Annotation Traditional genome annotation BLAST Similarities Traditional genome

Annotation
Traditional genome annotation
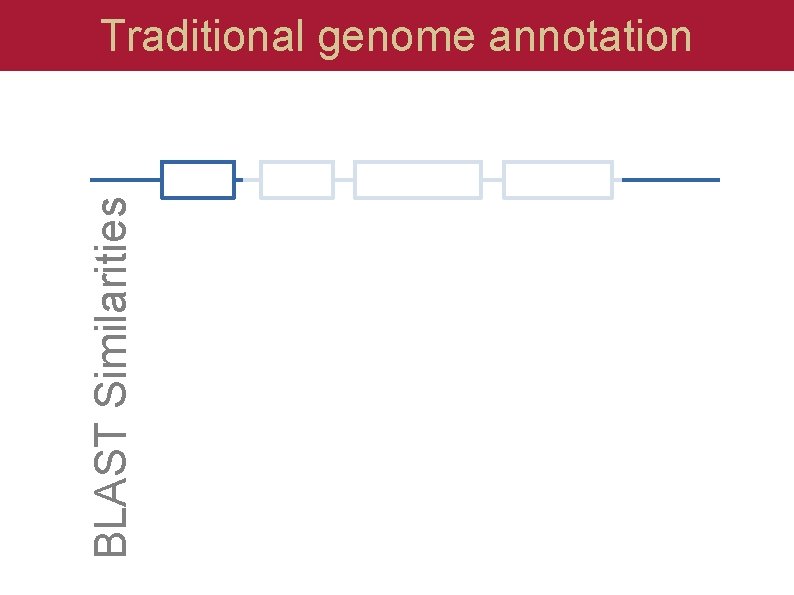
BLAST Similarities Traditional genome annotation

BLAST Similarities Traditional genome annotation
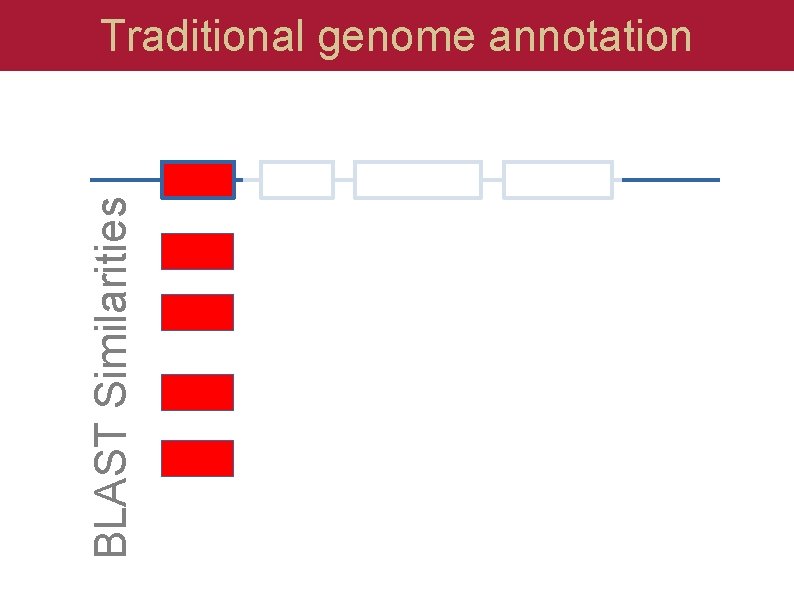
BLAST Similarities Traditional genome annotation
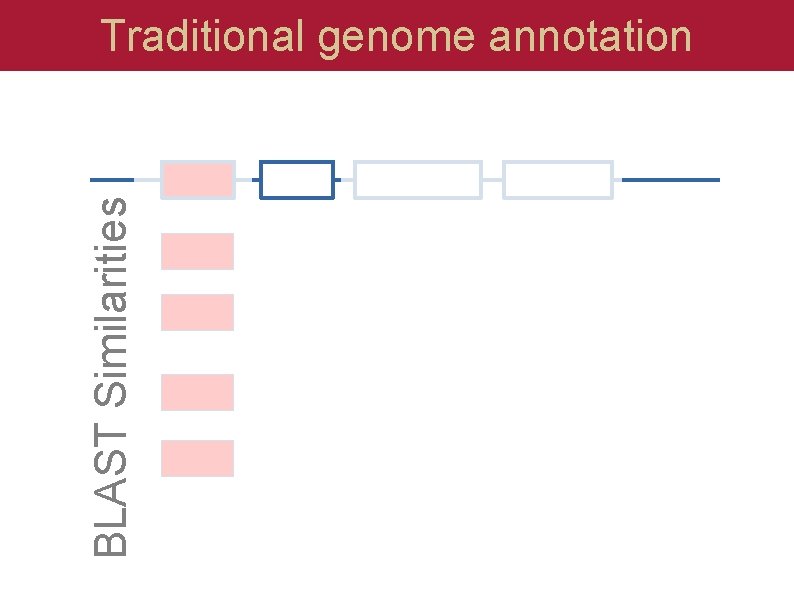
BLAST Similarities Traditional genome annotation
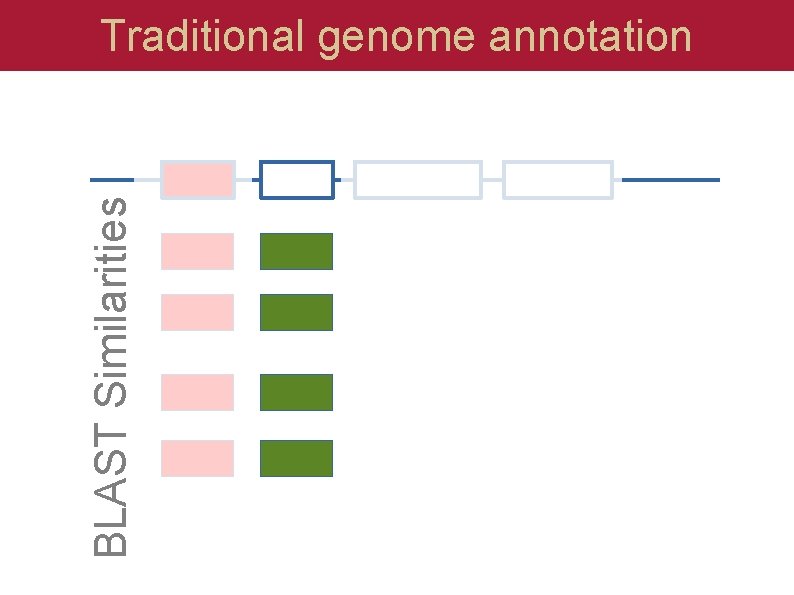
BLAST Similarities Traditional genome annotation
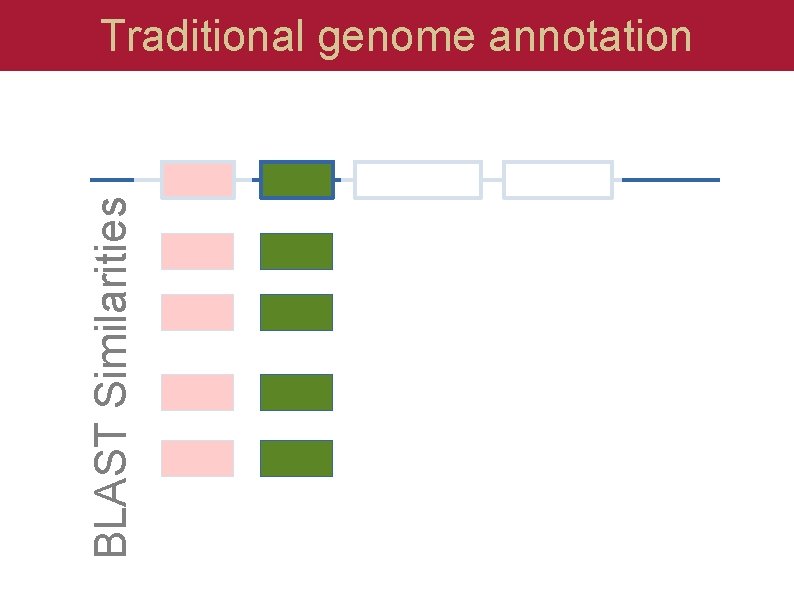
BLAST Similarities Traditional genome annotation
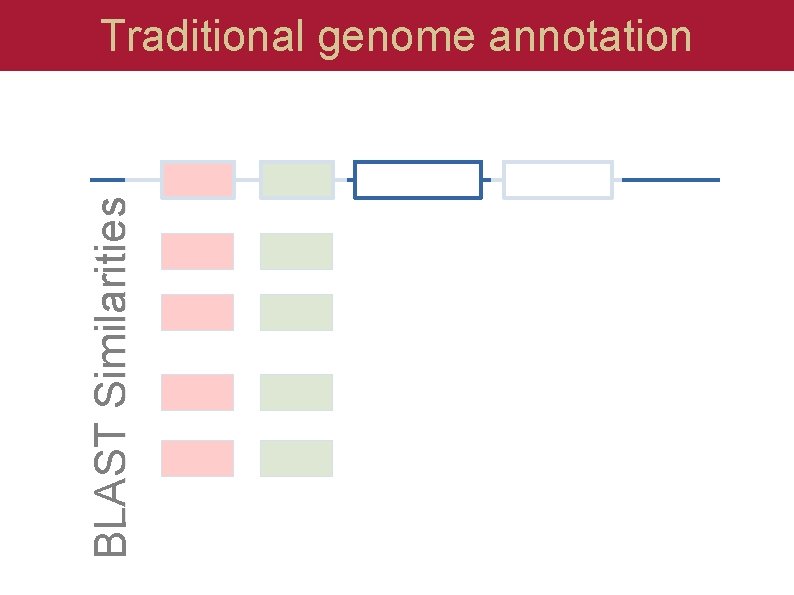
BLAST Similarities Traditional genome annotation
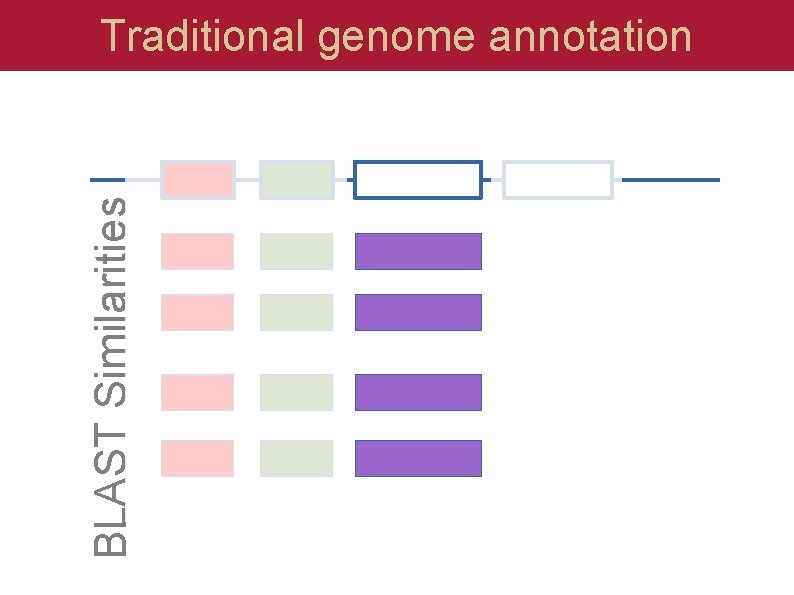
BLAST Similarities Traditional genome annotation
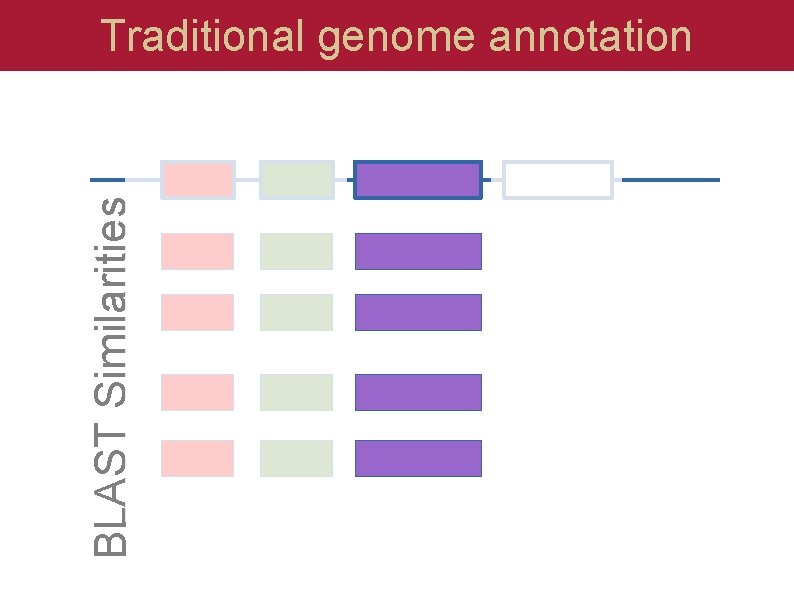
BLAST Similarities Traditional genome annotation
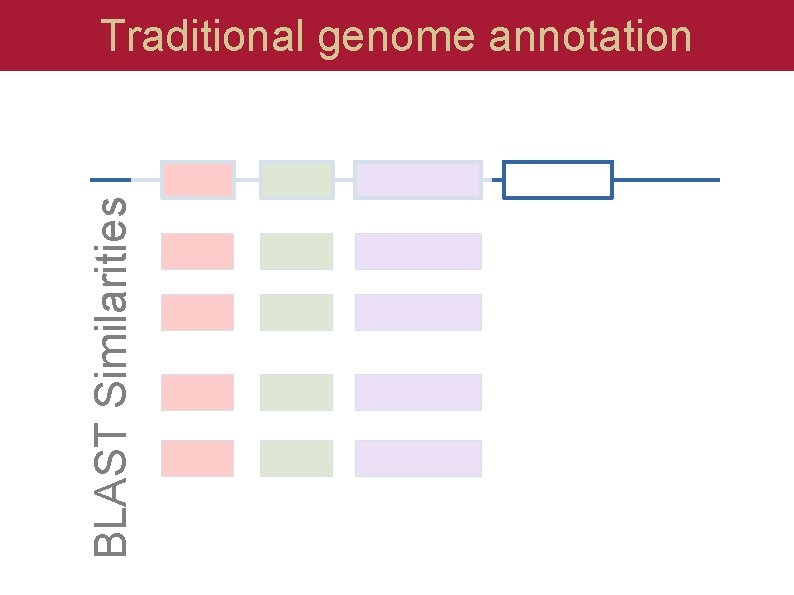
BLAST Similarities Traditional genome annotation
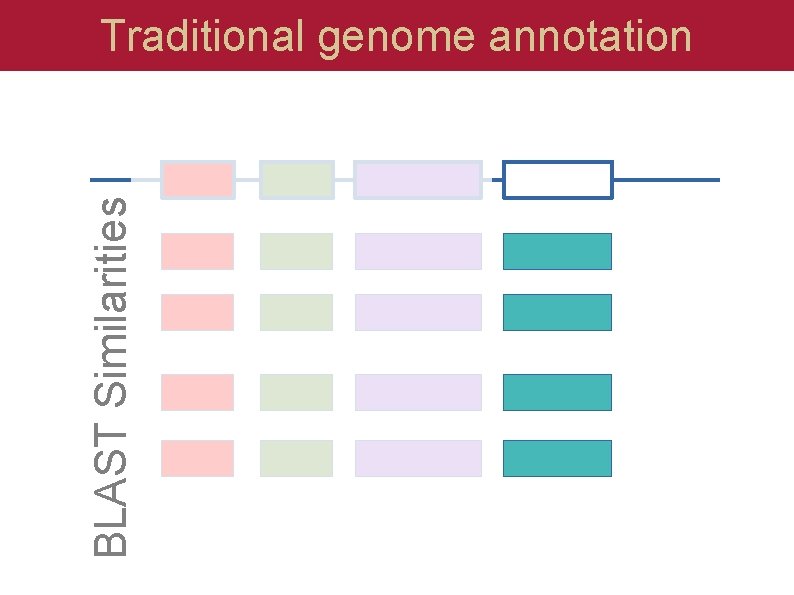
BLAST Similarities Traditional genome annotation
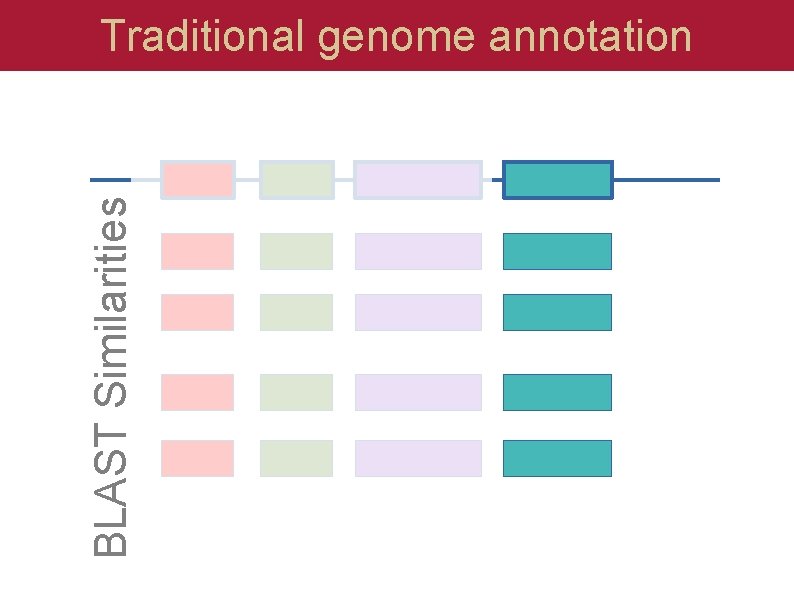
BLAST Similarities Traditional genome annotation
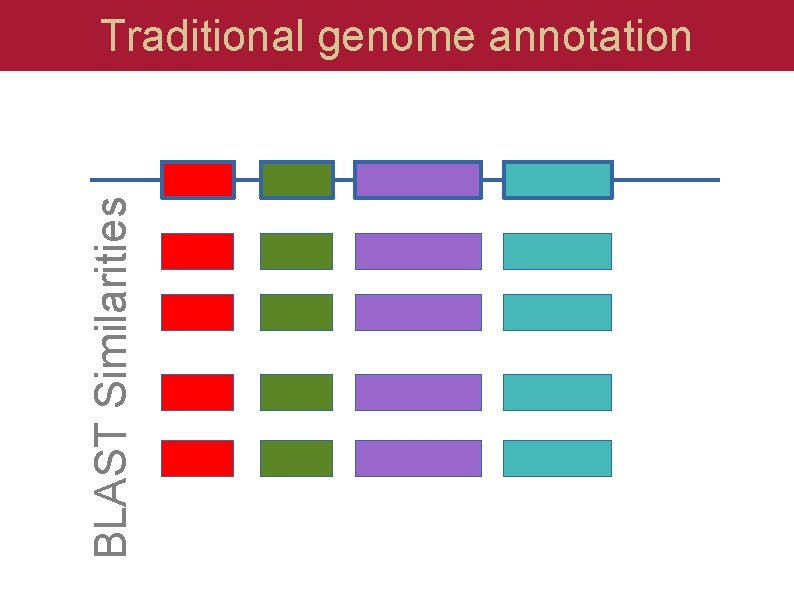
BLAST Similarities Traditional genome annotation

Protein Families
Protein Families
Protein Families
Protein Families

Gene Ontology A “hierarchy” of functions Does not need to be linear Directed Acyclic Graph Controlled Vocabulary Decides which words or phrases to use

GO Gene ontology A eukaryotic focus Drosophila Mus Saccharomyces Homo

GO Cellular component Molecular function The parts of a cell e. g. ligand binding Biological processes What things do
![GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate](http://slidetodoc.com/presentation_image_h/b5d203373f1b048759bd550a9b9437c3/image-23.jpg)
GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate kinase activity
![GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate](http://slidetodoc.com/presentation_image_h/b5d203373f1b048759bd550a9b9437c3/image-24.jpg)
GO Terms [GO ID, function] e. g: GO: 0004743 Ontology: molecular function Name: pyruvate kinase activity Mainly assigned by BLAST/HMMER/. . . etc

Directed Acyclic Graph Molecular function Catalytic activity Transferase activity, transferring phosphorous Kinase activity phosphotransferase activity, alcohol group as acceptor Pyruvate kinase activity

Problems Annotation by committee Eukaryotic focus Some efforts to counter that Owen White Arriane Toussaint Not very deep Strict controlled vocabulary

Alternatives
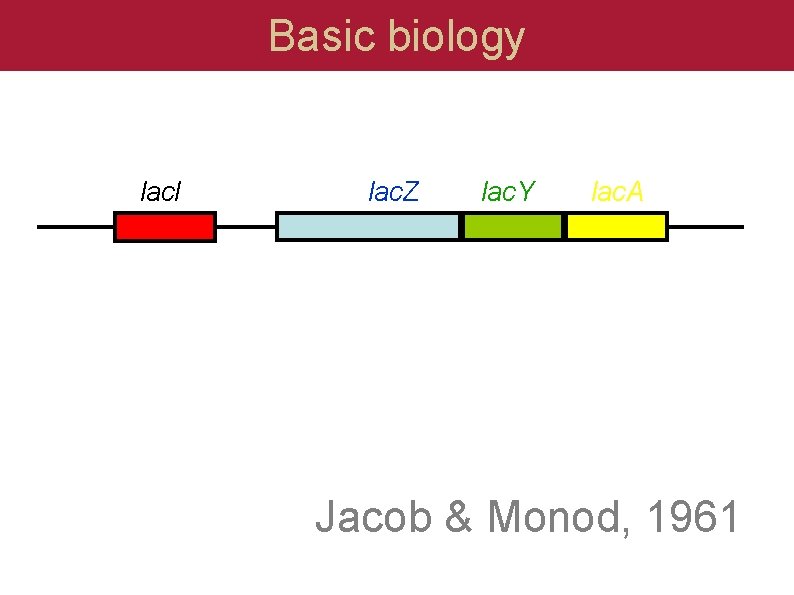
Basic biology lac. I lac. Z lac. Y lac. A Jacob & Monod, 1961

Basic biology lac. I lac. Z lac. Y lac. A
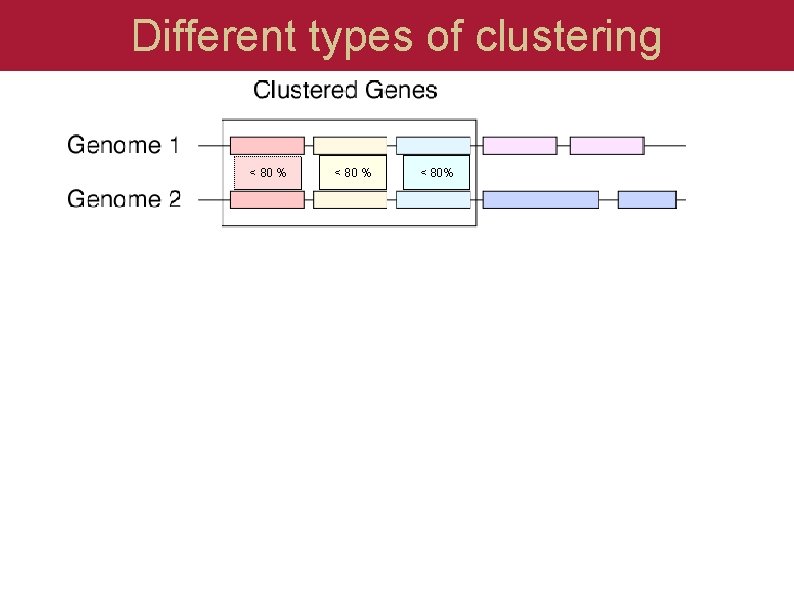
Different types of clustering < 80 % < 80%

Different types of clustering < 80 % < 80%
Purine metabolism

Different types of clustering < 80 % < 80%
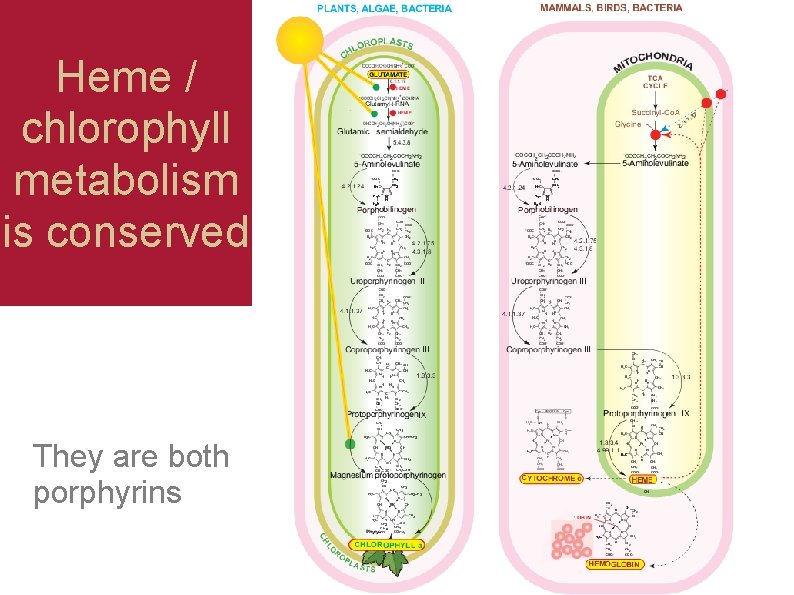
Heme / chlorophyll metabolism is conserved They are both porphyrins
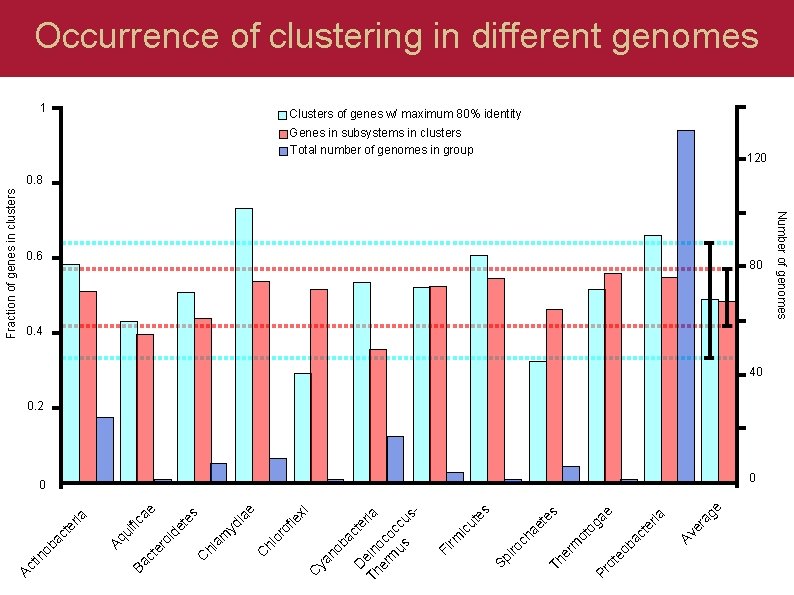
e ag ria er Av s e ga te ac eo b ot Pr ot o m er Th ae te ch ut es ic rm Fi iro Sp D te ria Th ein er oc m oc us cu s- ac no b ya C le xi of or hl C s ia e yd am hl C te ae ui fic id e er o ct Ba Aq ria te ac tin ob Fraction of genes in clusters 1 Genes in subsystems in clusters Total number of genomes in group 0. 6 80 0. 4 40 0. 2 0 0 Number of genomes Ac Occurrence of clustering in different genomes Clusters of genes w/ maximum 80% identity 120 0. 8
The Subsystems Approach to Annotation Subsystem is a generalization of “pathway” collection of functional roles jointly involved in a biological process or complex Functional Role is the abstract biological function of a gene product atomic, or user-defined, examples: 6 -phosphofructokinase (EC 2. 7. 1. 11) LSU ribosomal protein L 31 p Streptococcal virulence factors Should not contain “putative”, “thermostable”, etc Populated subsystem is complete spreadsheet of functions and roles
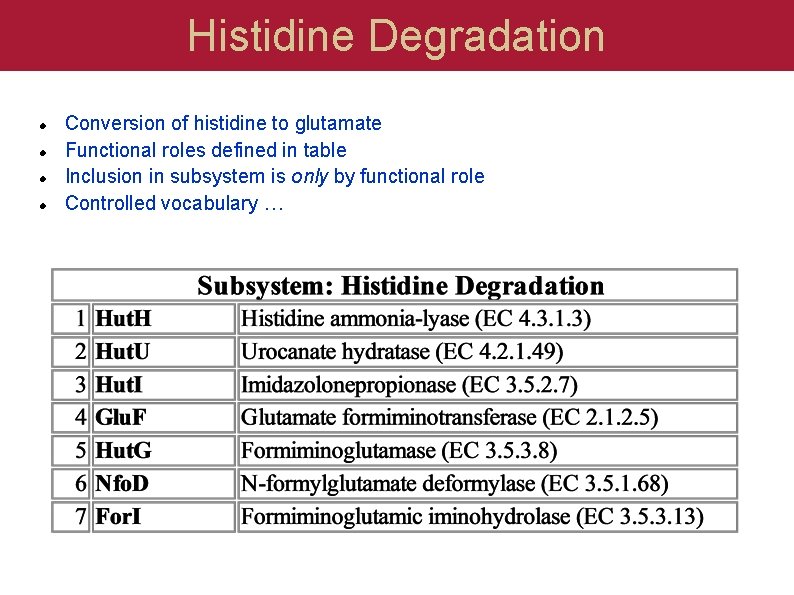
Histidine Degradation Conversion of histidine to glutamate Functional roles defined in table Inclusion in subsystem is only by functional role Controlled vocabulary …
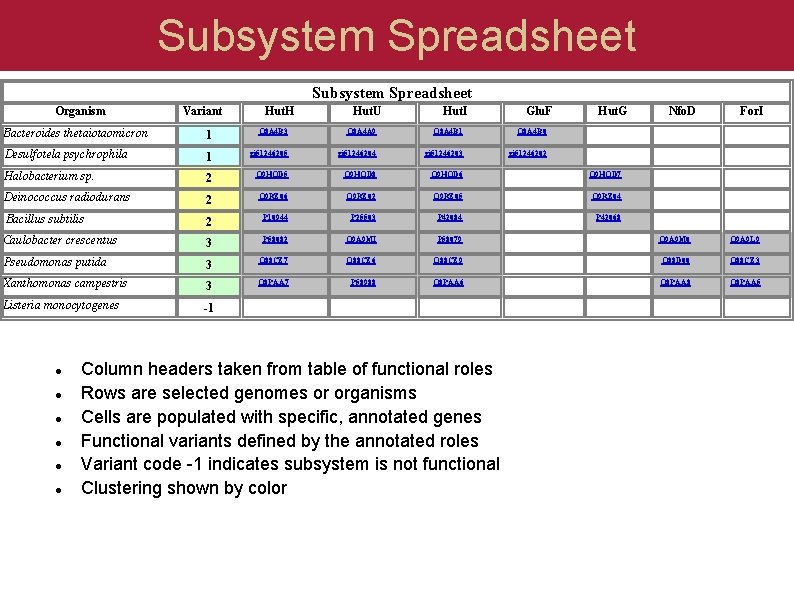
Subsystem Spreadsheet Organism Variant Hut. H Hut. U Hut. I Glu. F Hut. G Nfo. D For. I Bacteroides thetaiotaomicron 1 Q 8 A 4 B 3 Q 8 A 4 A 9 Q 8 A 4 B 1 Q 8 A 4 B 0 Desulfotela psychrophila 1 gi 51246205 gi 51246204 gi 51246203 gi 51246202 Halobacterium sp. 2 Q 9 HQD 5 Q 9 HQD 8 Q 9 HQD 6 Q 9 HQD 7 Deinococcus radiodurans 2 Q 9 RZ 06 Q 9 RZ 02 Q 9 RZ 05 Q 9 RZ 04 Bacillus subtilis 2 P 10944 P 25503 P 42084 P 42068 Caulobacter crescentus 3 P 58082 Q 9 A 9 MI P 58079 Q 9 A 9 M 0 Q 9 A 9 L 9 Pseudomonas putida 3 Q 88 CZ 7 Q 88 CZ 6 Q 88 CZ 9 Q 88 D 00 Q 88 CZ 3 Xanthomonas campestris 3 Q 8 PAA 7 P 58988 Q 8 PAA 6 Q 8 PAA 8 Q 8 PAA 5 Listeria monocytogenes -1 Column headers taken from table of functional roles Rows are selected genomes or organisms Cells are populated with specific, annotated genes Functional variants defined by the annotated roles Variant code -1 indicates subsystem is not functional Clustering shown by color
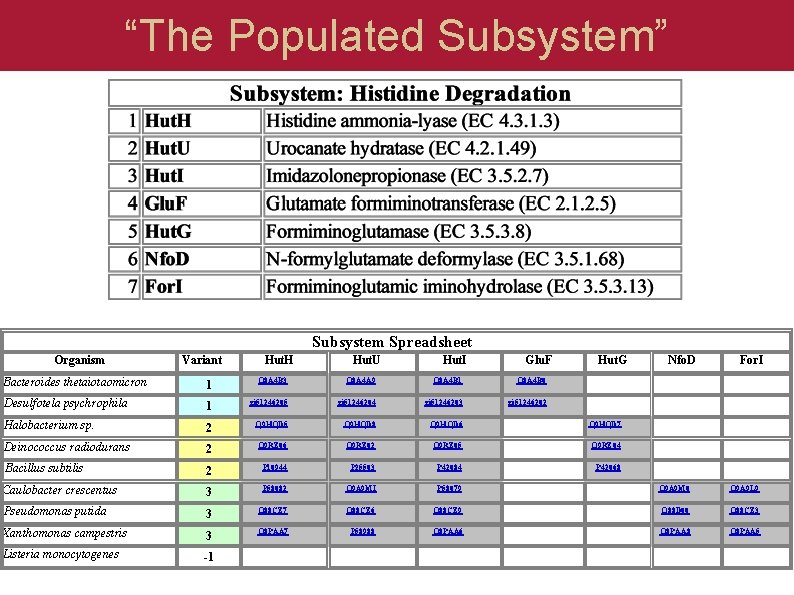
“The Populated Subsystem” Subsystem Spreadsheet Organism Variant Hut. H Hut. U Hut. I Glu. F Hut. G Nfo. D For. I Bacteroides thetaiotaomicron 1 Q 8 A 4 B 3 Q 8 A 4 A 9 Q 8 A 4 B 1 Q 8 A 4 B 0 Desulfotela psychrophila 1 gi 51246205 gi 51246204 gi 51246203 gi 51246202 Halobacterium sp. 2 Q 9 HQD 5 Q 9 HQD 8 Q 9 HQD 6 Q 9 HQD 7 Deinococcus radiodurans 2 Q 9 RZ 06 Q 9 RZ 02 Q 9 RZ 05 Q 9 RZ 04 Bacillus subtilis 2 P 10944 P 25503 P 42084 P 42068 Caulobacter crescentus 3 P 58082 Q 9 A 9 MI P 58079 Q 9 A 9 M 0 Q 9 A 9 L 9 Pseudomonas putida 3 Q 88 CZ 7 Q 88 CZ 6 Q 88 CZ 9 Q 88 D 00 Q 88 CZ 3 Xanthomonas campestris 3 Q 8 PAA 7 P 58988 Q 8 PAA 6 Q 8 PAA 8 Q 8 PAA 5 Listeria monocytogenes -1
Subsystems developed based on Wet lab Chromosomal context Metabolic context Phylogenetic context Microarray data Proteomics data …

About 2, 500 Subsystems Three level “hierarchy” • Amino Acids and Derivatives – Alanine, serine, and glycine • Serine Biosynthesis • Amino Acids and Derivatives – Lysine, threonine, methionine, and cysteine • Methionine Biosynthesis Make your own subsystems!
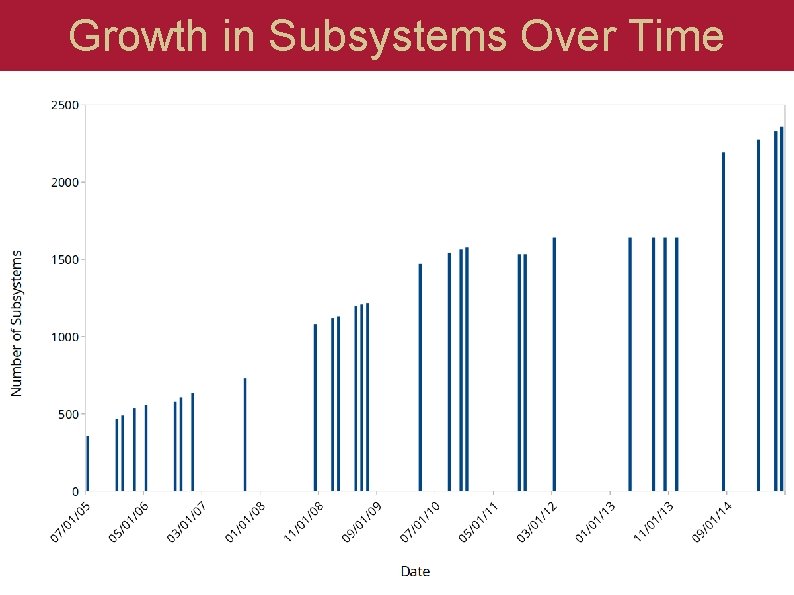
Growth in Subsystems Over Time
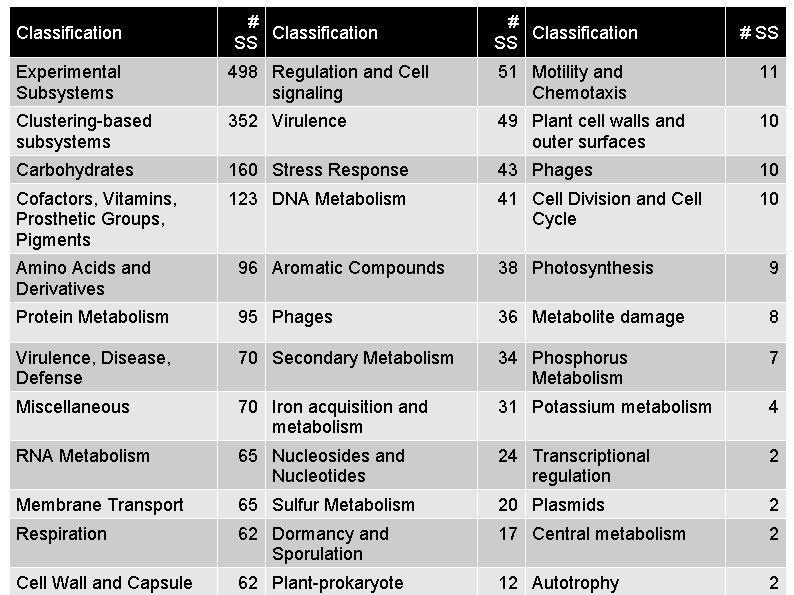
Classification # Classification SS # SS Experimental Subsystems 498 Regulation and Cell signaling 51 Motility and Chemotaxis 11 Clustering-based subsystems 352 Virulence 49 Plant cell walls and outer surfaces 10 Carbohydrates 160 Stress Response 43 Phages 10 Cofactors, Vitamins, Prosthetic Groups, Pigments 123 DNA Metabolism 41 Cell Division and Cell Cycle 10 Amino Acids and Derivatives 96 Aromatic Compounds 38 Photosynthesis 9 Protein Metabolism 95 Phages 36 Metabolite damage 8 Virulence, Disease, Defense 70 Secondary Metabolism 34 Phosphorus Metabolism 7 Miscellaneous 70 Iron acquisition and metabolism 31 Potassium metabolism 4 RNA Metabolism 65 Nucleosides and Nucleotides 24 Transcriptional regulation 2 Membrane Transport 65 Sulfur Metabolism 20 Plasmids 2 Respiration 62 Dormancy and Sporulation 17 Central metabolism 2 Cell Wall and Capsule 62 Plant-prokaryote 12 Autotrophy 2
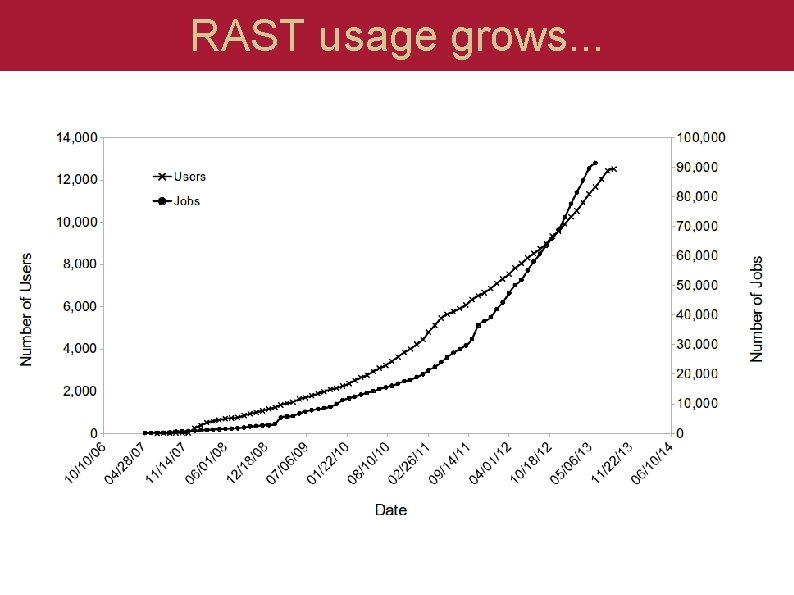
RAST usage grows. . .
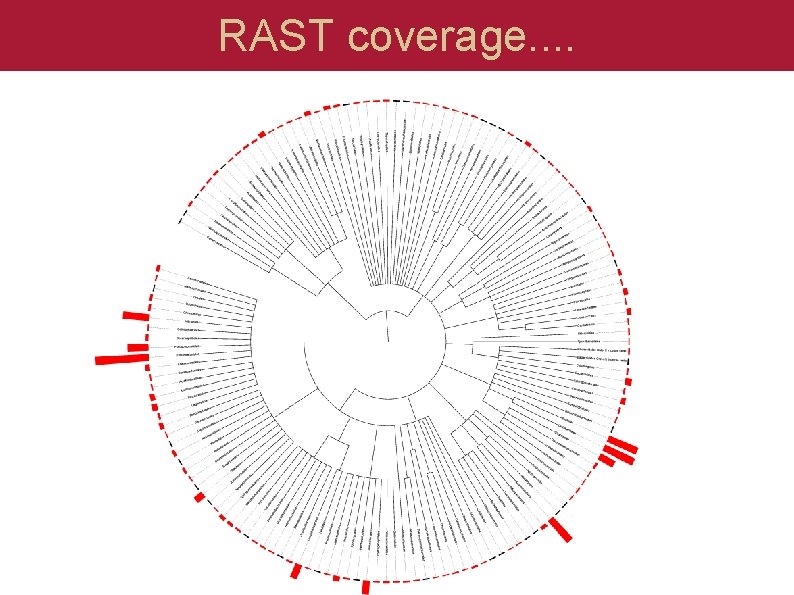
RAST coverage. .

RASTtk RAST 2. 0 Customizable choice of pipelines to run Same behind the scenes infrastructure
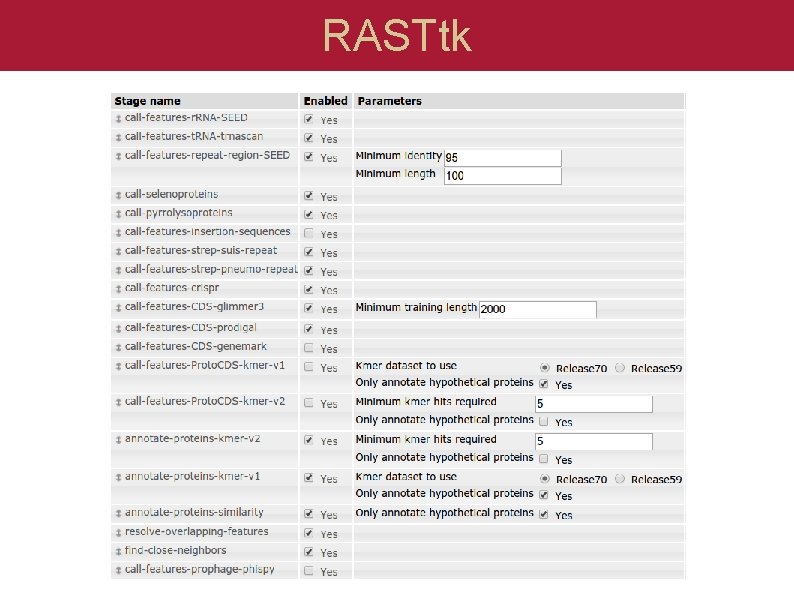
RASTtk
- Slides: 47