Using the models prediction deciding Peter Fox Data
![pairs(iris[1: 4], main = "Anderson's Iris Data -- 3 species”, pch = 21, bg pairs(iris[1: 4], main = "Anderson's Iris Data -- 3 species”, pch = 21, bg](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-6.jpg)
![splom extra! require(lattice) super. sym <- trellis. par. get("superpose. symbol") splom(~iris[1: 4], groups = splom extra! require(lattice) super. sym <- trellis. par. get("superpose. symbol") splom(~iris[1: 4], groups =](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-7.jpg)
![parallelplot(~iris[1: 4] | Species, iris) 8 parallelplot(~iris[1: 4] | Species, iris) 8](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-8.jpg)
![parallelplot(~iris[1: 4], iris, groups = Species, horizontal. axis = FALSE, scales = list(x = parallelplot(~iris[1: 4], iris, groups = Species, horizontal. axis = FALSE, scales = list(x =](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-9.jpg)



![> pfit. K<- prune(fit. K, cp= fit. K$cptable[which. min(fit. K$cptable[, "xerror"]), "CP"]) > plot(pfit. > pfit. K<- prune(fit. K, cp= fit. K$cptable[which. min(fit. K$cptable[, "xerror"]), "CP"]) > plot(pfit.](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-16.jpg)



![Bayes > cl <- kmeans(iris[, 1: 4], 3) > table(cl$cluster, iris[, 5]) setosa versicolor Bayes > cl <- kmeans(iris[, 1: 4], 3) > table(cl$cluster, iris[, 5]) setosa versicolor](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-27.jpg)
![Digging into iris classifier<-naive. Bayes(iris[, 1: 4], iris[, 5]) table(predict(classifier, iris[, -5]), iris[, 5], Digging into iris classifier<-naive. Bayes(iris[, 1: 4], iris[, 5]) table(predict(classifier, iris[, -5]), iris[, 5],](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-28.jpg)
![Digging into iris > classifier$apriori iris[, 5] setosa versicolor virginica 50 50 50 > Digging into iris > classifier$apriori iris[, 5] setosa versicolor virginica 50 50 50 >](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-29.jpg)

![House Votes 1984 > predict(model, House. Votes 84[1: 10, -1], type = "raw") democrat House Votes 1984 > predict(model, House. Votes 84[1: 10, -1], type = "raw") democrat](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-32.jpg)
![House Votes 1984 > pred <- predict(model, House. Votes 84[, -1]) > table(pred, House. House Votes 1984 > pred <- predict(model, House. Votes 84[, -1]) > table(pred, House.](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-33.jpg)




- Slides: 37

Using the models, prediction, deciding Peter Fox Data Analytics – ITWS-4963/ITWS-6965 Week 7 b, March 7, 2014 1

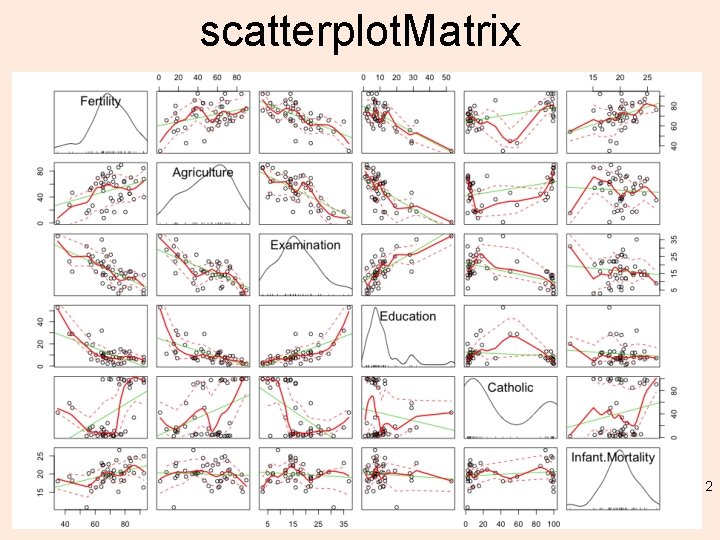
scatterplot. Matrix 2

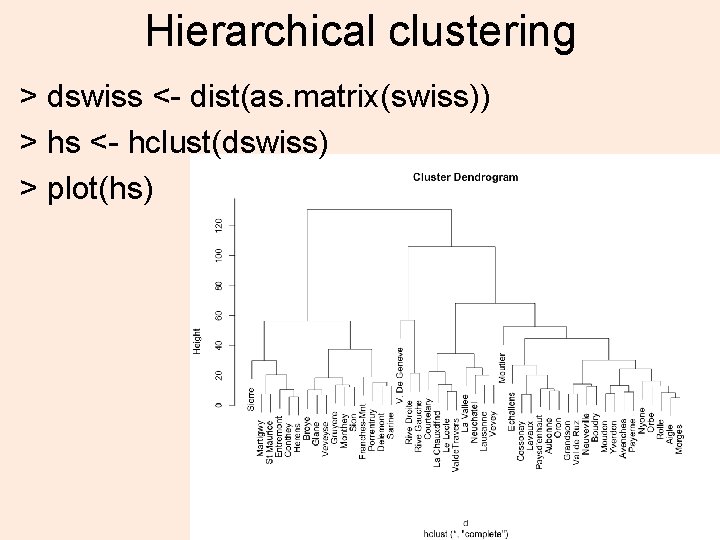
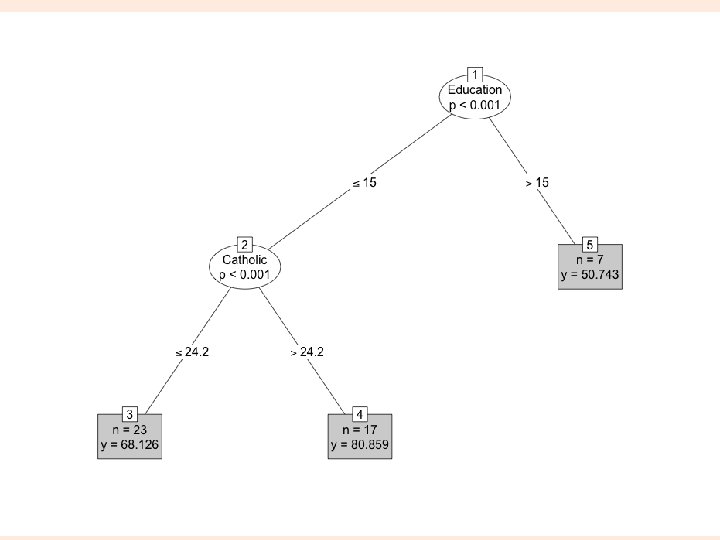
Hierarchical clustering > dswiss <- dist(as. matrix(swiss)) > hs <- hclust(dswiss) > plot(hs) 3

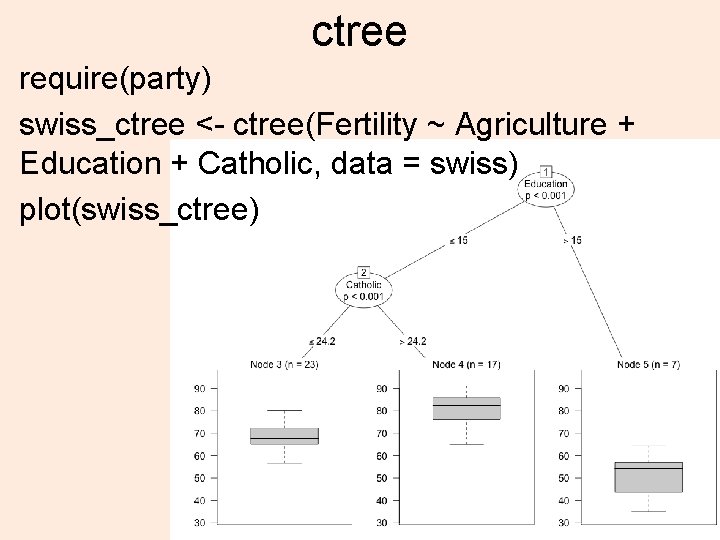
ctree require(party) swiss_ctree <- ctree(Fertility ~ Agriculture + Education + Catholic, data = swiss) plot(swiss_ctree) 4

5
![pairsiris1 4 main Andersons Iris Data 3 species pch 21 bg pairs(iris[1: 4], main = "Anderson's Iris Data -- 3 species”, pch = 21, bg](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-6.jpg)
pairs(iris[1: 4], main = "Anderson's Iris Data -- 3 species”, pch = 21, bg = c("red", "green 3", "blue")[unclass(iris$Species)]) 6
![splom extra requirelattice super sym trellis par getsuperpose symbol splomiris1 4 groups splom extra! require(lattice) super. sym <- trellis. par. get("superpose. symbol") splom(~iris[1: 4], groups =](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-7.jpg)
splom extra! require(lattice) super. sym <- trellis. par. get("superpose. symbol") splom(~iris[1: 4], groups = Species, data = iris, panel = panel. superpose, key = list(title = "Three Varieties of Iris", columns = 3, points = list(pch = super. sym$pch[1: 3], col = super. sym$col[1: 3]), text = list(c("Setosa", "Versicolor", "Virginica")))) splom(~iris[1: 3]|Species, data = iris, layout=c(2, 2), pscales = 0, varnames = c("Sepaln. Length", "Sepaln. Width", "Petaln. Length"), page = function(. . . ) { ltext(x = seq(. 6, . 8, length. out = 4), y = seq(. 9, . 6, length. out = 4), labels = c("Three", "Varieties", "of", "Iris"), cex = 2) }) 7
![parallelplotiris1 4 Species iris 8 parallelplot(~iris[1: 4] | Species, iris) 8](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-8.jpg)
parallelplot(~iris[1: 4] | Species, iris) 8
![parallelplotiris1 4 iris groups Species horizontal axis FALSE scales listx parallelplot(~iris[1: 4], iris, groups = Species, horizontal. axis = FALSE, scales = list(x =](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-9.jpg)
parallelplot(~iris[1: 4], iris, groups = Species, horizontal. axis = FALSE, scales = list(x = list(rot = 90))) 9

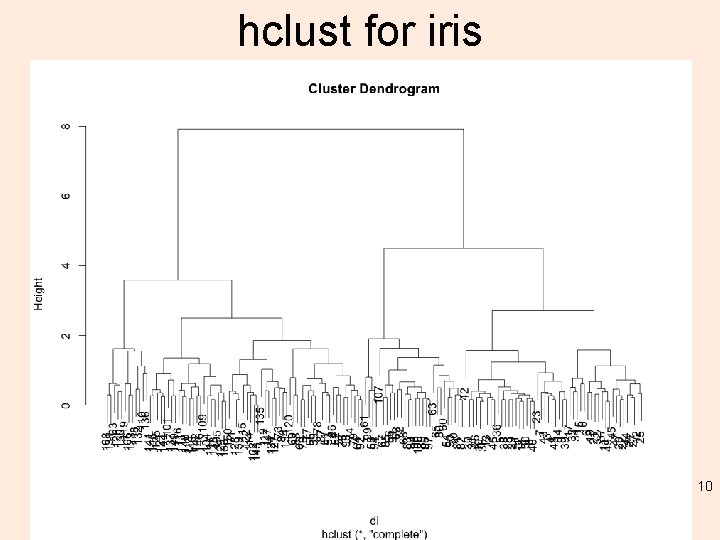
hclust for iris 10

plot(iris_ctree) 11

Ctree > iris_ctree <- ctree(Species ~ Sepal. Length + Sepal. Width + Petal. Length + Petal. Width, data=iris) > print(iris_ctree) Conditional inference tree with 4 terminal nodes Response: Species Inputs: Sepal. Length, Sepal. Width, Petal. Length, Petal. Width Number of observations: 150 1) Petal. Length <= 1. 9; criterion = 1, statistic = 140. 264 2)* weights = 50 1) Petal. Length > 1. 9 3) Petal. Width <= 1. 7; criterion = 1, statistic = 67. 894 4) Petal. Length <= 4. 8; criterion = 0. 999, statistic = 13. 865 5)* weights = 46 4) Petal. Length > 4. 8 6)* weights = 8 3) Petal. Width > 1. 7 7)* weights = 46 12

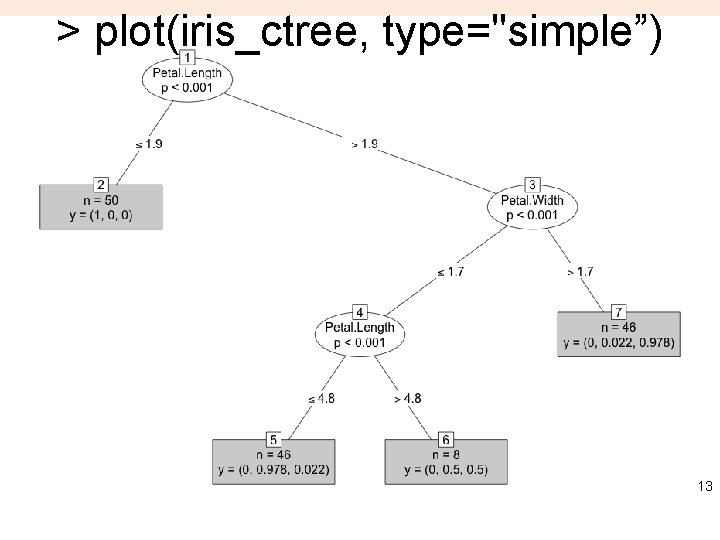
> plot(iris_ctree, type="simple”) 13

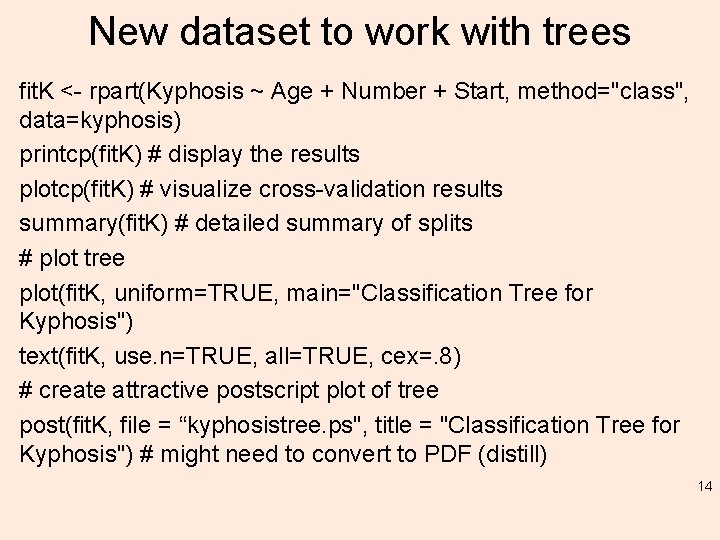
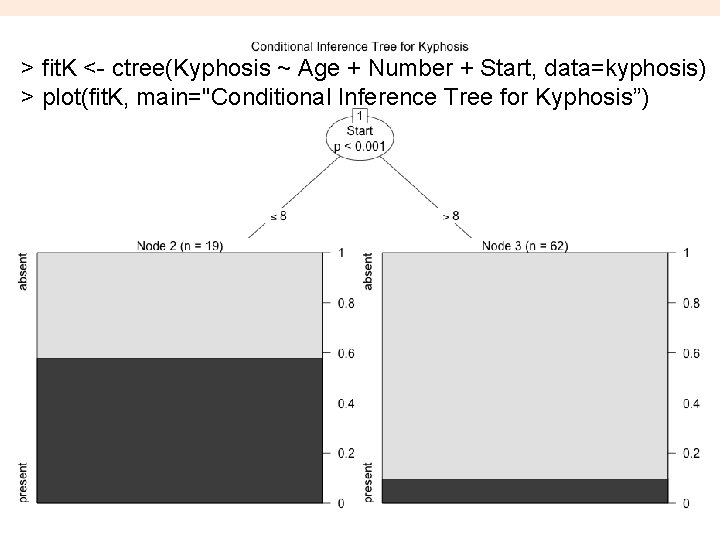
New dataset to work with trees fit. K <- rpart(Kyphosis ~ Age + Number + Start, method="class", data=kyphosis) printcp(fit. K) # display the results plotcp(fit. K) # visualize cross-validation results summary(fit. K) # detailed summary of splits # plot tree plot(fit. K, uniform=TRUE, main="Classification Tree for Kyphosis") text(fit. K, use. n=TRUE, all=TRUE, cex=. 8) # create attractive postscript plot of tree post(fit. K, file = “kyphosistree. ps", title = "Classification Tree for Kyphosis") # might need to convert to PDF (distill) 14

15
![pfit K prunefit K cp fit Kcptablewhich minfit Kcptable xerror CP plotpfit > pfit. K<- prune(fit. K, cp= fit. K$cptable[which. min(fit. K$cptable[, "xerror"]), "CP"]) > plot(pfit.](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-16.jpg)
> pfit. K<- prune(fit. K, cp= fit. K$cptable[which. min(fit. K$cptable[, "xerror"]), "CP"]) > plot(pfit. K, uniform=TRUE, main="Pruned Classification Tree for Kyphosis") 16 > text(pfit. K, use. n=TRUE, all=TRUE, cex=. 8) > post(pfit. K, file = “ptree. ps", title = "Pruned Classification Tree for Kyphosis”)

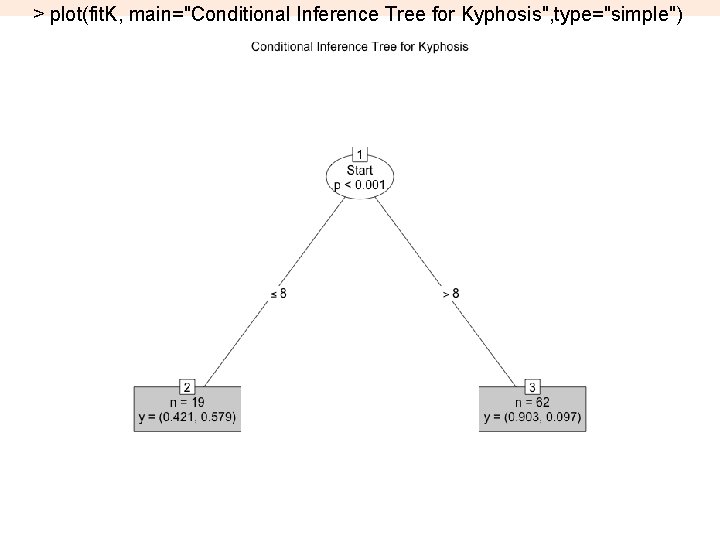
> fit. K <- ctree(Kyphosis ~ Age + Number + Start, data=kyphosis) > plot(fit. K, main="Conditional Inference Tree for Kyphosis”) 17

> plot(fit. K, main="Conditional Inference Tree for Kyphosis", type="simple") 18

random. Forest > require(random. Forest) > fit. KF <- random. Forest(Kyphosis ~ Age + Number + Start, data=kyphosis) > print(fit. KF) # view results Call: random. Forest(formula = Kyphosis ~ Age + Number + Start, data = kyphosis) Type of random forest: classification Number of trees: 500 No. of variables tried at each split: 1 OOB estimate of error rate: 20. 99% Confusion matrix: absent present class. error absent 59 5 0. 0781250 present 12 5 0. 7058824 > importance(fit. KF) # importance of each predictor Mean. Decrease. Gini Age 8. 654112 Number 5. 584019 Start 10. 168591 Random forests improve predictive accuracy by generating a large number of bootstrapped trees (based on random samples of variables), classifying a case using each tree in this new "forest", and deciding a final predicted outcome by combining the results across all of the trees 19 (an average in regression, a majority vote in classification).

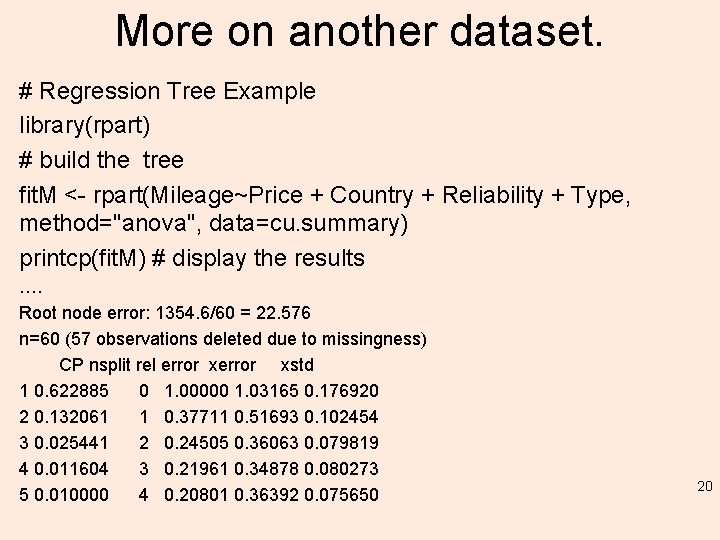
More on another dataset. # Regression Tree Example library(rpart) # build the tree fit. M <- rpart(Mileage~Price + Country + Reliability + Type, method="anova", data=cu. summary) printcp(fit. M) # display the results …. Root node error: 1354. 6/60 = 22. 576 n=60 (57 observations deleted due to missingness) CP nsplit rel error xstd 1 0. 622885 0 1. 00000 1. 03165 0. 176920 2 0. 132061 1 0. 37711 0. 51693 0. 102454 3 0. 025441 2 0. 24505 0. 36063 0. 079819 4 0. 011604 3 0. 21961 0. 34878 0. 080273 5 0. 010000 4 0. 20801 0. 36392 0. 075650 20

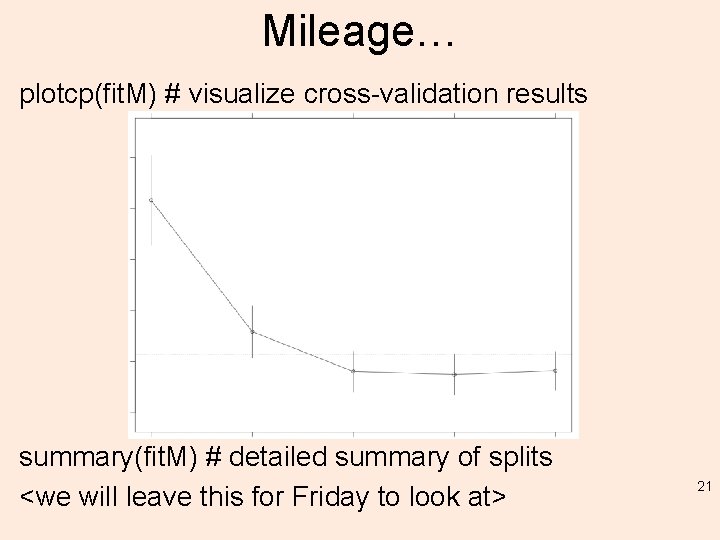
Mileage… plotcp(fit. M) # visualize cross-validation results summary(fit. M) # detailed summary of splits <we will leave this for Friday to look at> 21

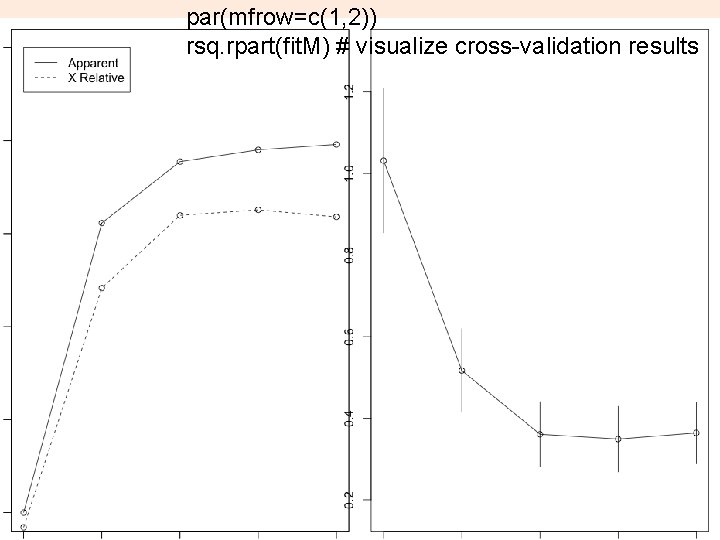
par(mfrow=c(1, 2)) rsq. rpart(fit. M) # visualize cross-validation results 22

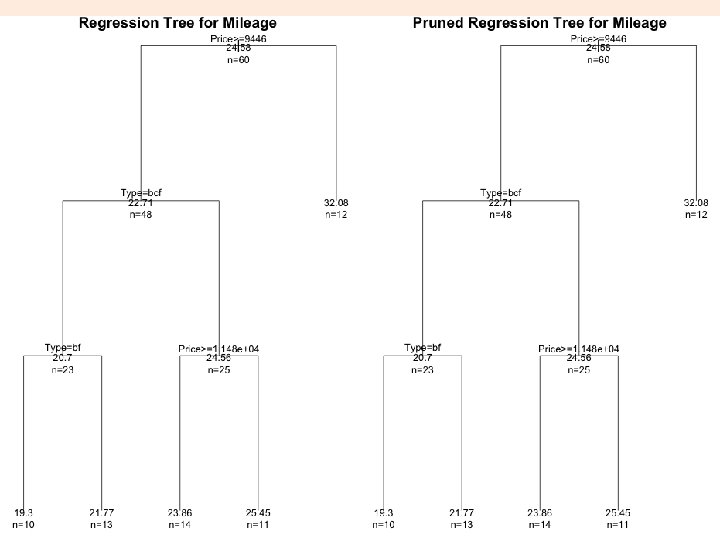
# plot tree plot(fit. M, uniform=TRUE, main="Regression Tree for Mileage ") text(fit. M, use. n=TRUE, all=TRUE, cex=. 8) # prune the tree pfit. M<- prune(fit. M, cp=0. 01160389) # from cptable # plot the pruned tree plot(pfit. M, uniform=TRUE, main="Pruned Regression Tree for Mileage") text(pfit. M, use. n=TRUE, all=TRUE, cex=. 8) post(pfit. M, file = ”ptree 2. ps", title = "Pruned Regression Tree for Mileage”) 23

24

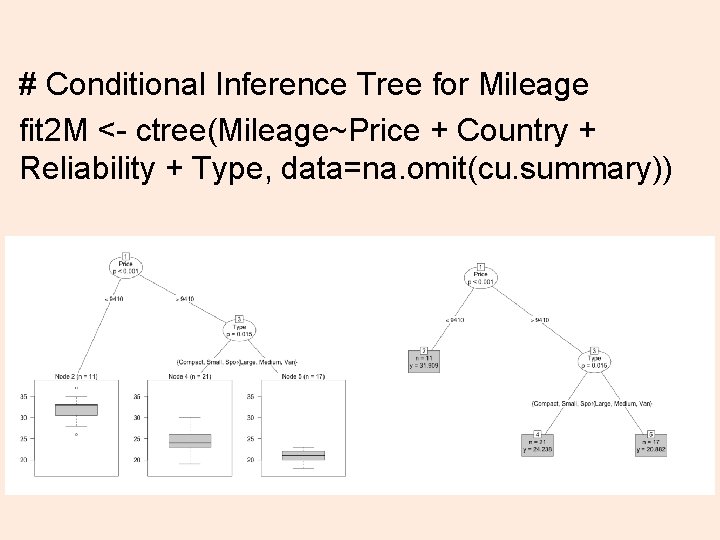
# Conditional Inference Tree for Mileage fit 2 M <- ctree(Mileage~Price + Country + Reliability + Type, data=na. omit(cu. summary)) 25

Enough of trees! 26
![Bayes cl kmeansiris 1 4 3 tableclcluster iris 5 setosa versicolor Bayes > cl <- kmeans(iris[, 1: 4], 3) > table(cl$cluster, iris[, 5]) setosa versicolor](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-27.jpg)
Bayes > cl <- kmeans(iris[, 1: 4], 3) > table(cl$cluster, iris[, 5]) setosa versicolor virginica 2 0 2 36 1 0 48 14 3 50 0 0 # > m <- naive. Bayes(iris[, 1: 4], iris[, 5]) > table(predict(m, iris[, 1: 4]), iris[, 5]) setosa versicolor virginica setosa 50 0 0 versicolor 0 47 3 virginica 0 3 47 pairs(iris[1: 4], main="Iris Data (red=setosa, green=versicolor , blue=virginica)", pch=21, bg=c("red", "green 3", "blue")[u nclass(iris$Species)]) 27
![Digging into iris classifiernaive Bayesiris 1 4 iris 5 tablepredictclassifier iris 5 iris 5 Digging into iris classifier<-naive. Bayes(iris[, 1: 4], iris[, 5]) table(predict(classifier, iris[, -5]), iris[, 5],](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-28.jpg)
Digging into iris classifier<-naive. Bayes(iris[, 1: 4], iris[, 5]) table(predict(classifier, iris[, -5]), iris[, 5], dnn=list('predicted', 'actual')) actual predicted setosa versicolor virginica setosa 50 0 0 versicolor 0 47 3 virginica 0 3 47 28
![Digging into iris classifierapriori iris 5 setosa versicolor virginica 50 50 50 Digging into iris > classifier$apriori iris[, 5] setosa versicolor virginica 50 50 50 >](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-29.jpg)
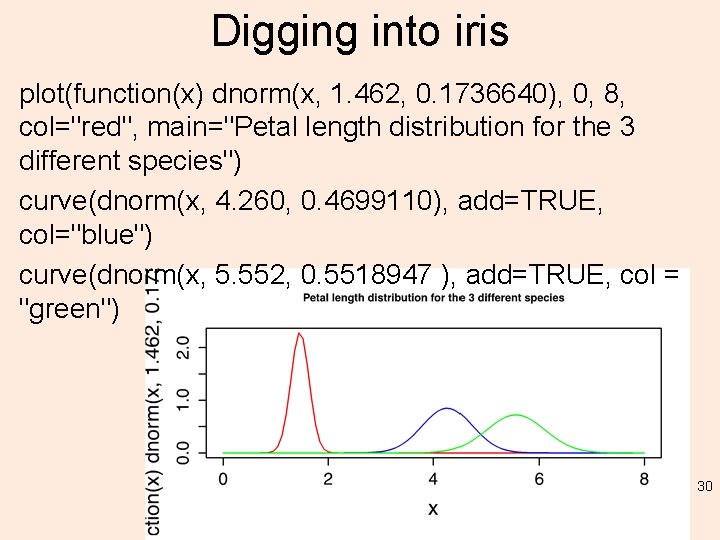
Digging into iris > classifier$apriori iris[, 5] setosa versicolor virginica 50 50 50 > classifier$tables$Petal. Length iris[, 5] [, 1] [, 2] setosa 1. 462 0. 1736640 versicolor 4. 260 0. 4699110 virginica 5. 552 0. 5518947 29

Digging into iris plot(function(x) dnorm(x, 1. 462, 0. 1736640), 0, 8, col="red", main="Petal length distribution for the 3 different species") curve(dnorm(x, 4. 260, 0. 4699110), add=TRUE, col="blue") curve(dnorm(x, 5. 552, 0. 5518947 ), add=TRUE, col = "green") 30

http: //www. ugrad. stat. ubc. ca/R/library/mlb ench/html/House. Votes 84. html > require(mlbench) > data(House. Votes 84) > model <- naive. Bayes(Class ~. , data = House. Votes 84) > predict(model, House. Votes 84[1: 10, -1]) [1] republican democrat republican [10] democrat Levels: democrat republican 31
![House Votes 1984 predictmodel House Votes 841 10 1 type raw democrat House Votes 1984 > predict(model, House. Votes 84[1: 10, -1], type = "raw") democrat](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-32.jpg)
House Votes 1984 > predict(model, House. Votes 84[1: 10, -1], type = "raw") democrat republican [1, ] 1. 029209 e-07 9. 999999 e-01 [2, ] 5. 820415 e-08 9. 999999 e-01 [3, ] 5. 684937 e-03 9. 943151 e-01 [4, ] 9. 985798 e-01 1. 420152 e-03 [5, ] 9. 666720 e-01 3. 332802 e-02 [6, ] 8. 121430 e-01 1. 878570 e-01 [7, ] 1. 751512 e-04 9. 998248 e-01 [8, ] 8. 300100 e-06 9. 999917 e-01 [9, ] 8. 277705 e-08 9. 999999 e-01 32 [10, ] 1. 000000 e+00 5. 029425 e-11
![House Votes 1984 pred predictmodel House Votes 84 1 tablepred House House Votes 1984 > pred <- predict(model, House. Votes 84[, -1]) > table(pred, House.](https://slidetodoc.com/presentation_image_h2/9bb37255a6968f9149012fac6f1c9d12/image-33.jpg)
House Votes 1984 > pred <- predict(model, House. Votes 84[, -1]) > table(pred, House. Votes 84$Class) pred democrat republican democrat 238 13 republican 29 155 33

So now you could complete this: > data(Hair. Eye. Color) > mosaicplot(Hair. Eye. Color) > margin. table(Hair. Eye. Color, 3) Sex Male Female 279 313 > margin. table(Hair. Eye. Color, c(1, 3)) Sex Hair Male Female Black 56 52 Brown 143 Red 34 37 Blond 46 81 Construct a naïve Bayes classifier and test. 34

Assignments to come… • Term project (A 6). Due ~ week 13. 30% (25% written, 5% oral; individual). • Assignment 7: Predictive and Prescriptive Analytics. Due ~ week 9/10. 20% (15% written and 5% oral; individual); 35

Coming weeks • I will be out of town Friday March 21 and 28 • On March 21 you will have a lab – attendance will be taken – to work on assignments (term (6) and assignment 7). Your project proposals (Assignment 5) are on March 18. • On March 28 you will have a lecture on SVM, thus the Tuesday March 25 will be a lab. • Back to regular schedule in April (except 18 th) 36

Admin info (keep/ print this slide) • • • Class: ITWS-4963/ITWS 6965 Hours: 12: 00 pm-1: 50 pm Tuesday/ Friday Location: SAGE 3101 Instructor: Peter Fox Instructor contact: pfox@cs. rpi. edu, 518. 276. 4862 (do not leave a msg) Contact hours: Monday** 3: 00 -4: 00 pm (or by email appt) Contact location: Winslow 2120 (sometimes Lally 207 A announced by email) TA: Lakshmi Chenicheri chenil@rpi. edu Web site: http: //tw. rpi. edu/web/courses/Data. Analytics/2014 – Schedule, lectures, syllabus, reading, assignments, etc. 37