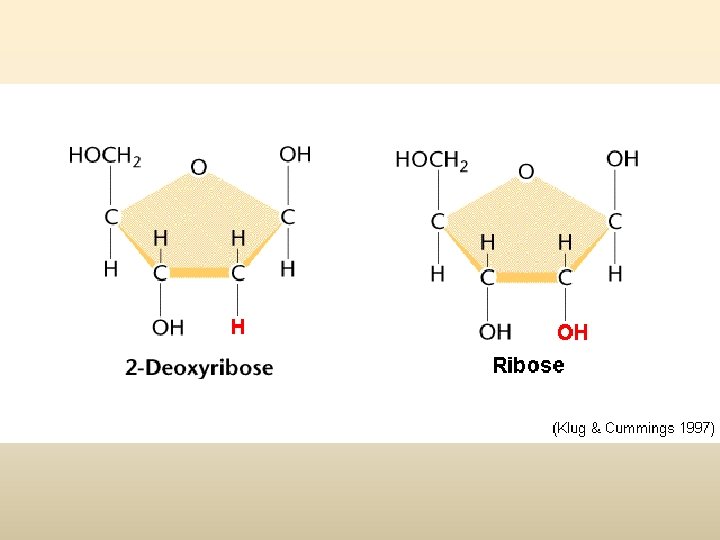
Nucleosome base 1 Pyrimidine base one ring base

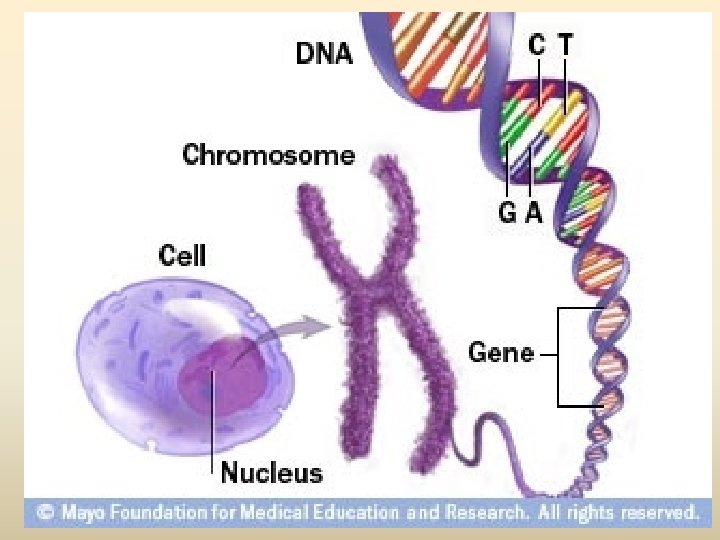

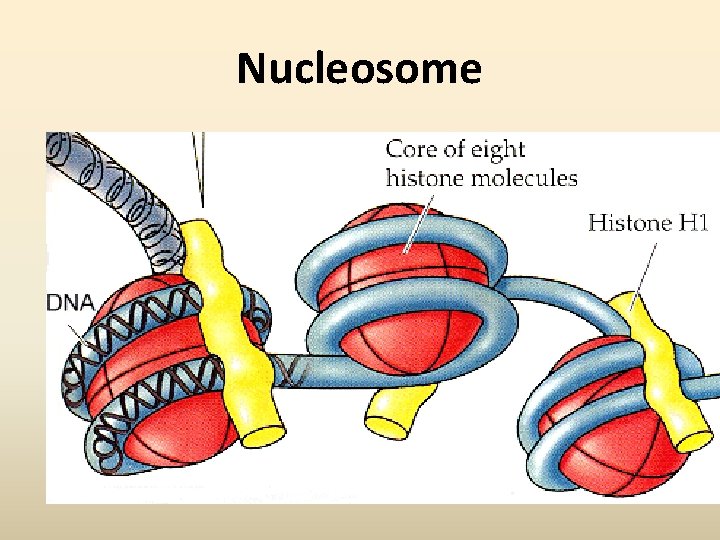
Nucleosome




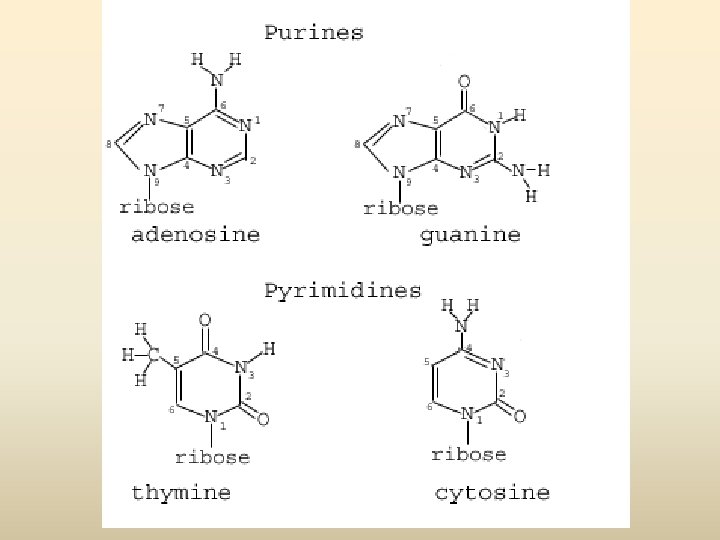
เบส (base) 1. Pyrimidine base (one ring base) มสองชนด (1) thymine (T) พบใน DNA เทานนสวนใน RNA จะพบ uracil (U) ซงมโครงสรางคลายกบ thymine (2) cytosine (C) 2. Purine two ring base มสองชนด คอ (1) Adinine (A) (2) Guanine (G)

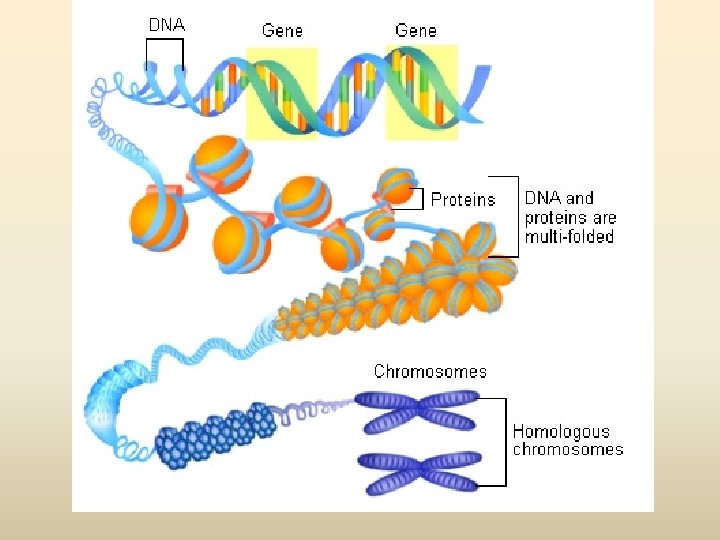
DNA (Deoxyribonucleic acid) • องคประกอบ - deoxyribose sugar - Base (1) pyrimidine (thymine ; T, cytosine; C) (2) purine (adinine: A, guanine; G) - phosphoric acid, phosphate





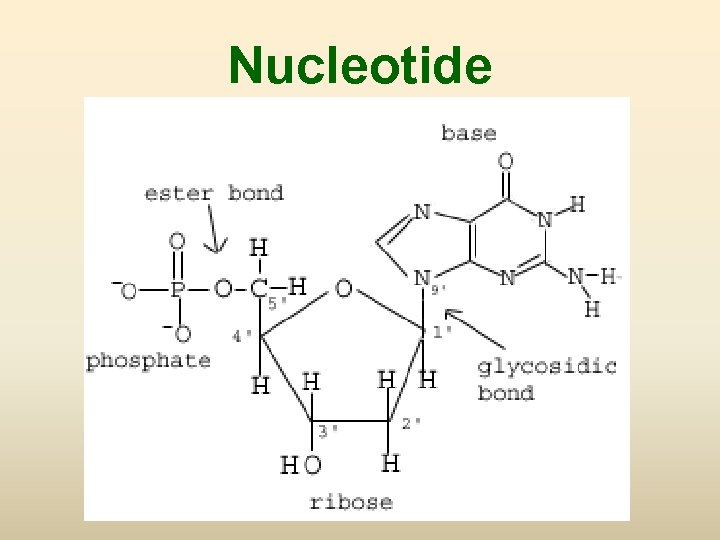
Nucleotide
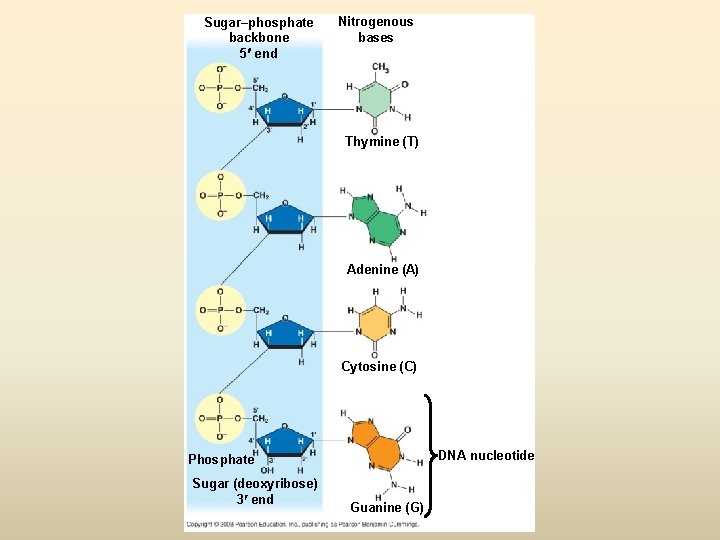
Sugar–phosphate backbone 5 end Nitrogenous bases Thymine (T) Adenine (A) Cytosine (C) DNA nucleotide Phosphate Sugar (deoxyribose) 3 end Guanine (G)
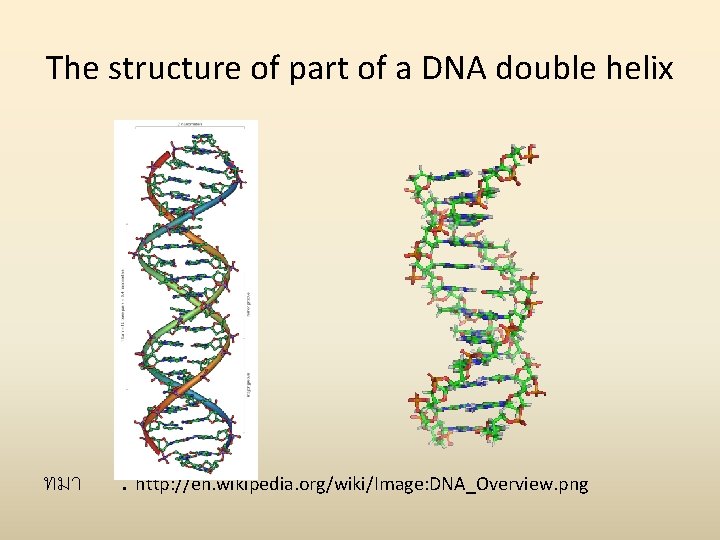
The structure of part of a DNA double helix ทมา : http: //en. wikipedia. org/wiki/Image: DNA_Overview. png
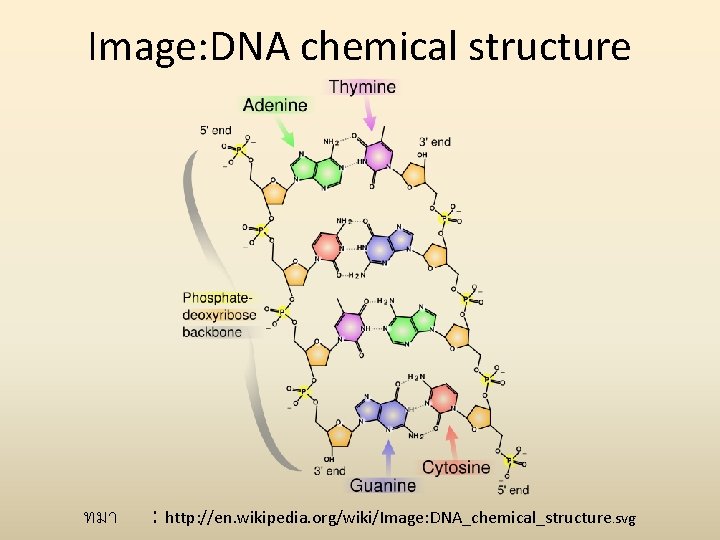
Image: DNA chemical structure ทมา : http: //en. wikipedia. org/wiki/Image: DNA_chemical_structure. svg
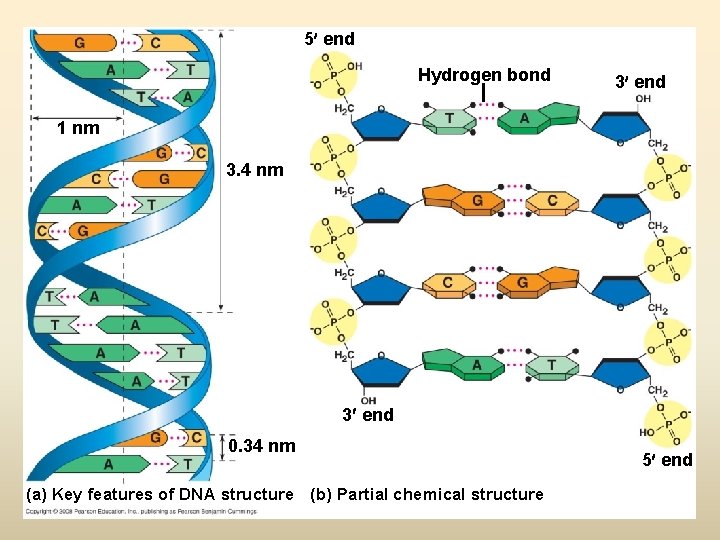
5 end Hydrogen bond 3 end 1 nm 3. 4 nm 3 end 0. 34 nm (a) Key features of DNA structure (b) Partial chemical structure 5 end
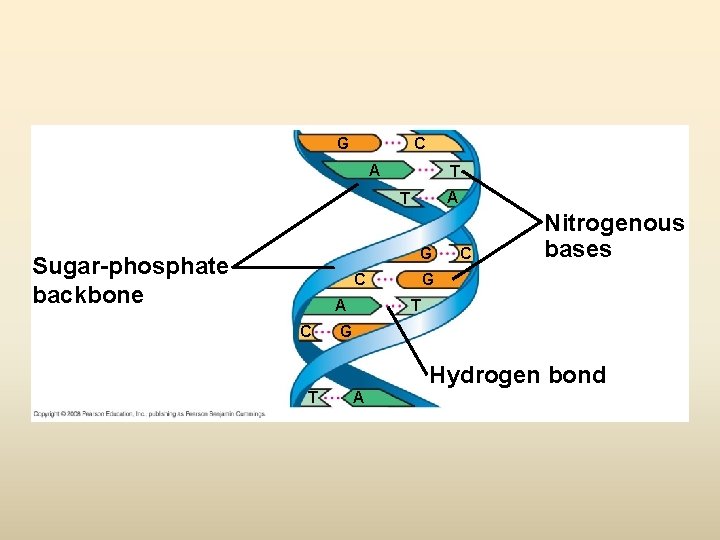
G C A T G Sugar-phosphate backbone C A C C Nitrogenous bases G T G Hydrogen bond T A
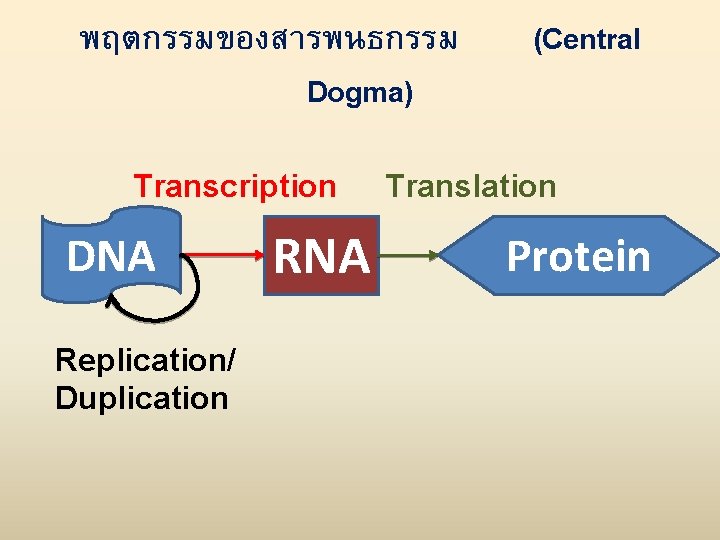
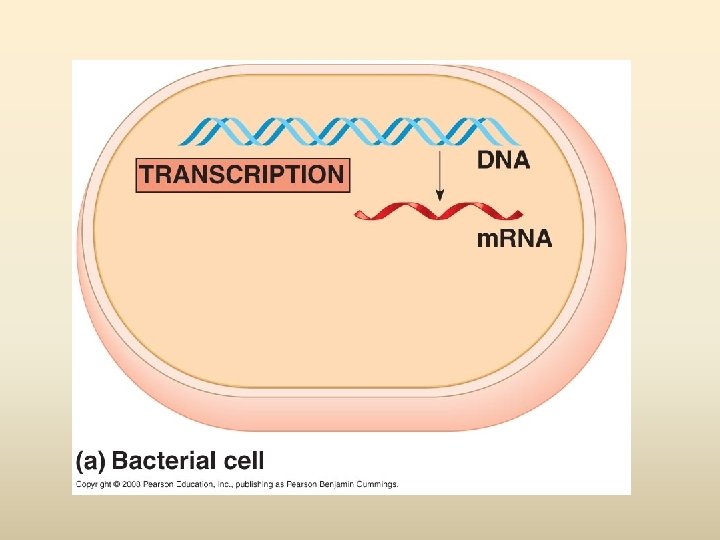
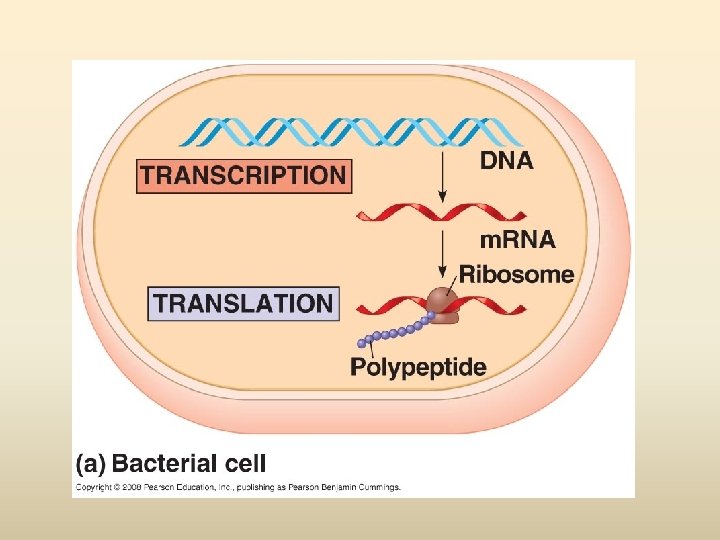
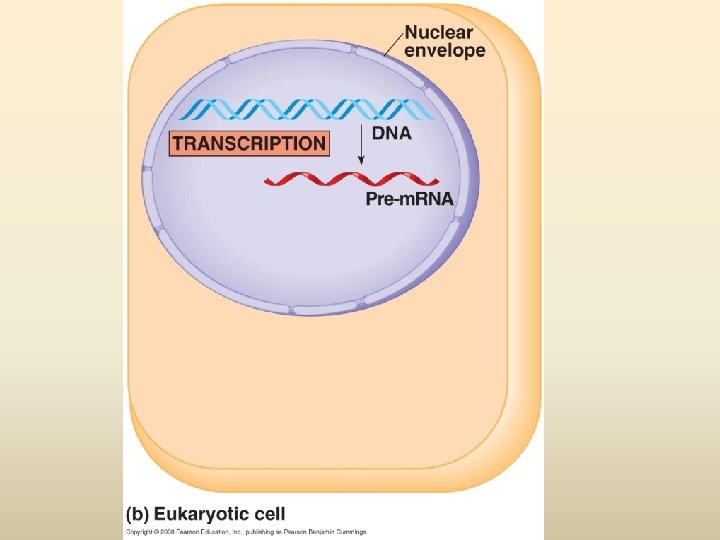
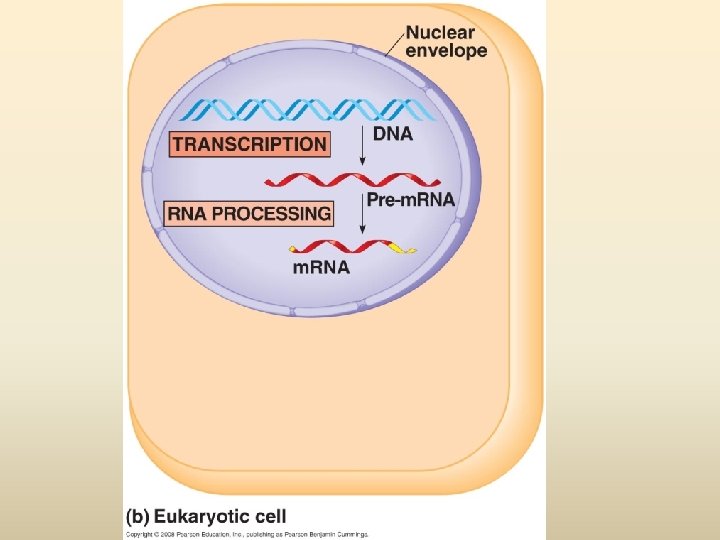
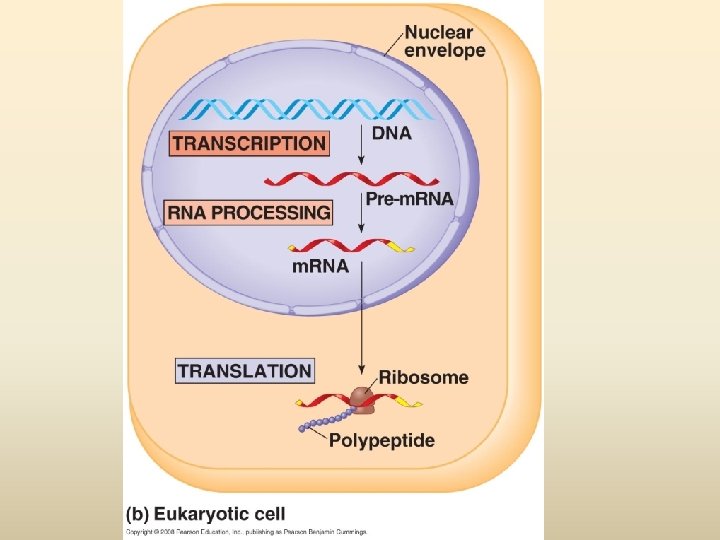
พฤตกรรมของสารพนธกรรม Dogma) Transcription DNA Replication/ Duplication RNA (Central Translation Protein

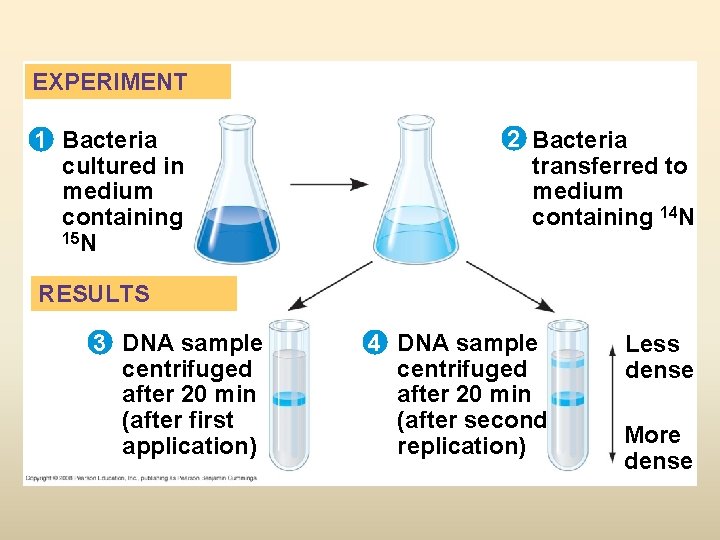
EXPERIMENT 1 Bacteria cultured in medium containing 15 N 2 Bacteria transferred to medium containing 14 N RESULTS 3 DNA sample centrifuged after 20 min (after first application) 4 DNA sample centrifuged after 20 min (after second replication) Less dense More dense
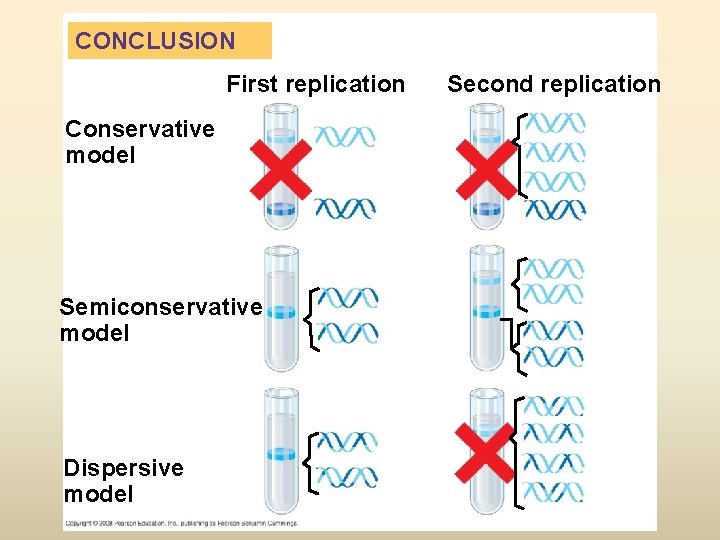
CONCLUSION First replication Conservative model Semiconservative model Dispersive model Second replication

A T A T C G C G T A T A T G C G C (a) Parent molecule (b) Separation of strands (c) “Daughter” DNA molecules, each consisting of one parental strand one new strand
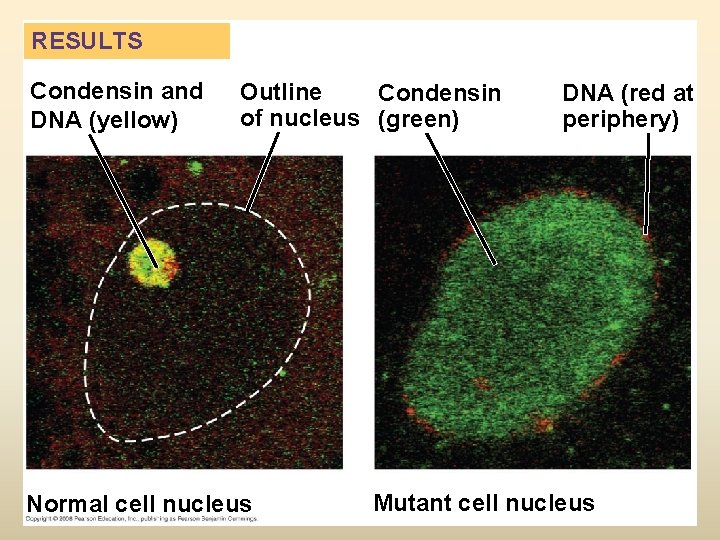
RESULTS Condensin and DNA (yellow) Outline Condensin of nucleus (green) Normal cell nucleus DNA (red at periphery) Mutant cell nucleus

Getting Started Replication begins at special sites called origins of replication, where the two DNA strands are separated, opening up a replication “bubble” A eukaryotic chromosome may have hundreds or even thousands of origins of replication Replication proceeds in both directions from each origin, until the entire molecule is copied Animation: Origins of Replication Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
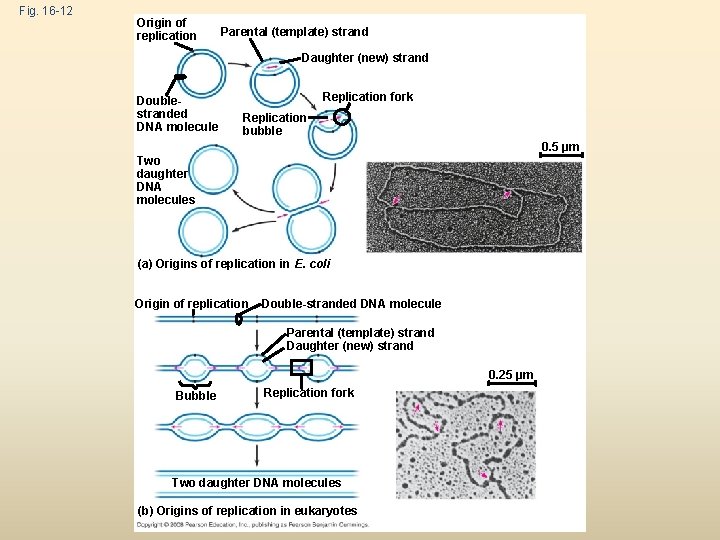
Fig. 16 -12 Origin of replication Parental (template) strand Daughter (new) strand Doublestranded DNA molecule Replication fork Replication bubble 0. 5 µm Two daughter DNA molecules (a) Origins of replication in E. coli Origin of replication Double-stranded DNA molecule Parental (template) strand Daughter (new) strand 0. 25 µm Bubble Replication fork Two daughter DNA molecules (b) Origins of replication in eukaryotes

At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating Helicases are enzymes that untwist the double helix at the replication forks Single-strand binding protein binds to and stabilizes single-stranded DNA until it can be used as a template Topoisomerase corrects “overwinding” ahead of replication forks by breaking, swiveling, and rejoining DNA strands Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
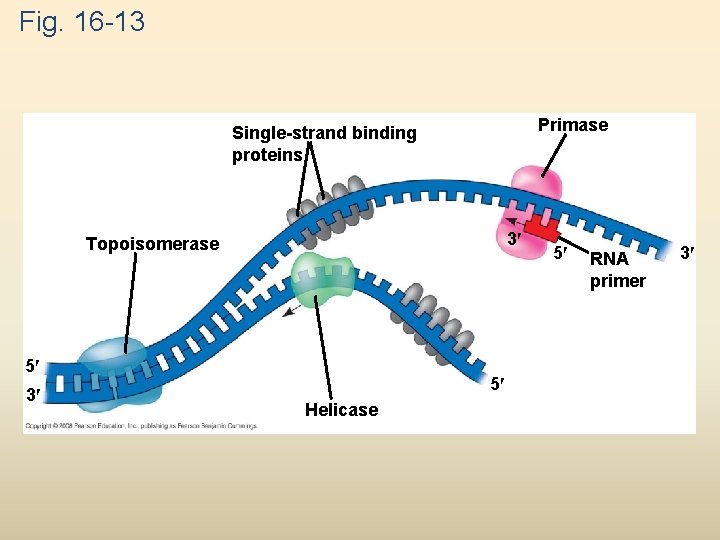
Fig. 16 -13 Primase Single-strand binding proteins 3 Topoisomerase 5 3 5 Helicase 5 RNA primer 3
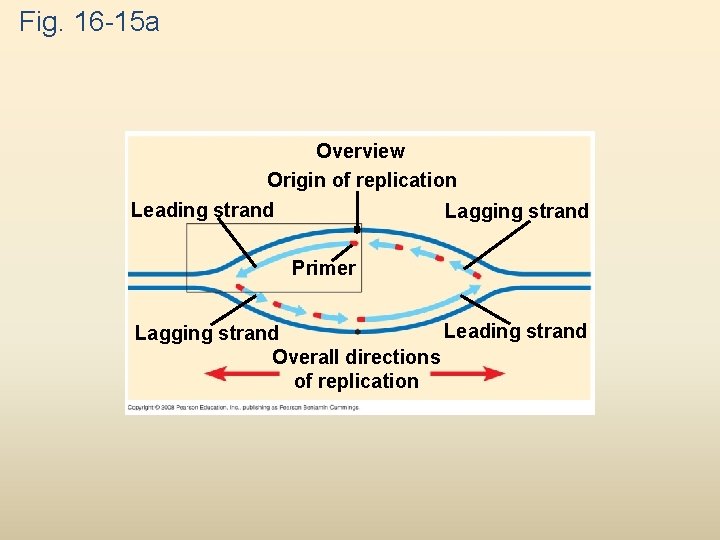
Fig. 16 -15 a Overview Origin of replication Leading strand Lagging strand Primer Leading strand Lagging strand Overall directions of replication
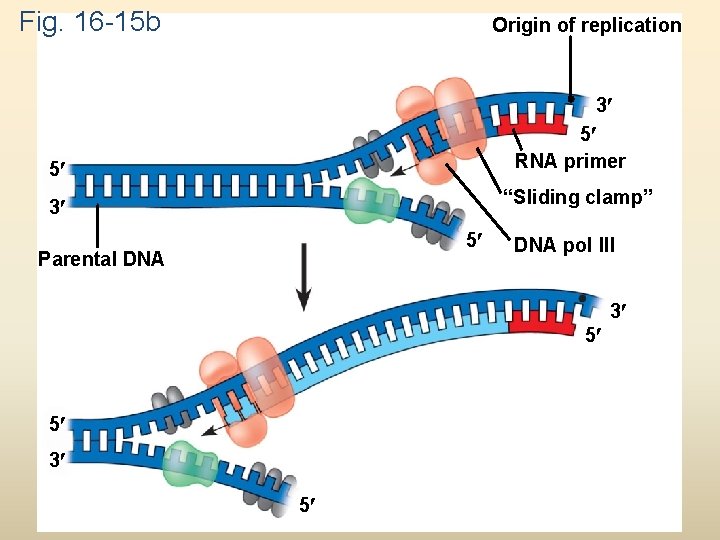
Fig. 16 -15 b Origin of replication 3 5 RNA primer 5 “Sliding clamp” 3 5 Parental DNA pol III 3 5 5 3 5

To elongate the other new strand, called the lagging strand, DNA polymerase must work in the direction away from the replication fork The lagging strand is synthesized as a series of segments called Okazaki fragments, which are joined together by DNA ligase Animation: Lagging Strand Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
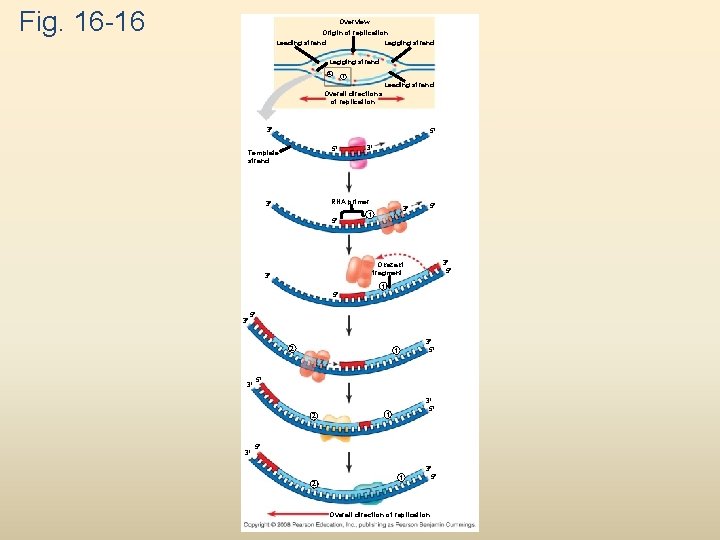
Fig. 16 -16 Overview Origin of replication Lagging strand Leading strand Lagging strand 2 1 Leading strand Overall directions of replication 3 5 5 Template strand 3 RNA primer 3 5 3 1 5 3 5 Okazaki fragment 3 1 5 3 5 2 3 3 5 1 5 2 1 3 5 Overall direction of replication
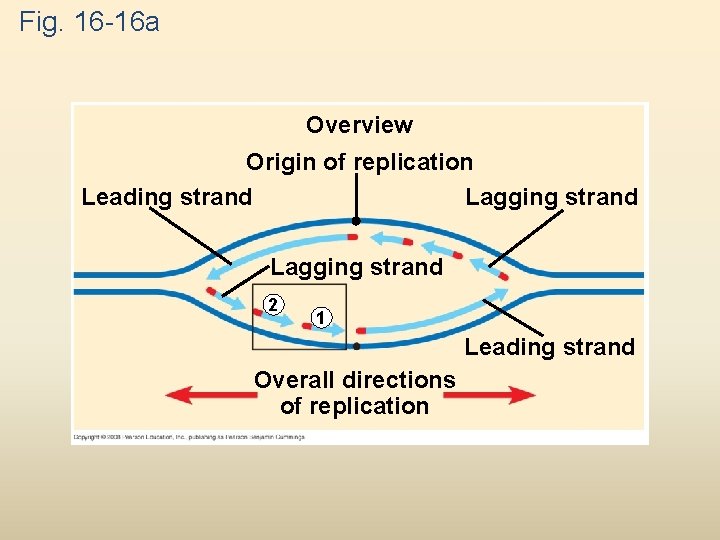
Fig. 16 -16 a Overview Origin of replication Leading strand Lagging strand 2 1 Leading strand Overall directions of replication
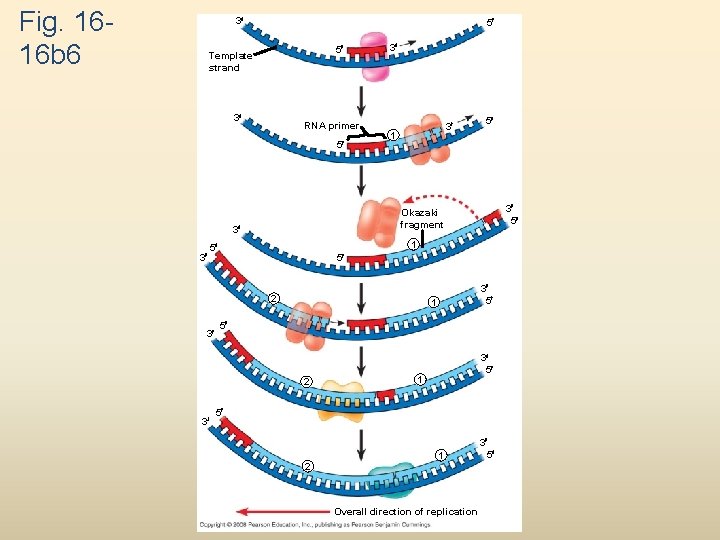
Fig. 1616 b 6 3 5 5 Template strand 3 RNA primer 5 3 1 5 2 3 5 1 5 2 3 3 5 1 5 3 5 Okazaki fragment 3 3 3 3 5 1 5 2 1 Overall direction of replication 3 5
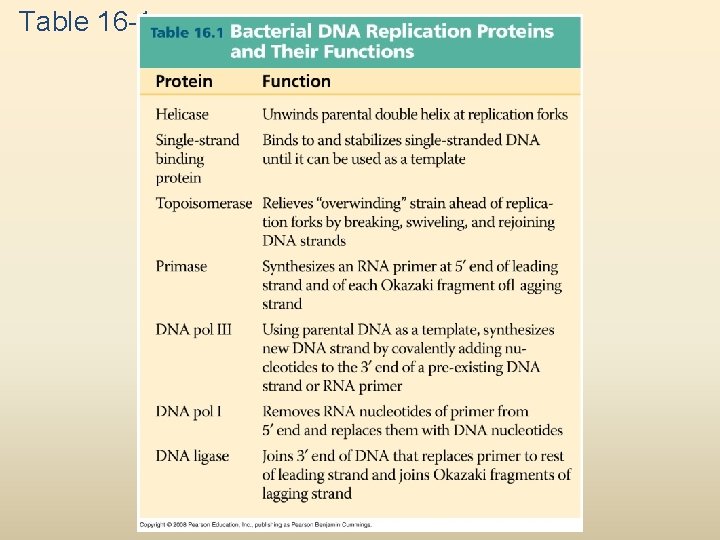
Table 16 -1
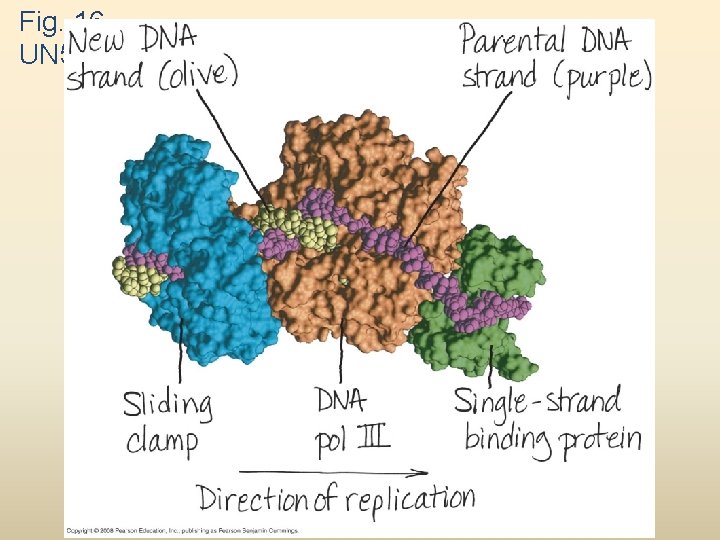
Fig. 16 UN 5
- Slides: 50