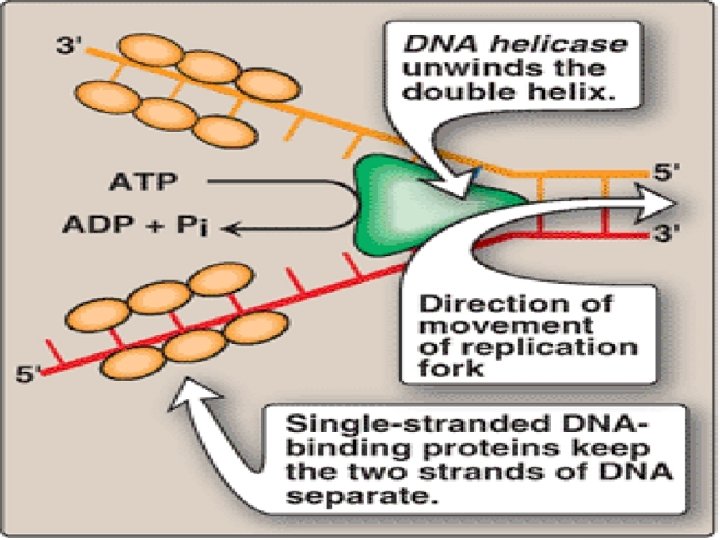
• Dna. A protein binds specific nucleotide sequence( Dna. A boxes/Replication recognition sequence), within the origin of replication • Causing AT rich region in ori to melt (ATP dependant process) • Results in strand separation with formation of SSDNA • DNA Helicase binds to SSDNA near replication fork, forcing strands apart (Energy requiring process, ATP)

• Dna. B is principle helicase in E coli , requires Dna. C also for binding to DNA • SSDBPs ( single stranded DNA binding proteins) binds to SSDNA and keeps two strands of DNA separated


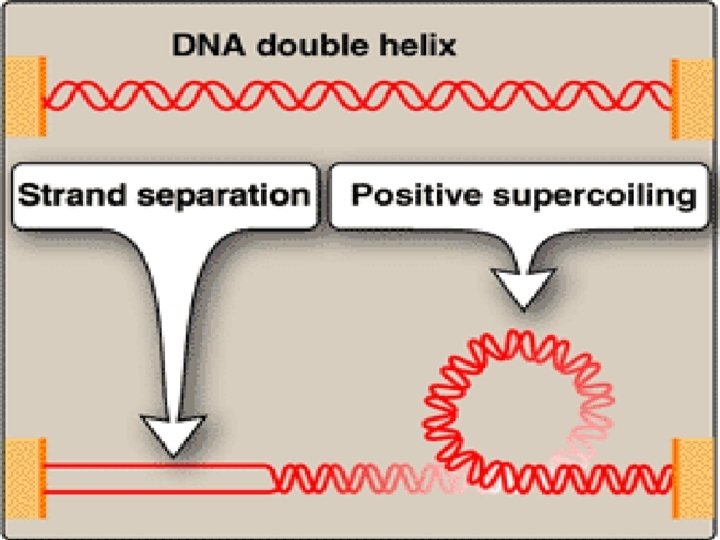
Solving the problem of supercoils • Upon separation of two strands , problem is encountered • Positive supercoils: in the region of DNA ahead of replication fork as a result of overwinding • Negative supercoils: in the region behind the fork • This problem interferes with further unwinding of DNA double helix

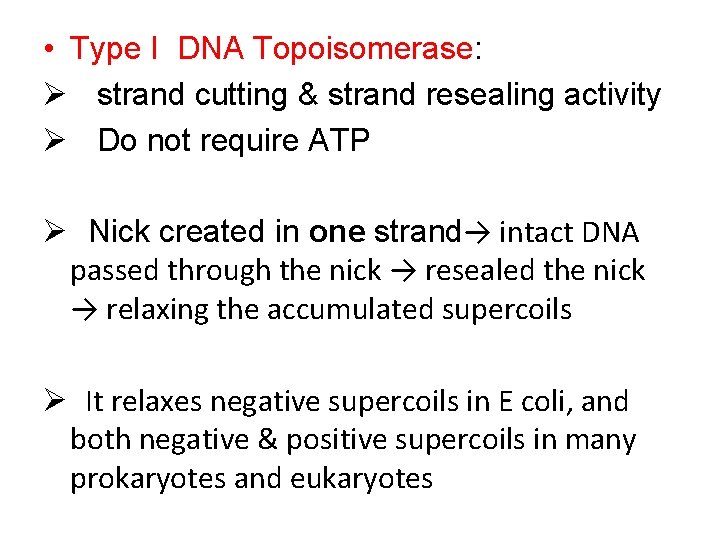
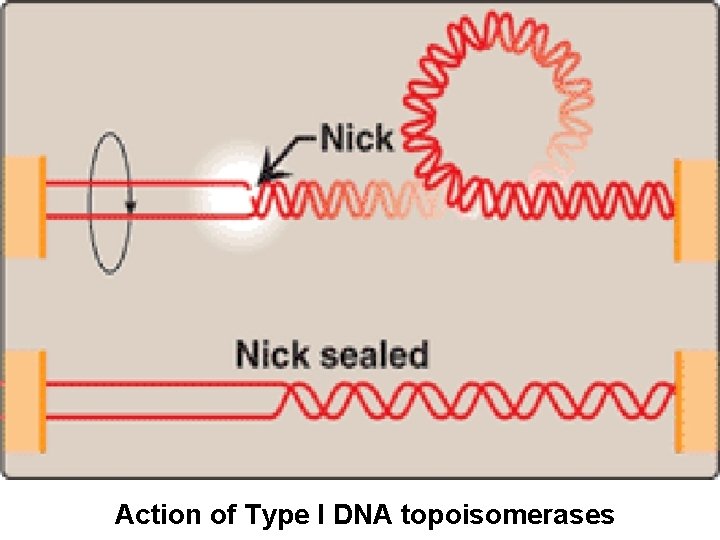
• Type I DNA Topoisomerase: Ø strand cutting & strand resealing activity Ø Do not require ATP Ø Nick created in one strand→ intact DNA passed through the nick → resealed the nick → relaxing the accumulated supercoils Ø It relaxes negative supercoils in E coli, and both negative & positive supercoils in many prokaryotes and eukaryotes

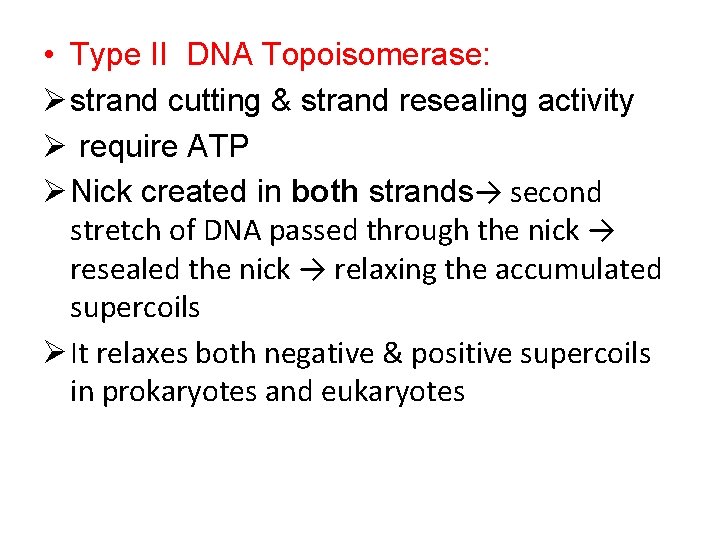
• Type II DNA Topoisomerase: Ø strand cutting & strand resealing activity Ø require ATP Ø Nick created in both strands→ second stretch of DNA passed through the nick → resealed the nick → relaxing the accumulated supercoils Ø It relaxes both negative & positive supercoils in prokaryotes and eukaryotes


Action of Type I DNA topoisomerases

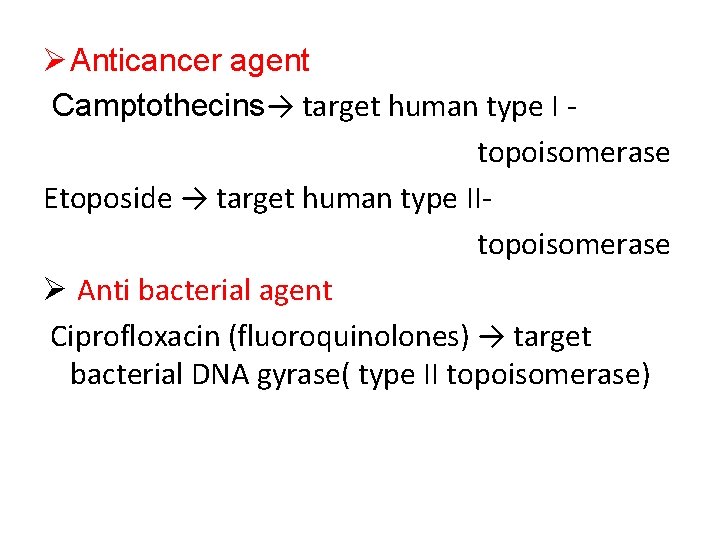
Ø Anticancer agent Camptothecins→ target human type I topoisomerase Etoposide → target human type IItopoisomerase Ø Anti bacterial agent Ciprofloxacin (fluoroquinolones) → target bacterial DNA gyrase( type II topoisomerase)

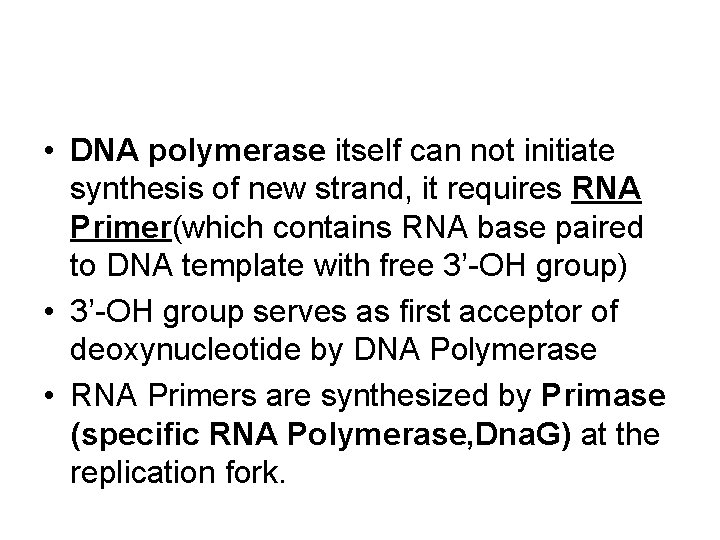
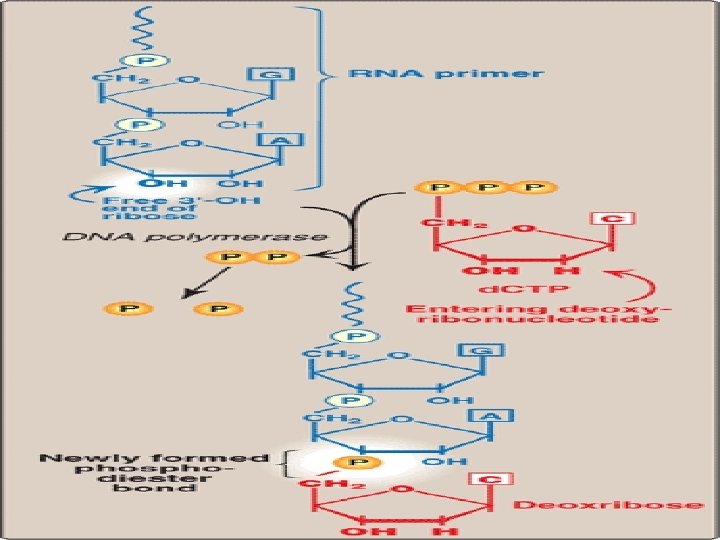
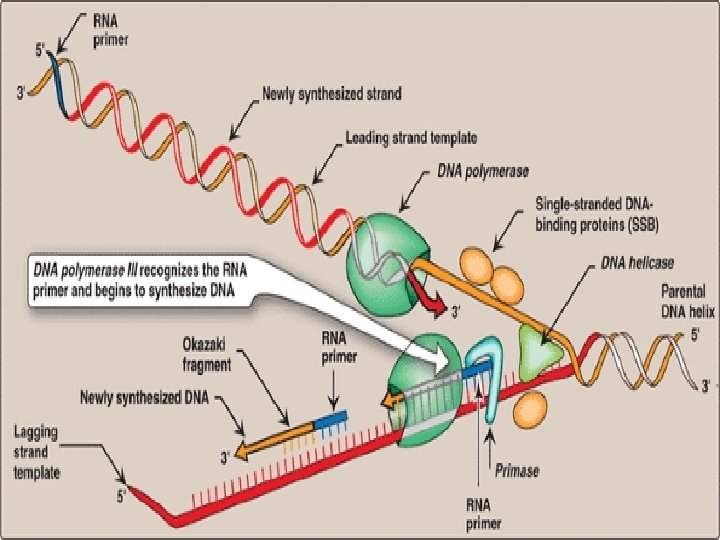
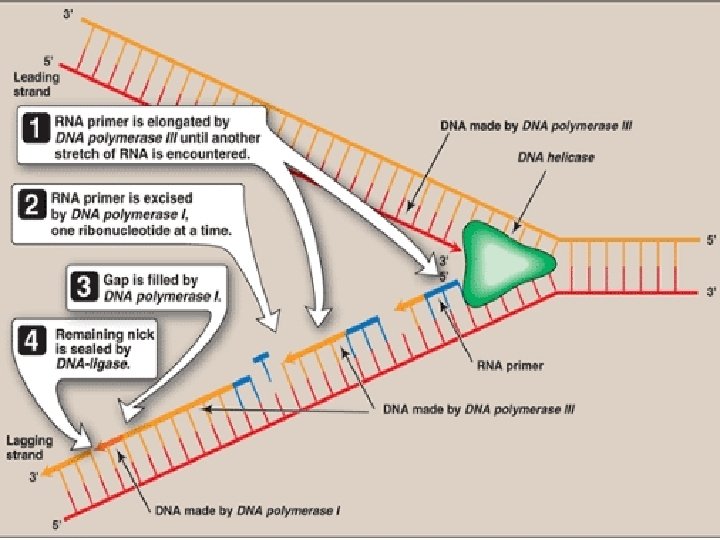
• DNA polymerase itself can not initiate synthesis of new strand, it requires RNA Primer(which contains RNA base paired to DNA template with free 3’-OH group) • 3’-OH group serves as first acceptor of deoxynucleotide by DNA Polymerase • RNA Primers are synthesized by Primase (specific RNA Polymerase, Dna. G) at the replication fork.


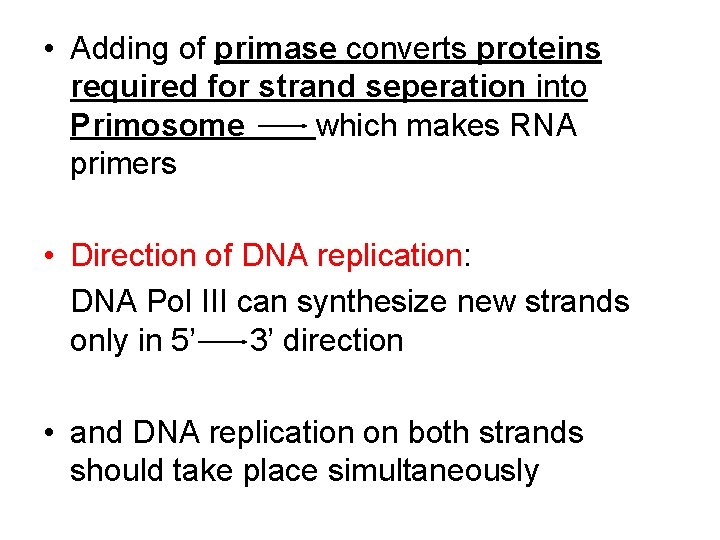
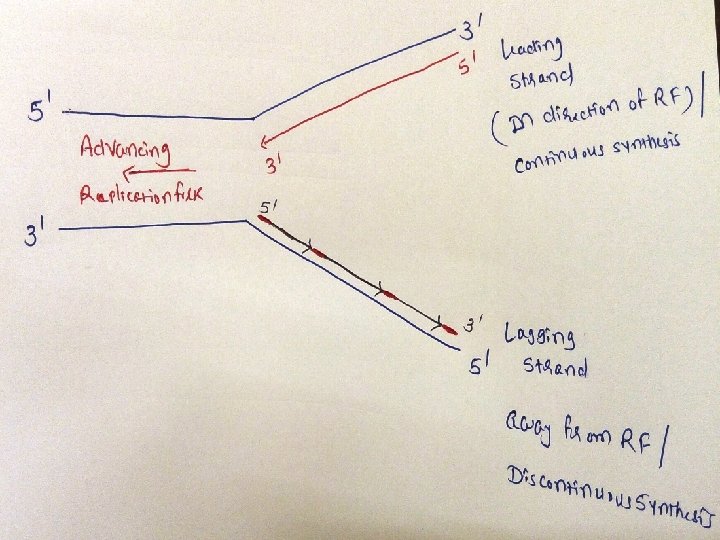
• Adding of primase converts proteins required for strand seperation into Primosome which makes RNA primers • Direction of DNA replication: DNA Pol III can synthesize new strands only in 5’ 3’ direction • and DNA replication on both strands should take place simultaneously

• So, strand which is being copied in direction of advancing replication fork is c/a leading strand, which is synthesized continuously • Strand which is being copied in direction away from replication fork is c/a lagging strand, which is synthesized discontinuously • Lagging strand requires multiple RNA primers • With small stretches of DNA being copied near replication fork. these small stretches of discontinuous DNA on lagging strand, are termed as Okazaki fragments


Elongation

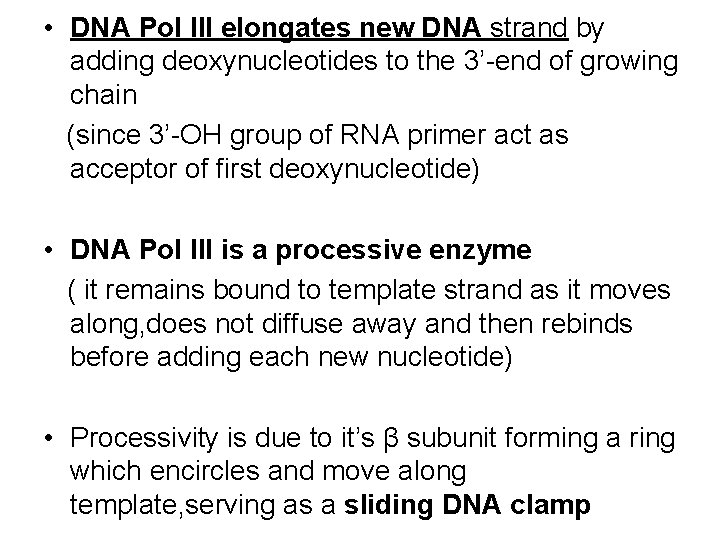
• DNA Pol III elongates new DNA strand by adding deoxynucleotides to the 3’-end of growing chain (since 3’-OH group of RNA primer act as acceptor of first deoxynucleotide) • DNA Pol III is a processive enzyme ( it remains bound to template strand as it moves along, does not diffuse away and then rebinds before adding each new nucleotide) • Processivity is due to it’s β subunit forming a ring which encircles and move along template, serving as a sliding DNA clamp

• With addition of each new nucleotide to the growing chain, PPi is released • Hydrolysis of PPi to 2 Pi gives energy to drive the reactions forward


• For survival of organism, it is important to synthesize new strand with less no of errors, since misreading of template sequence can reduce deletorious lethal mutations

• DNA Pol III has proof reading activity (3’ 5’ exonuclease), which removes mismatched nucleotides Example: C=A instead of C=G 3’ 5’ exonuclease removes mismatched nucleotide A 5’ 3’ polymerase activity replaces it with correct nucleotide containing G (excision must be in reverse direction from that of synthesis)


• Excision of RNA primer and their replacement by DNA • DNA polymerase I : • has a 5′→ 3′ exonuclease activity that is able to hydrolytically remove the RNA primer • In addition it has 5′→ 3′ polymerase activity that synthesizes DNA, • and the 3′→ 5′ exonuclease activity that proofreads

• First, 5′→ 3′ exonuclease activity of DNA Pol I is able to hydrolytically remove the RNA primer • Gap is created • Gap is filled by 5′→ 3′ polymerase activity of DNA Pol I that synthesizes DNA, • To remove the errors in newly synthesized strand, 3′→ 5′ exonuclease activity of DNA Pol I proofreads the new chain


• This removal /synthesis /proofreading continues untill RNA primer is totally degraded, and gap is filled with DNA • Nick is present between two stretches of DNA in leading strand, between multiple stretches of DNA in lagging strand • Nick is sealed by DNA Ligase

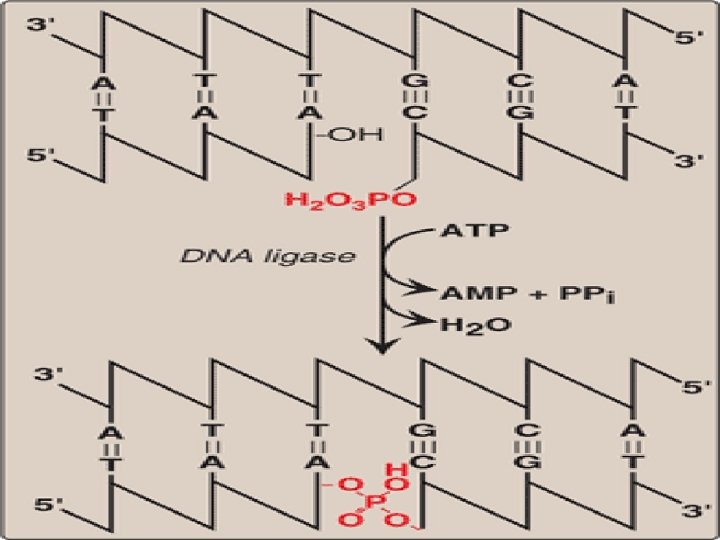
DNA ligase • The final phosphodiester linkage between the 5′-phosphate group on the DNA chain synthesized by DNA polymerase III and the 3′-hydroxyl group on the chain made by DNA polymerase I is catalyzed by DNA ligase • This joining requires energy from hydrolysis of ATP to AMP and Ppi



Termination • Binding of protein TUS(terminus utilization substance) to replication termination site(ter sites) on the DNA, • Stopping the movement of DNA Polymerase

Termination • Mediated by sequence specific binding of protein, Tus(Terminus utilization substance) to replication termination site(ter sites) on DNA , stopping the movement of DNA Polymerase

Eukaryotic DNA Replication

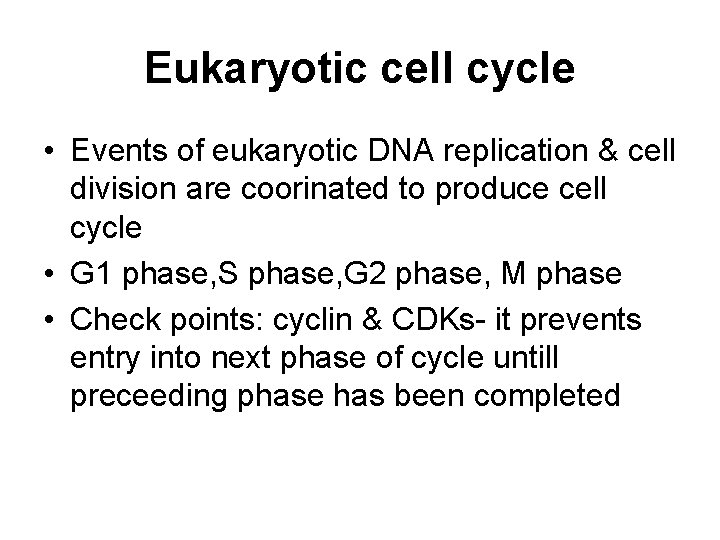
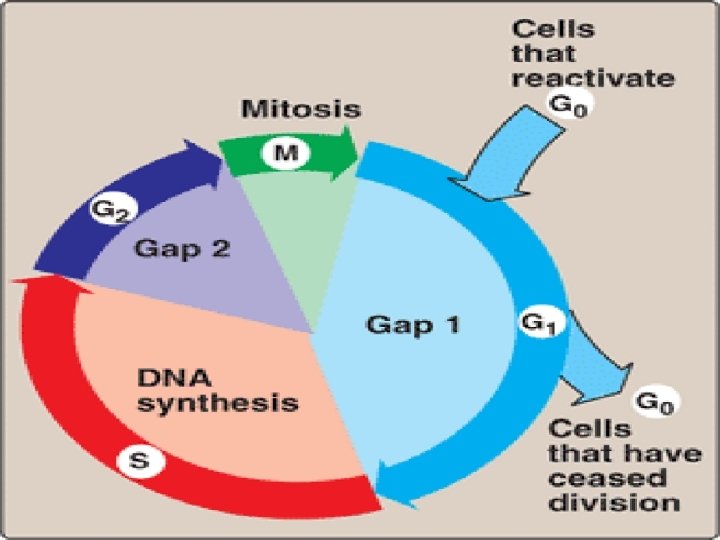
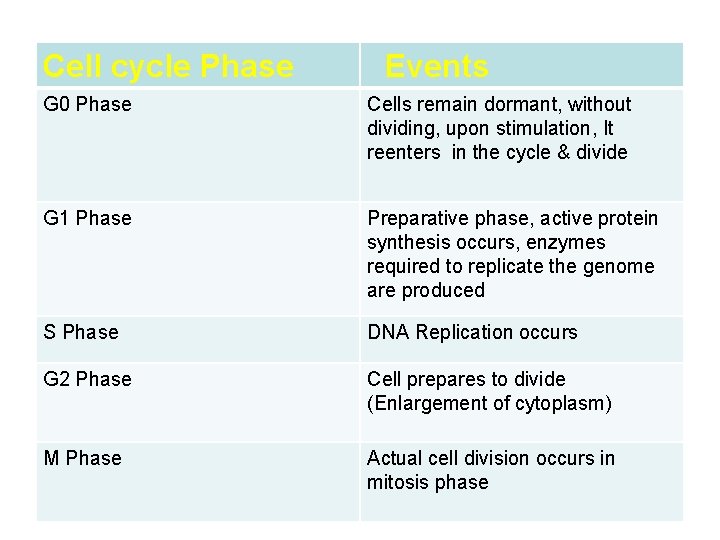
Eukaryotic cell cycle • Events of eukaryotic DNA replication & cell division are coorinated to produce cell cycle • G 1 phase, S phase, G 2 phase, M phase • Check points: cyclin & CDKs- it prevents entry into next phase of cycle untill preceeding phase has been completed


Cell cycle Phase Events G 0 Phase Cells remain dormant, without dividing, upon stimulation, It reenters in the cycle & divide G 1 Phase Preparative phase, active protein synthesis occurs, enzymes required to replicate the genome are produced S Phase DNA Replication occurs G 2 Phase Cell prepares to divide (Enlargement of cytoplasm) M Phase Actual cell division occurs in mitosis phase

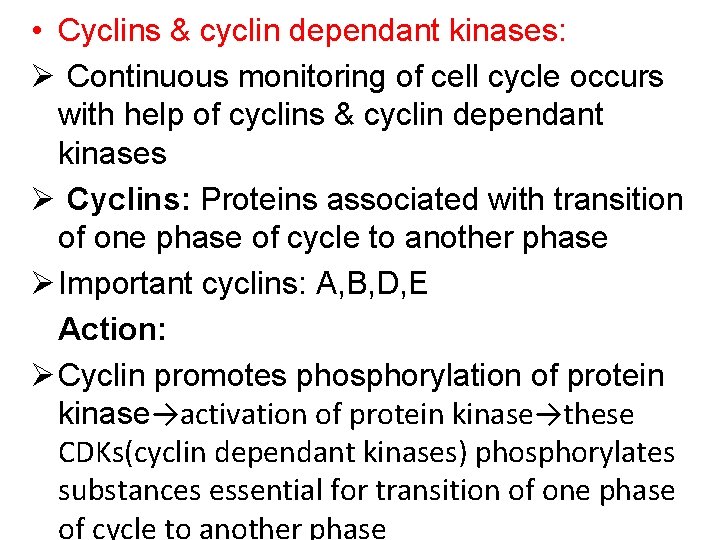
• Cyclins & cyclin dependant kinases: Ø Continuous monitoring of cell cycle occurs with help of cyclins & cyclin dependant kinases Ø Cyclins: Proteins associated with transition of one phase of cycle to another phase Ø Important cyclins: A, B, D, E Action: Ø Cyclin promotes phosphorylation of protein kinase→activation of protein kinase→these CDKs(cyclin dependant kinases) phosphorylates substances essential for transition of one phase of cycle to another phase

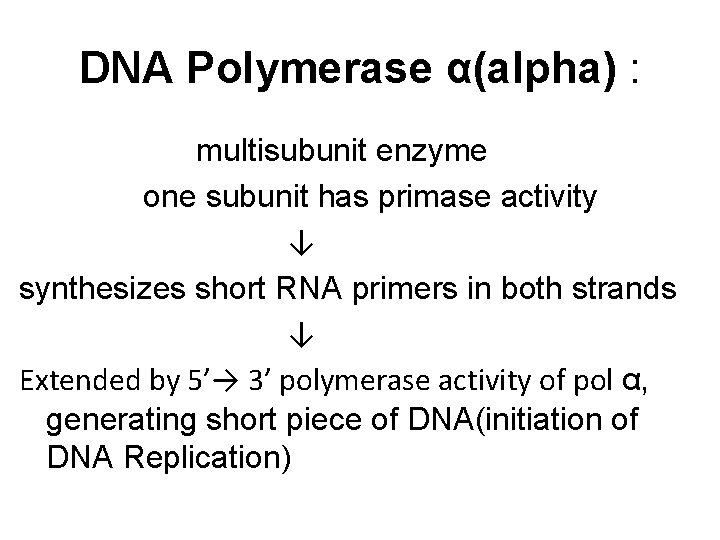
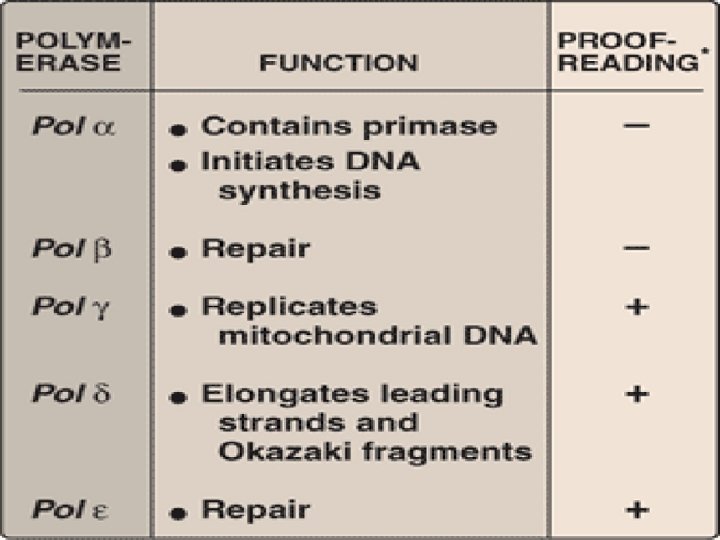
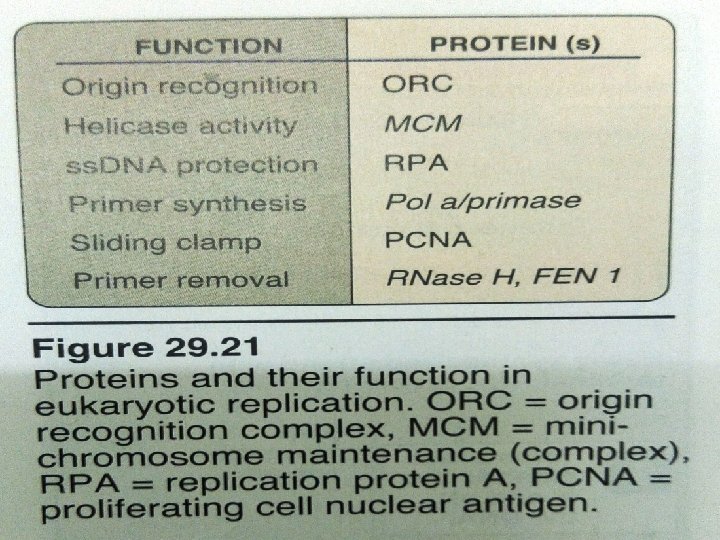
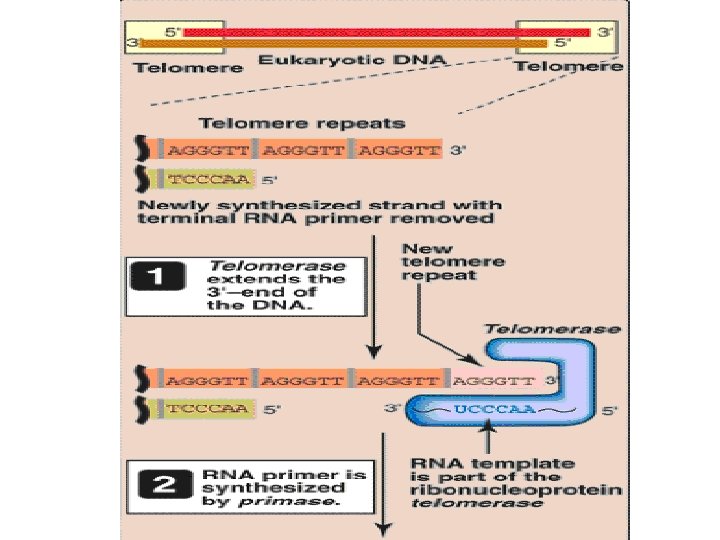
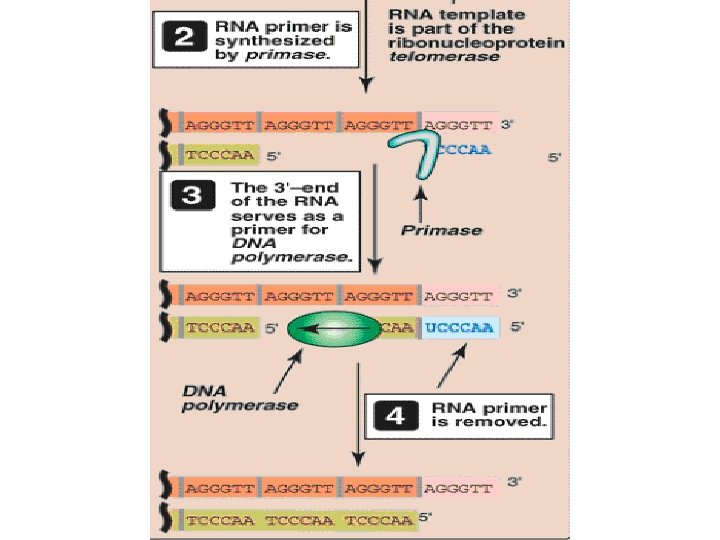
Eukaryotic DNA Replication • Multiple origins of replication • RNA primers are removed by RNase H and FEN 1 rather than by a DNA polymerase. • At least five key eukaryotic DNA polymerases have been identified • Telomeres • Occurs during S phase of cell cycle

DNA Polymerase α(alpha) : multisubunit enzyme one subunit has primase activity ↓ synthesizes short RNA primers in both strands ↓ Extended by 5’→ 3’ polymerase activity of pol α, generating short piece of DNA(initiation of DNA Replication)

• Pol ε(epsilon) : Completes DNA synthesis on leading strand, proofreading activity by 3’→ 5’ exonuclease activity • Pol δ(delta) : elongates okazaki fragments on lagging strand, proofreading activity by 3’→ 5’ exonuclease activity

• Pol β(beta) – Gap filling in DNA repair • Pol γ(gamma)- replicates mitochondrial DNA, proofreading activity by 3’→ 5’ exonuclease activity



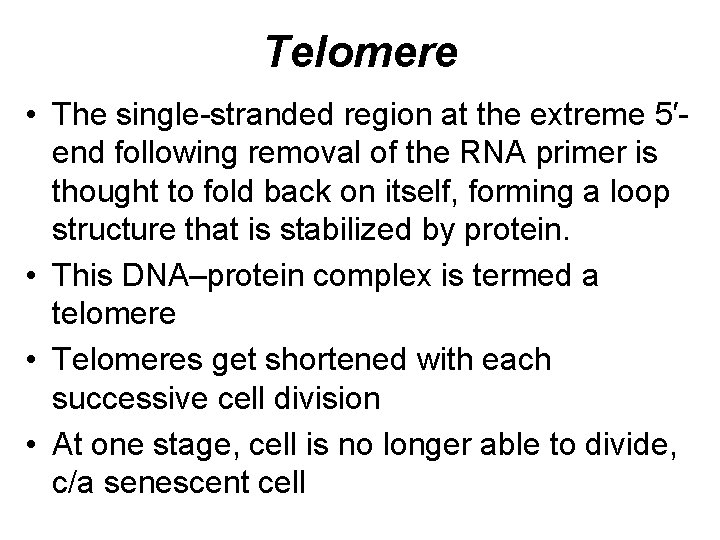
Telomere • The single-stranded region at the extreme 5′end following removal of the RNA primer is thought to fold back on itself, forming a loop structure that is stabilized by protein. • This DNA–protein complex is termed a telomere • Telomeres get shortened with each successive cell division • At one stage, cell is no longer able to divide, c/a senescent cell

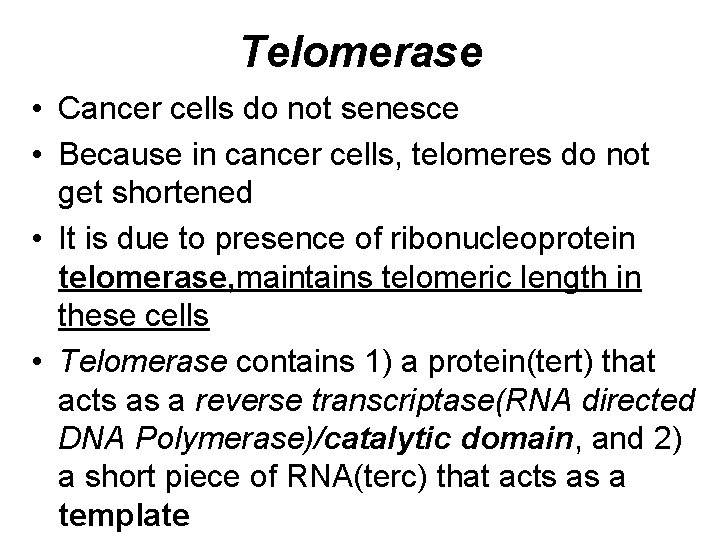
Telomerase • Cancer cells do not senesce • Because in cancer cells, telomeres do not get shortened • It is due to presence of ribonucleoprotein telomerase, maintains telomeric length in these cells • Telomerase contains 1) a protein(tert) that acts as a reverse transcriptase(RNA directed DNA Polymerase)/catalytic domain, and 2) a short piece of RNA(terc) that acts as a template



Reverse transcriptase HIV virus contain SSRNA genome When HIV virus infects host cell, HIV viral RNA serves as template for 5’ to 3’ synthesis of DNA by using reverse transcriptase (viral Infected viral DNA enzyme) incorporates into host genome, resulting in AIDS

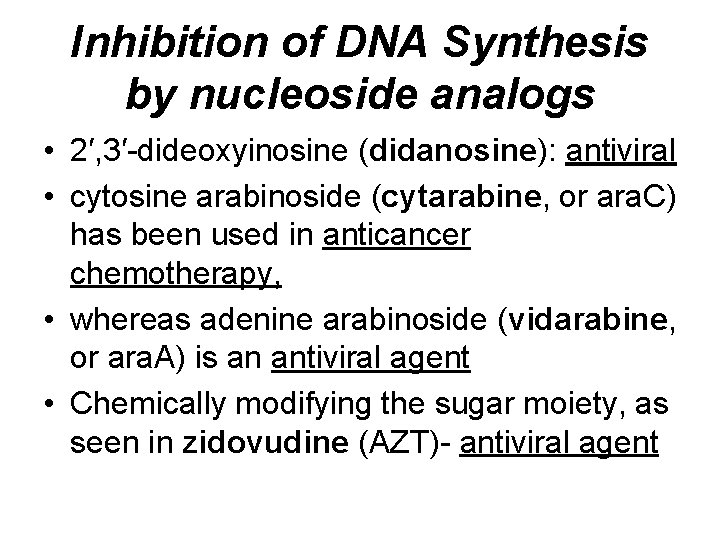
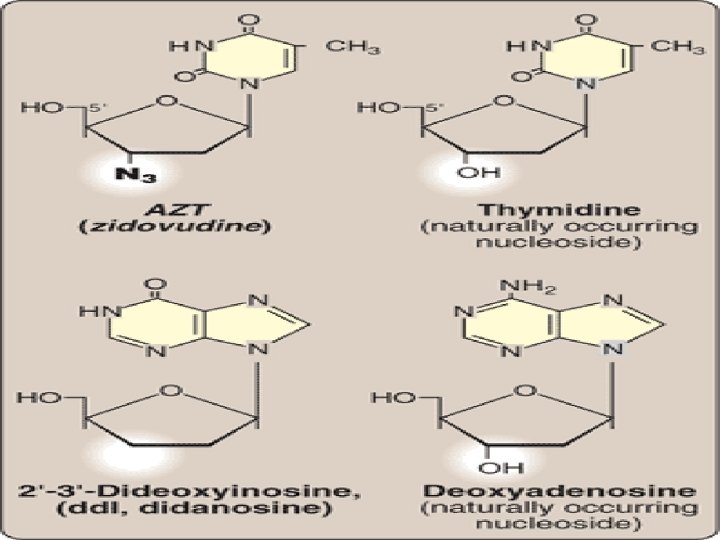
Inhibition of DNA Synthesis by nucleoside analogs • 2′, 3′-dideoxyinosine (didanosine): antiviral • cytosine arabinoside (cytarabine, or ara. C) has been used in anticancer chemotherapy, • whereas adenine arabinoside (vidarabine, or ara. A) is an antiviral agent • Chemically modifying the sugar moiety, as seen in zidovudine (AZT)- antiviral agent


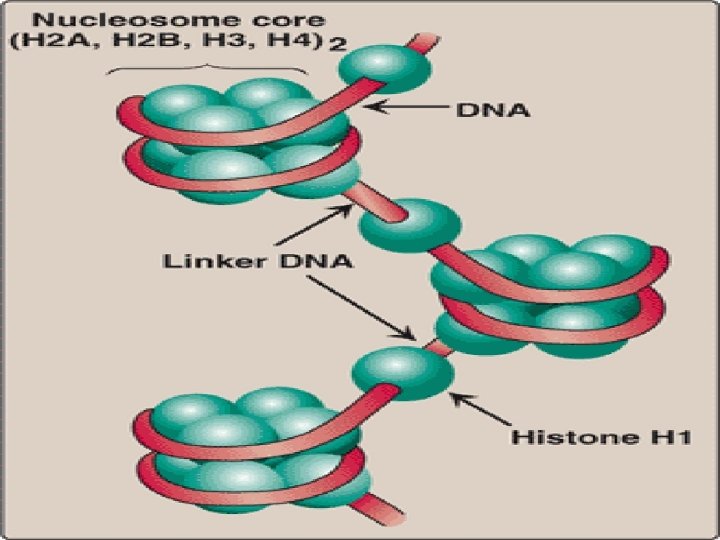
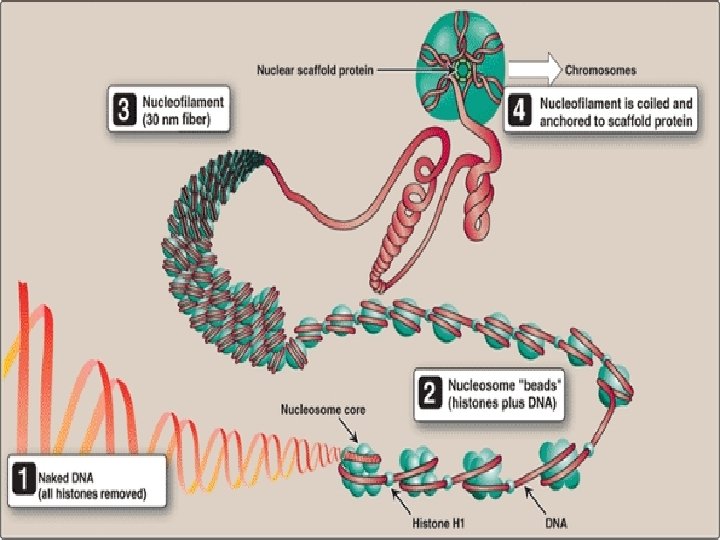
Organization of Eukaryotic DNA • Eukaryotic DNA is associated with tightly bound basic proteins, called histones. • These serve to order the DNA into basic structural units, called nucleosomes, that resemble beads on a string



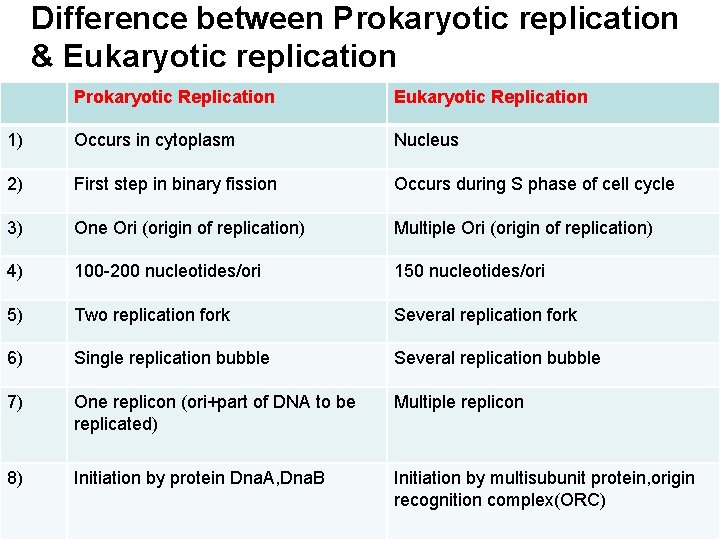
Difference between Prokaryotic replication & Eukaryotic replication Prokaryotic Replication Eukaryotic Replication 1) Occurs in cytoplasm Nucleus 2) First step in binary fission Occurs during S phase of cell cycle 3) One Ori (origin of replication) Multiple Ori (origin of replication) 4) 100 -200 nucleotides/ori 150 nucleotides/ori 5) Two replication fork Several replication fork 6) Single replication bubble Several replication bubble 7) One replicon (ori+part of DNA to be replicated) Multiple replicon 8) Initiation by protein Dna. A, Dna. B Initiation by multisubunit protein, origin recognition complex(ORC)

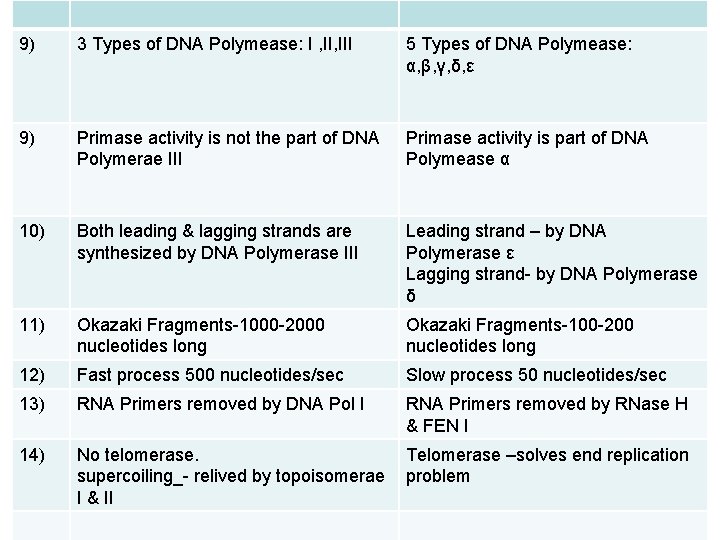
9) 3 Types of DNA Polymease: I , III 5 Types of DNA Polymease: α, β, γ, δ, ε 9) Primase activity is not the part of DNA Polymerae III Primase activity is part of DNA Polymease α 10) Both leading & lagging strands are synthesized by DNA Polymerase III Leading strand – by DNA Polymerase ε Lagging strand- by DNA Polymerase δ 11) Okazaki Fragments-1000 -2000 nucleotides long Okazaki Fragments-100 -200 nucleotides long 12) Fast process 500 nucleotides/sec Slow process 50 nucleotides/sec 13) RNA Primers removed by DNA Pol I RNA Primers removed by RNase H & FEN I 14) No telomerase. supercoiling_- relived by topoisomerae I & II Telomerase –solves end replication problem


DNA damage The damaging agents can be either chemicals, radiation, ultraviolet light ↓ Damages the DNA ↓ If damage is not repaired, permanent change is introduced ↓ It can lead to loss of control over proliferation of mutated cell, leading to cancer

DNA Repair Cells can repair these DNA damage ↓ repair system involves recognition of damage on the DNA ↓ removal/excision of the damage ↓ replacement /filling the gap left by excision , using sister strand as a template for DNA synthesis and ligation

Types of DNA Repair 1) 2) 3) 4) Mismatch repair Nucleotide excision repair Base excision repair Repair of double strand breaks

Mismatch repair


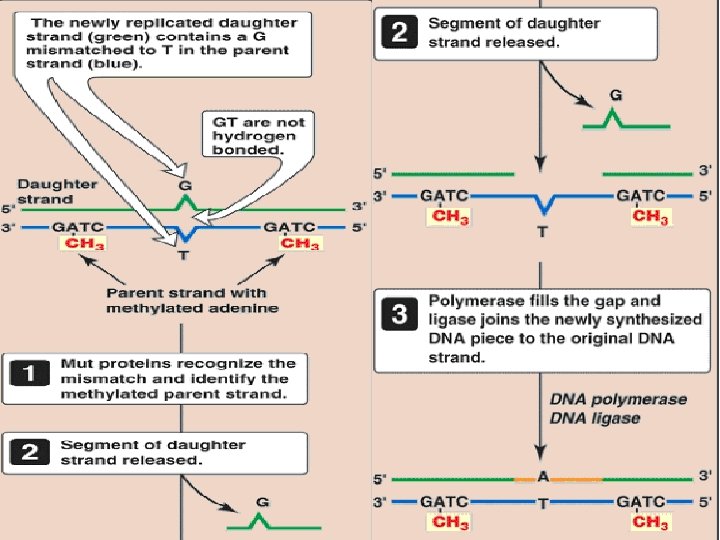
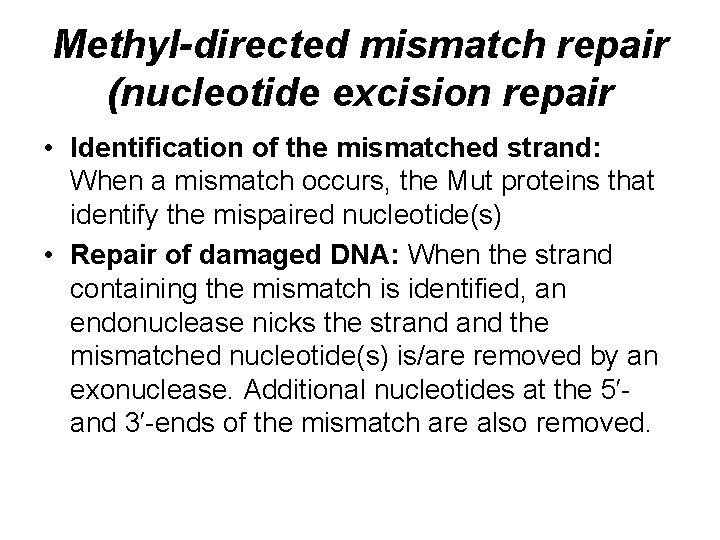
Methyl-directed mismatch repair (nucleotide excision repair • Identification of the mismatched strand: When a mismatch occurs, the Mut proteins that identify the mispaired nucleotide(s) • Repair of damaged DNA: When the strand containing the mismatch is identified, an endonuclease nicks the strand the mismatched nucleotide(s) is/are removed by an exonuclease. Additional nucleotides at the 5′and 3′-ends of the mismatch are also removed.

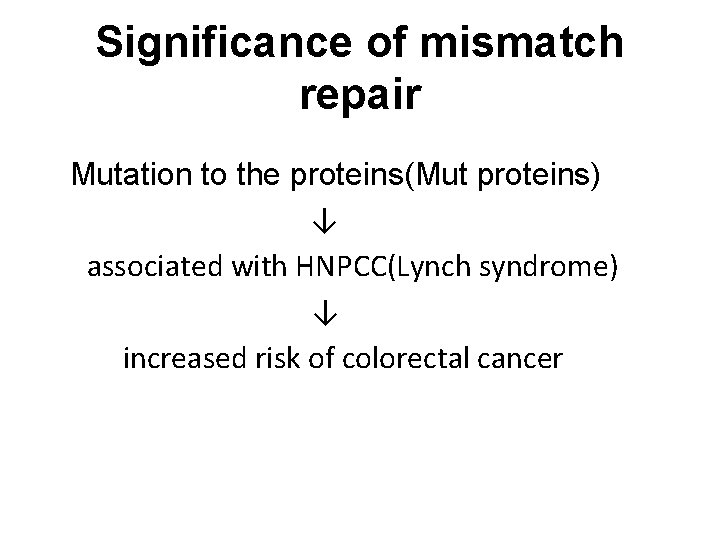
Significance of mismatch repair Mutation to the proteins(Mut proteins) ↓ associated with HNPCC(Lynch syndrome) ↓ increased risk of colorectal cancer

Repair of damage caused by ultraviolet (UV) light


Repair of damage caused by ultraviolet (UV) light • Exposure of a cell to UV light can result in the covalent joining of two adjacent pyrimidines (usually thymines), producing a dimer • First, a UV-specific endonuclease (called uvr. ABC excinuclease) recognizes the dimer, and cleaves the damaged strand on both the 5′-side and 3′-side of the dimer

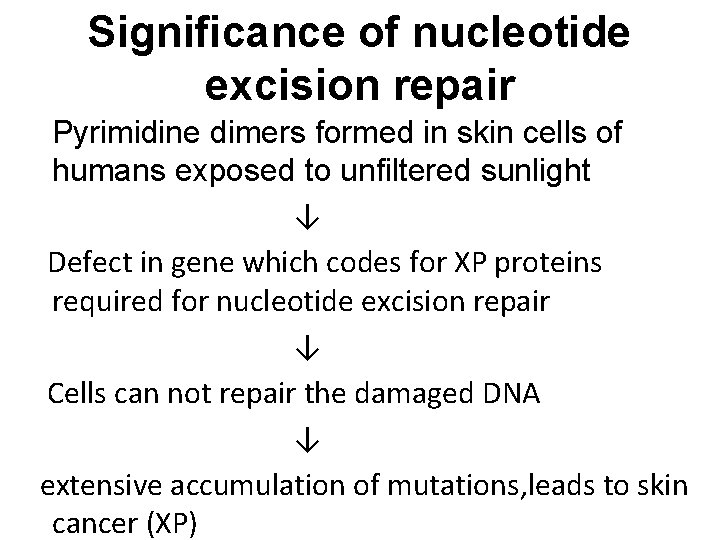
Significance of nucleotide excision repair Pyrimidine dimers formed in skin cells of humans exposed to unfiltered sunlight ↓ Defect in gene which codes for XP proteins required for nucleotide excision repair ↓ Cells can not repair the damaged DNA ↓ extensive accumulation of mutations, leads to skin cancer (XP)

Patient with xeroderma pigmentosum

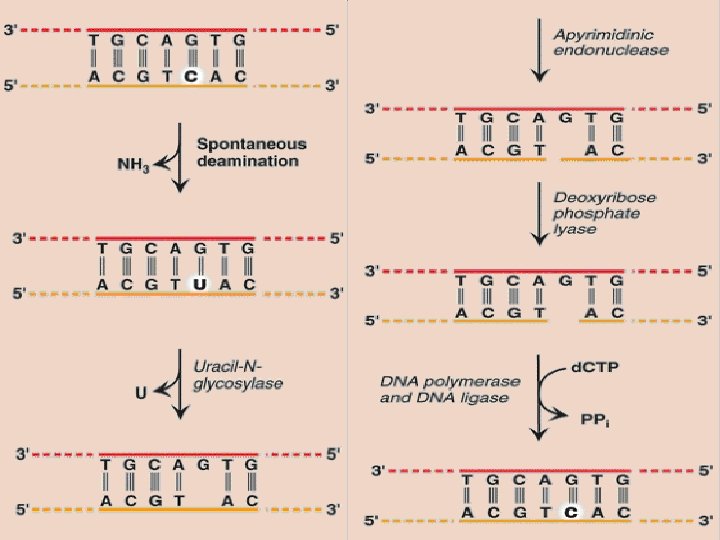
Repair of base alteration(base excision repair) • Bases of DNA can be altered spontaneously, as is the case with cytosine, which slowly undergoes deamination (the loss of its amino group) to form uracil • Bases can also be lost by action of deaminating/alkylating agents

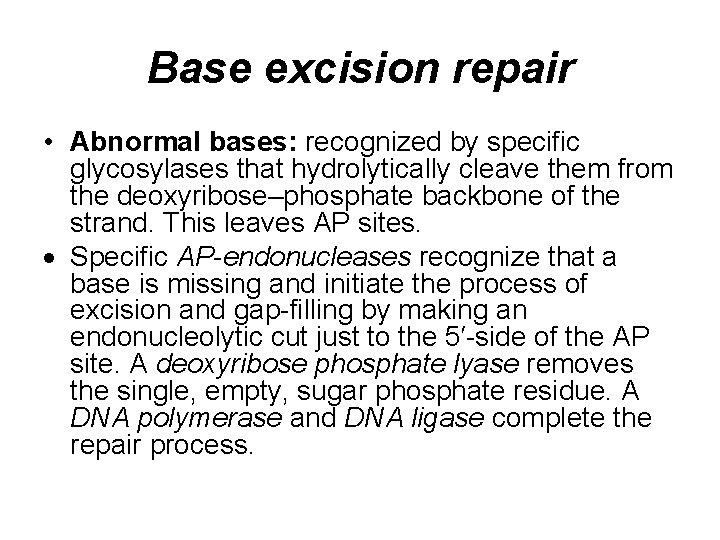
Base excision repair • Abnormal bases: recognized by specific glycosylases that hydrolytically cleave them from the deoxyribose–phosphate backbone of the strand. This leaves AP sites. Specific AP-endonucleases recognize that a base is missing and initiate the process of excision and gap-filling by making an endonucleolytic cut just to the 5′-side of the AP site. A deoxyribose phosphate lyase removes the single, empty, sugar phosphate residue. A DNA polymerase and DNA ligase complete the repair process.


Repair of double-strand breaks (ionizing radiation/oxidative free radicals ) 1) Non-homologous end-joining repair : ü group of proteins mediate recognition, processing & ligation of ends of two DNA fragments ü Defects in this repair system are associated with a predisposition to cancer and immunodeficiency syndromes (DNA is lost during repair)

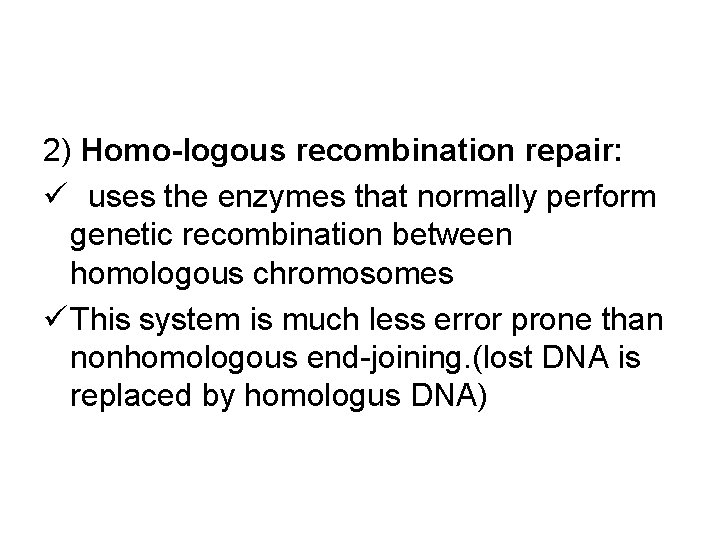
2) Homo-logous recombination repair: ü uses the enzymes that normally perform genetic recombination between homologous chromosomes ü This system is much less error prone than nonhomologous end-joining. (lost DNA is replaced by homologus DNA)

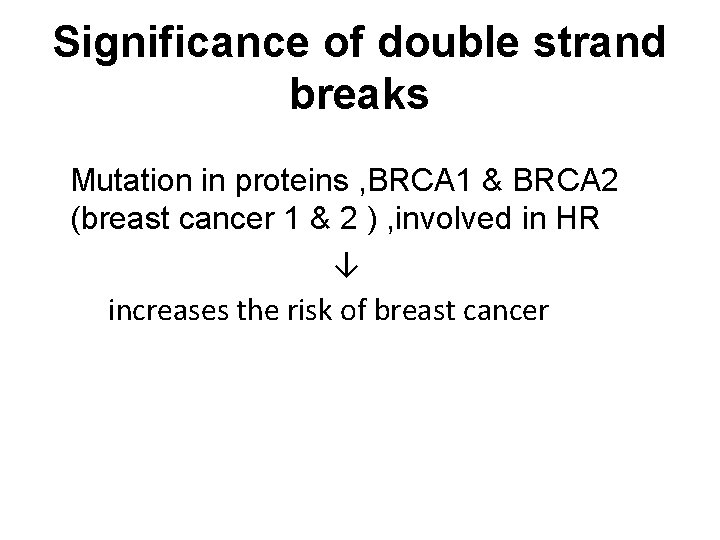
Significance of double strand breaks Mutation in proteins , BRCA 1 & BRCA 2 (breast cancer 1 & 2 ) , involved in HR ↓ increases the risk of breast cancer
