Fig 16 17 Overview Origin of replication Lagging





- Slides: 20

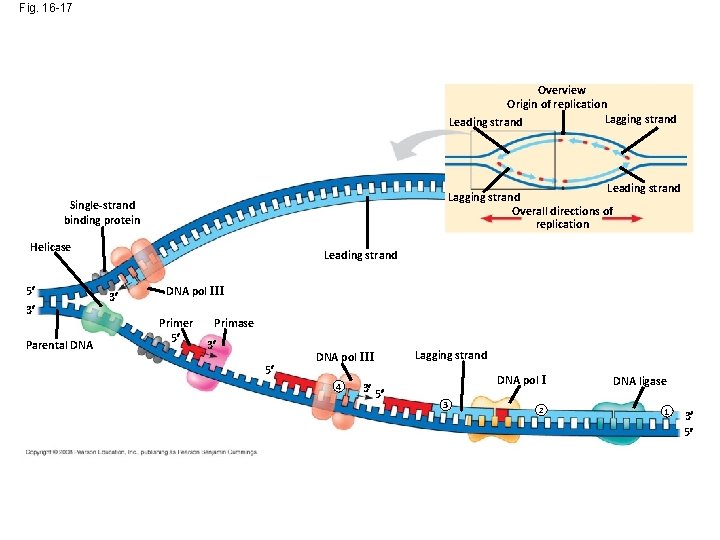
Fig. 16 -17 Overview Origin of replication Lagging strand Leading strand Lagging strand Overall directions of replication Single-strand binding protein Helicase 5 3 Parental DNA Leading strand 3 DNA pol III Primer 5 Primase 3 5 DNA pol III 4 3 5 Lagging strand DNA pol I 3 2 DNA ligase 1 3 5

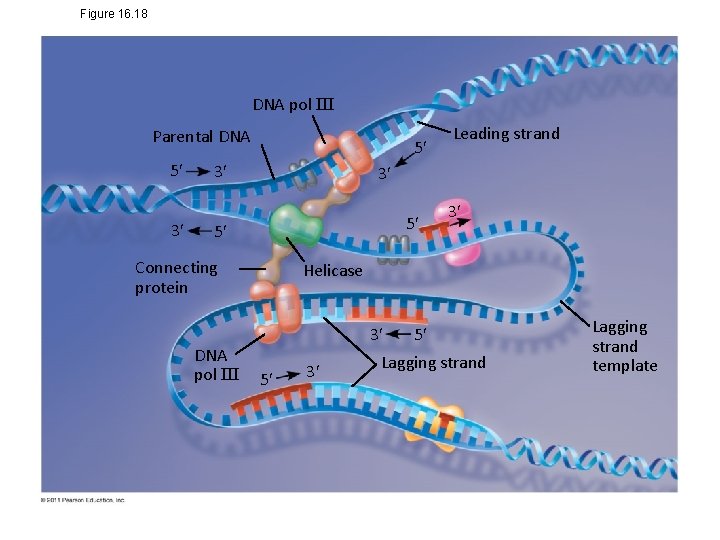
Figure 16. 18 DNA pol III Parental DNA 5 3 5 3 3 5 5 Connecting protein DNA pol III Leading strand 3 Helicase 3 5 3 5 Lagging strand template

Cool animation of DNA replication (and other stuff) http: //www. ted. com/talks/drew_berry_animations_of_unseeable_biology. html

How do we know all this? -That the lagging strand is made in fragments -That DNA ligase joins these fragments together

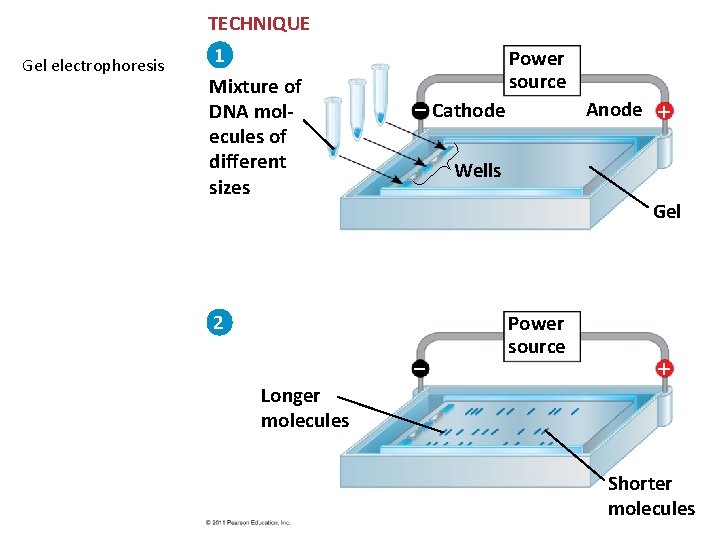
TECHNIQUE Gel electrophoresis 1 Mixture of DNA molecules of different sizes Power source Cathode Anode Wells Gel 2 Power source Longer molecules Shorter molecules

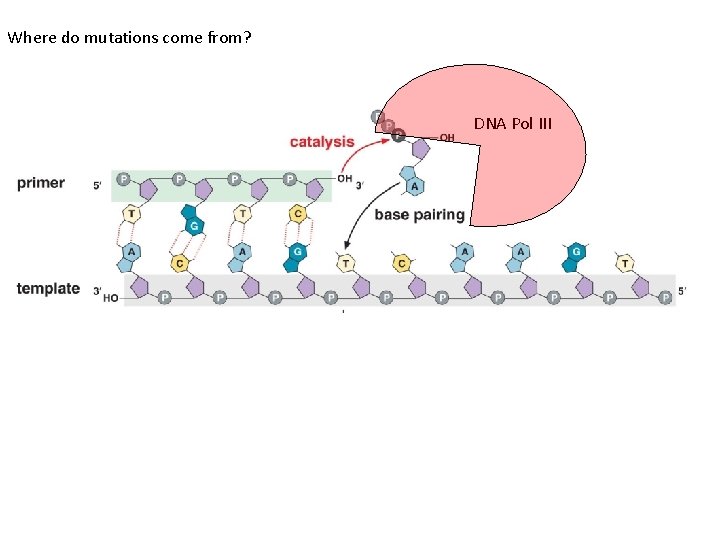
Where do mutations come from? DNA Pol III

How often do mutations occur? ~1 “error” for every 1010 bases replicated E. Coli genome: 4. 6 x 10^6 b. p. H. Sapiens genome (diploid): 6 x 10^9 b. p. DNA pol III adds the wrong base every 105 bases Extra fidelity comes from: 1. “Proofreading” by DNA pol III (and pol I) 2. Mismatch repair pathway

How often do mutations occur? ~1 “error” for every 1010 bases replicated E. Coli genome: 4. 6 x 10^6 b. p. H. Sapiens genome (diploid): 6 x 10^9 b. p. DNA pol III adds the wrong base every 105 bases

How often do mutations occur? ~1 “error” for every 1010 bases replicated E. Coli genome: 4. 6 x 10^6 b. p. H. Sapiens genome (diploid): 6 x 10^9 b. p. DNA pol III adds the wrong base every 105 bases Extra fidelity comes from: 1. “Proofreading” by DNA pol III (and pol I) 2. Mismatch repair pathway Chemically damaged DNA can lead to much higher rates of mutation

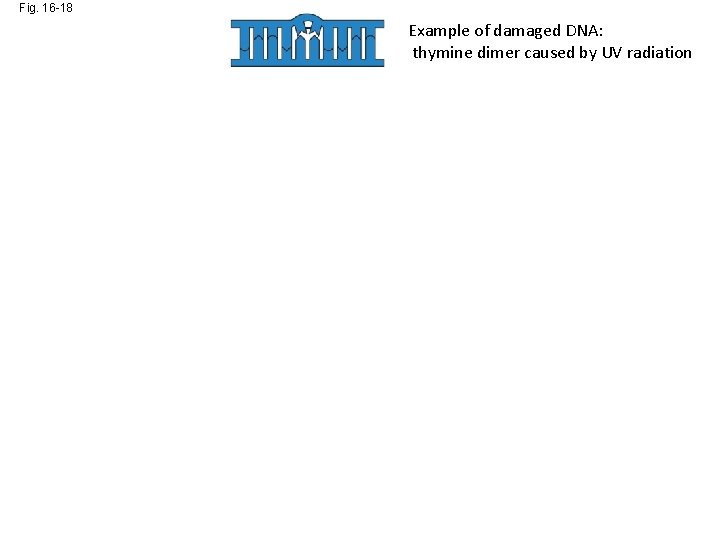
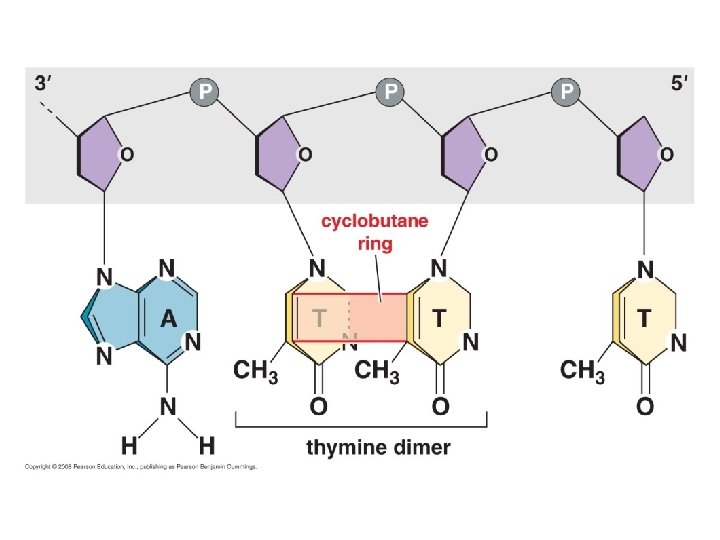
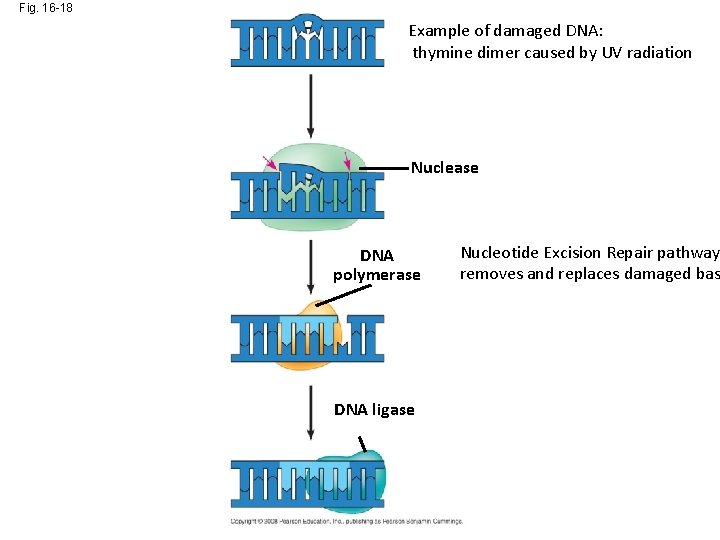
Fig. 16 -18 Example of damaged DNA: thymine dimer caused by UV radiation Nuclease DNA polymerase DNA ligase Nucleotide Excision Repair pathway has removed and replaced damaged bases


Fig. 16 -18 Example of damaged DNA: thymine dimer caused by UV radiation Nuclease DNA polymerase DNA ligase Nucleotide Excision Repair pathway removes and replaces damaged bas

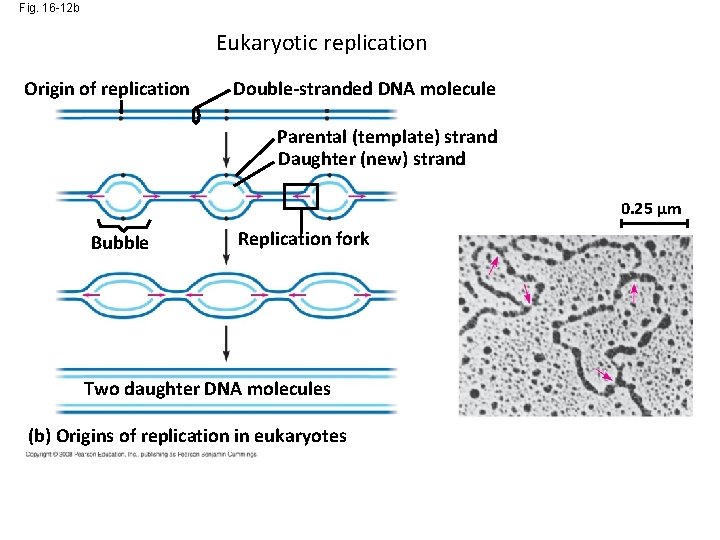
Fig. 16 -12 b Eukaryotic replication Origin of replication Double-stranded DNA molecule Parental (template) strand Daughter (new) strand 0. 25 µm Bubble Replication fork Two daughter DNA molecules (b) Origins of replication in eukaryotes

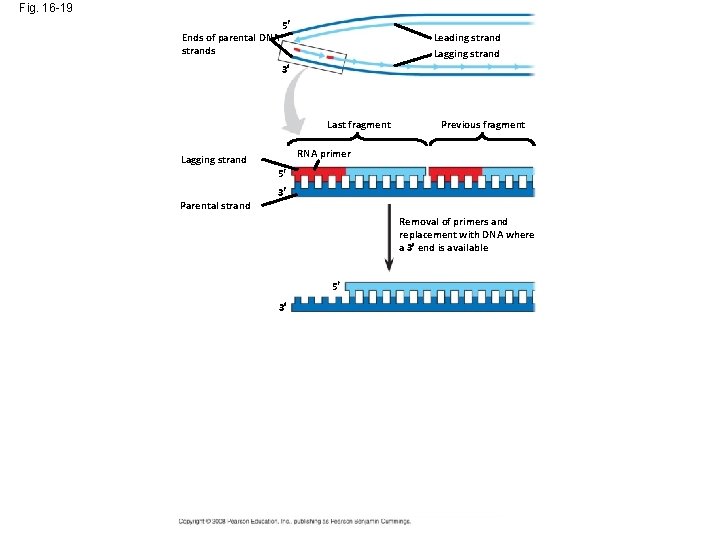
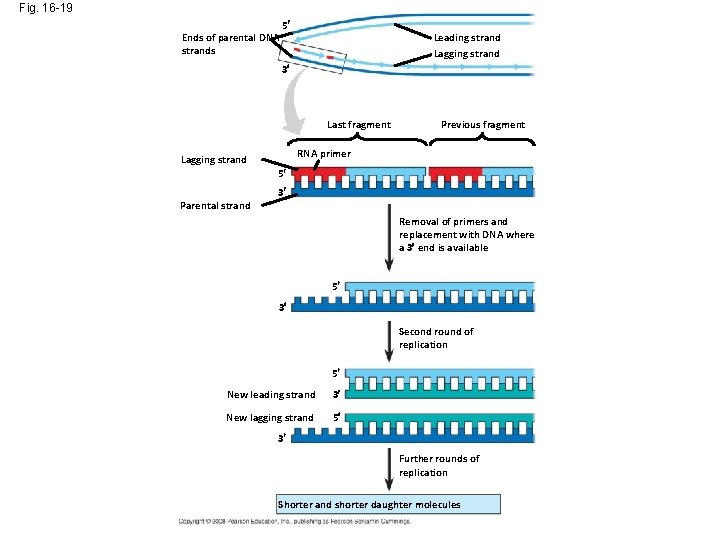
Fig. 16 -19 Ends of parental DNA strands 5 Leading strand Lagging strand 3 Last fragment Previous fragment RNA primer Lagging strand 5 Parental strand 3 Removal of primers and replacement with DNA where a 3 end is available 5 3 Second round of replication 5 New leading strand 3 New lagging strand 5 3 Further rounds of replication Shorter and shorter daughter molecules

Fig. 16 -19 Ends of parental DNA strands 5 Leading strand Lagging strand 3 Last fragment Previous fragment RNA primer Lagging strand 5 Parental strand 3 Removal of primers and replacement with DNA where a 3 end is available 5 3 Second round of replication 5 New leading strand 3 New lagging strand 5 3 Further rounds of replication Shorter and shorter daughter molecules

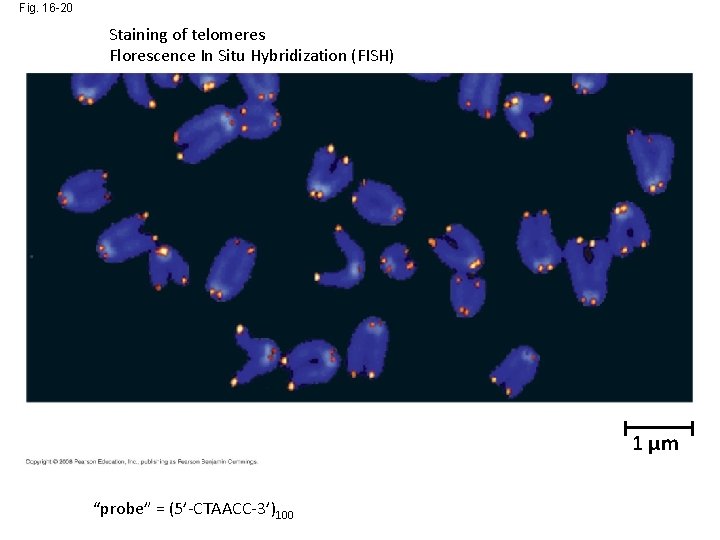
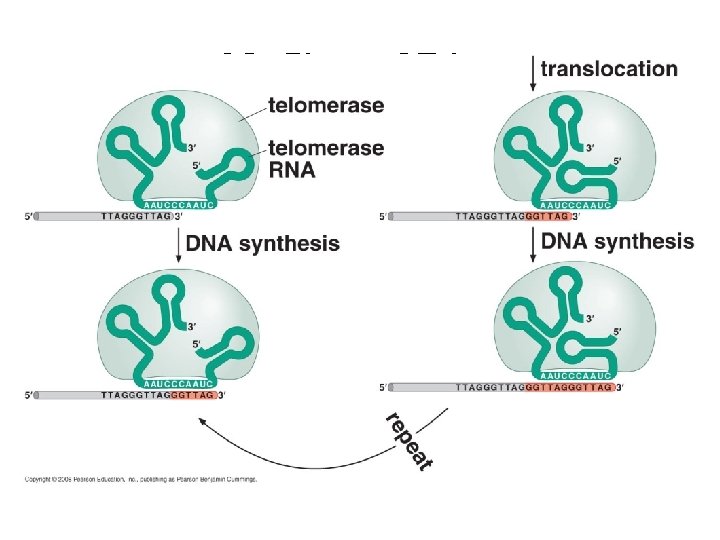
Fig. 16 -20 Staining of telomeres Florescence In Situ Hybridization (FISH) 1 µm “probe” = (5’-CTAACC-3’)100

08_Figure 37. jpg

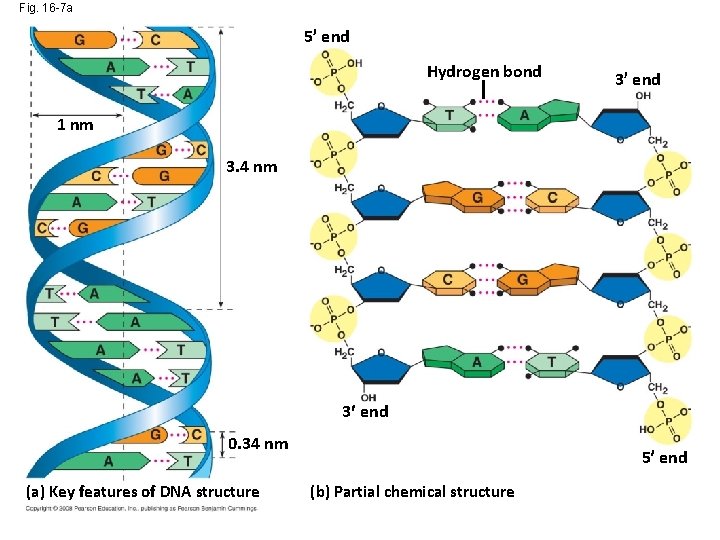
Fig. 16 -7 a 5 end Hydrogen bond 3 end 1 nm 3. 4 nm 3 end 0. 34 nm (a) Key features of DNA structure 5 end (b) Partial chemical structure

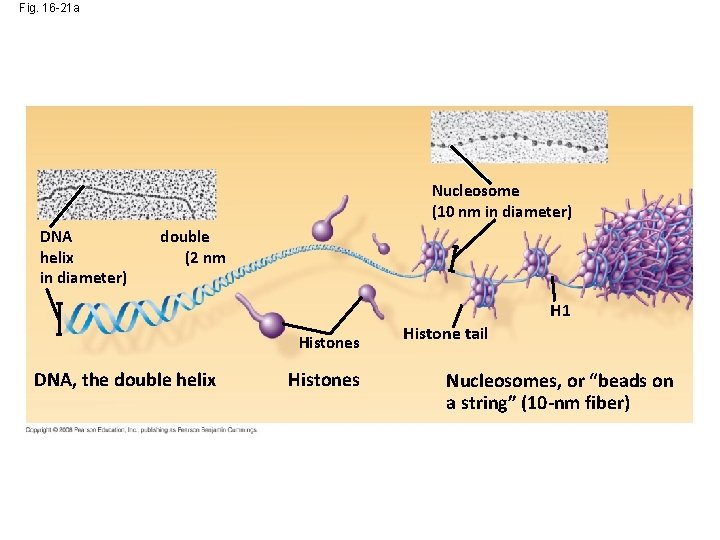
Fig. 16 -21 a Nucleosome (10 nm in diameter) DNA helix in diameter) double (2 nm H 1 Histones DNA, the double helix Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)

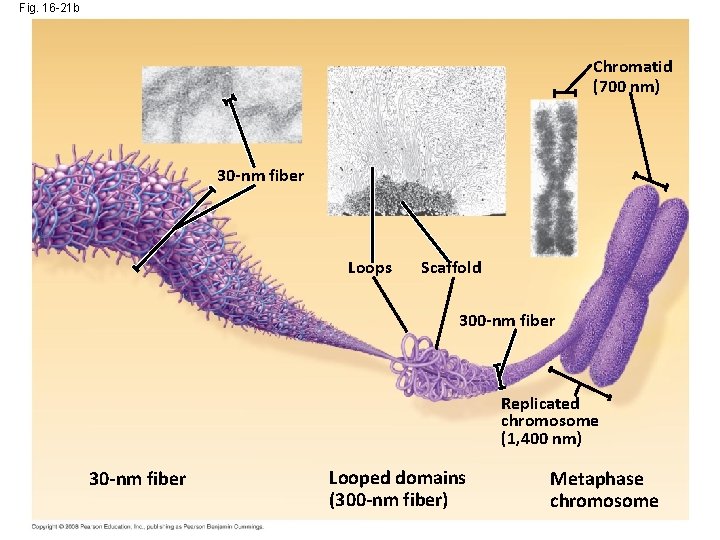
Fig. 16 -21 b Chromatid (700 nm) 30 -nm fiber Loops Scaffold 300 -nm fiber Replicated chromosome (1, 400 nm) 30 -nm fiber Looped domains (300 -nm fiber) Metaphase chromosome