Stats 330 Lecture 23 Department of Statistics 2012
























- Slides: 45

Stats 330: Lecture 23 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 1

Plan of the day In today’s lecture we continue our discussion of the multiple logistic regression model Topics covered – Models and submodels – Residuals for Multiple Logistic Regression – Diagnostics in Multiple Logistic Regression – No analogue of R 2 Reference: Coursebook, sections 5. 2. 3, 5. 2. 3 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 2

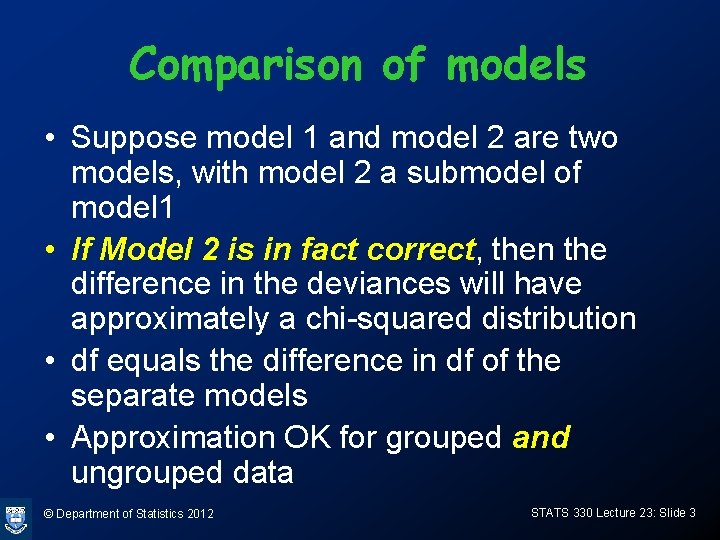
Comparison of models • Suppose model 1 and model 2 are two models, with model 2 a submodel of model 1 • If Model 2 is in fact correct, then the difference in the deviances will have approximately a chi-squared distribution • df equals the difference in df of the separate models • Approximation OK for grouped and ungrouped data © Department of Statistics 2012 STATS 330 Lecture 23: Slide 3

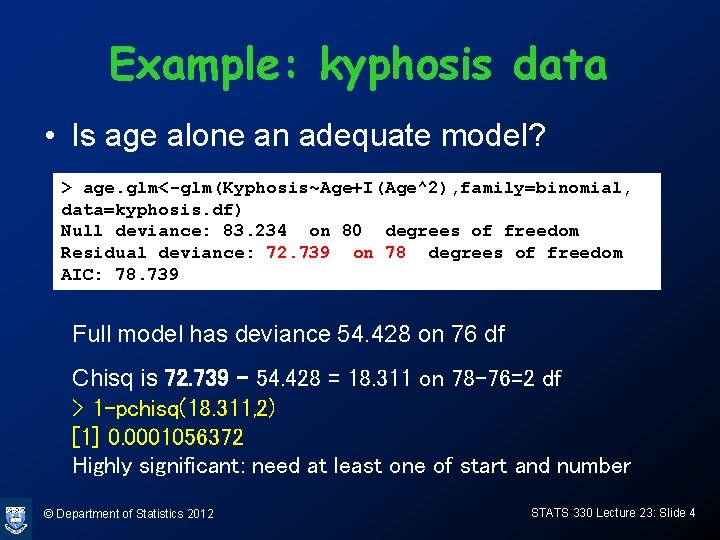
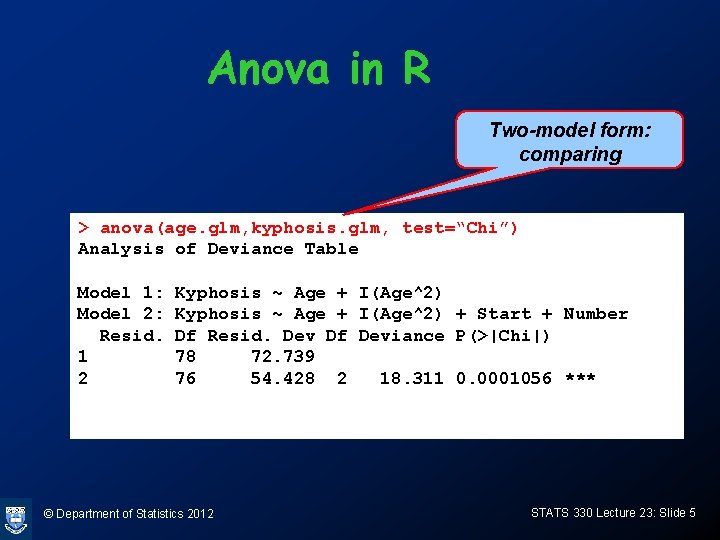
Example: kyphosis data • Is age alone an adequate model? > age. glm<-glm(Kyphosis~Age+I(Age^2), family=binomial, data=kyphosis. df) Null deviance: 83. 234 on 80 degrees of freedom Residual deviance: 72. 739 on 78 degrees of freedom AIC: 78. 739 Full model has deviance 54. 428 on 76 df Chisq is 72. 739 - 54. 428 = 18. 311 on 78 -76=2 df > 1 -pchisq(18. 311, 2) [1] 0. 0001056372 Highly significant: need at least one of start and number © Department of Statistics 2012 STATS 330 Lecture 23: Slide 4

Anova in R Two-model form: comparing > anova(age. glm, kyphosis. glm, test=“Chi”) Analysis of Deviance Table Model 1: Model 2: Resid. 1 2 Kyphosis ~ Age + I(Age^2) + Start + Number Df Resid. Dev Df Deviance P(>|Chi|) 78 72. 739 76 54. 428 2 18. 311 0. 0001056 *** © Department of Statistics 2012 STATS 330 Lecture 23: Slide 5

Residuals • Two kinds of residuals – Pearson residuals • useful for grouped data only • similar to residuals in linear regression, actual minus fitted value – Deviance residuals • useful for grouped and ungrouped data • Measure contribution of each covariate pattern to the deviance © Department of Statistics 2012 STATS 330 Lecture 23: Slide 6

Pearson residuals Pearson residual for pattern i is Probability predicted by model Standardized to have approximately unit variance, so big if more than 2 in absolute value © Department of Statistics 2012 STATS 330 Lecture 23: Slide 7

Deviance residuals (i) • For grouped data, the deviance is © Department of Statistics 2012 STATS 330 Lecture 23: Slide 8

Deviance residuals (i) • Thus, the deviance can be written as the sum of squares of M quantities d 1, …, d. M , one for each covariate pattern • Each di is the contribution to the deviance from the ith covariate pattern • If deviance residual is big (more than about 2 in magnitude), then the covariate pattern has a big influence on the likelihood, and hence the estimates © Department of Statistics 2012 STATS 330 Lecture 23: Slide 9

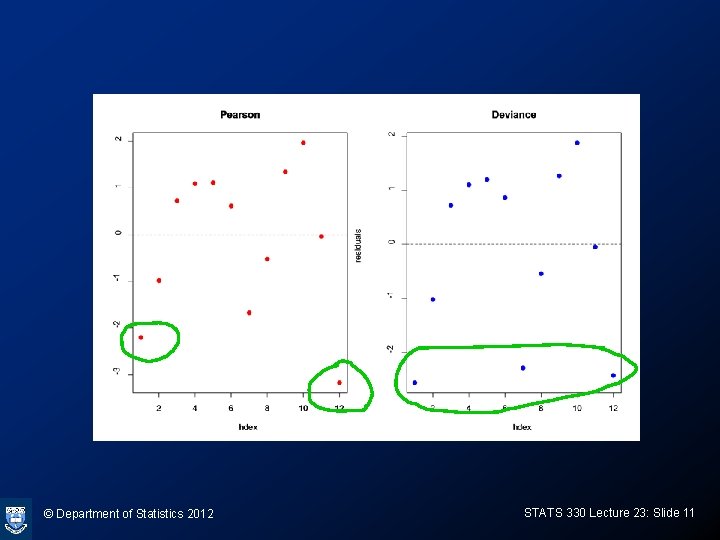
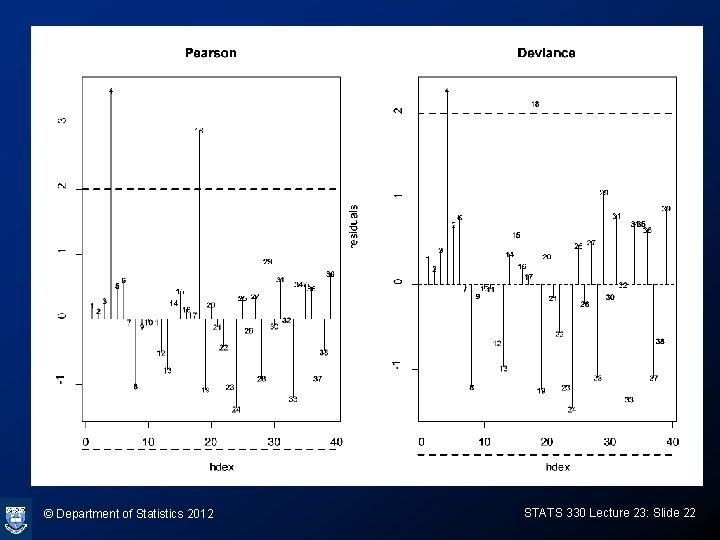
Calculating residuals > pearson. residuals<-residuals(budworm. glm, type="pearson") > deviance. residuals<-residuals(budworm. glm, type="deviance") > par(mfrow=c(1, 2)) > plot(pearson. residuals, ylab="residuals", main="Pearson") > abline(h=0, lty=2) > plot(deviance. residuals, ylab="residuals", main="Deviance") > abline(h=0, lty=2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 10

© Department of Statistics 2012 STATS 330 Lecture 23: Slide 11

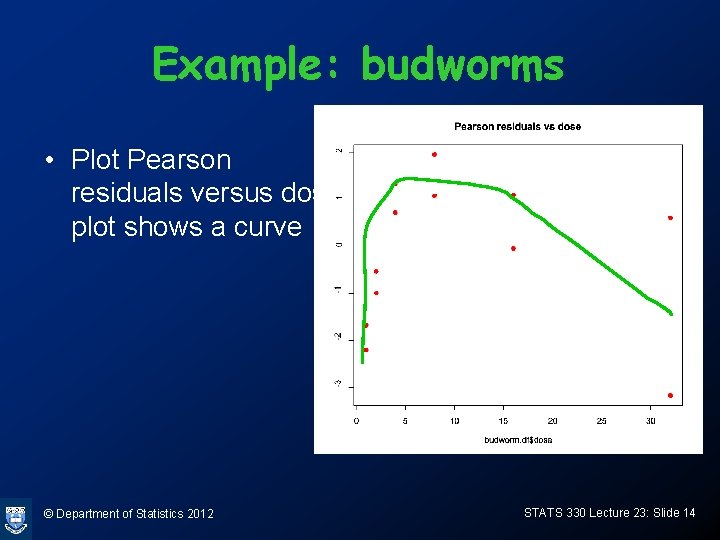
Diagnostics: outlier detection • Large residuals indicate covariate patterns poorly fitted by the model • Large Pearson residuals indicate a poor match between the “maximum model probabilities” and the logistic model probabilities, for grouped data • Large deviance residuals indicate influential points • Example: budworm data © Department of Statistics 2012 STATS 330 Lecture 23: Slide 12

Diagnostics: detecting nonlinear regression functions • For a single x, plot the logits of the maximal model probabilities against x • For multiple x’s, plot Pearson residuals against fitted probabilities, against individual x’s • If the data has most ni’s equal to 1, so can’t be grouped, try gam (cf kyphosis data) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 13

Example: budworms • Plot Pearson residuals versus dose, plot shows a curve © Department of Statistics 2012 STATS 330 Lecture 23: Slide 14

Diagnostics: influential points Will look at 3 diagnostics – Hat matrix diagonals – Cook’s distance – Leave-one-out Deviance Change © Department of Statistics 2012 STATS 330 Lecture 23: Slide 15

Example: vaso-constriction data Data from study of reflex vaso-constriction (narrowing of the blood vessels) of the skin of the fingers – Can be caused by sharp intake of breath © Department of Statistics 2012 STATS 330 Lecture 23: Slide 16

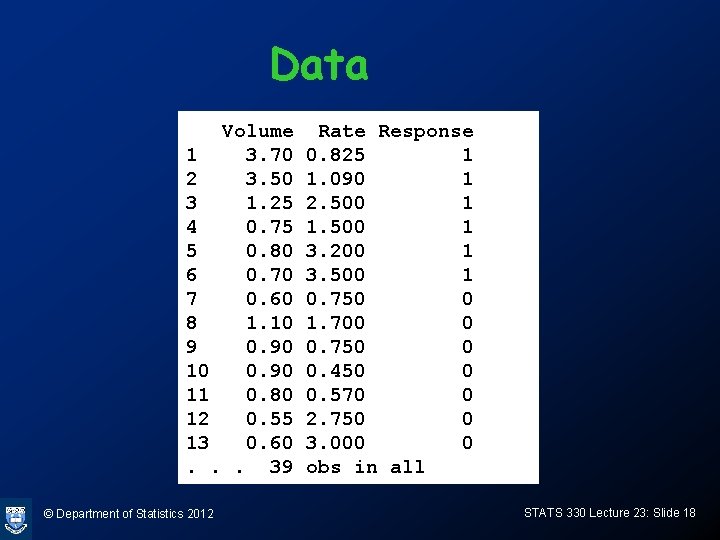
Example: vaso-constriction data Variables measured: Response = 0/1 1=vaso-constriction occurs, 0 = doesn’t occur Volume: volume of air breathed in Rate: rate of intake of breath © Department of Statistics 2012 STATS 330 Lecture 23: Slide 17

Data Volume 1 3. 70 2 3. 50 3 1. 25 4 0. 75 5 0. 80 6 0. 70 7 0. 60 8 1. 10 9 0. 90 10 0. 90 11 0. 80 12 0. 55 13 0. 60. . . 39 © Department of Statistics 2012 Rate Response 0. 825 1 1. 090 1 2. 500 1 1. 500 1 3. 200 1 3. 500 1 0. 750 0 1. 700 0 0. 750 0 0. 450 0 0. 570 0 2. 750 0 3. 000 0 obs in all STATS 330 Lecture 23: Slide 18

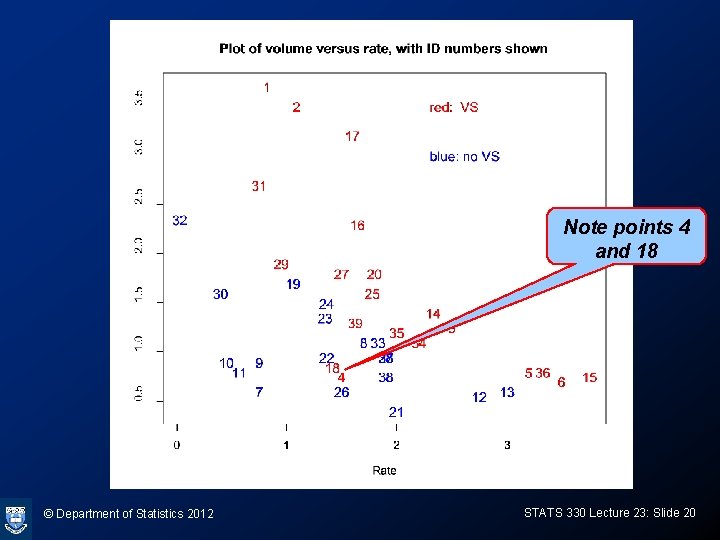
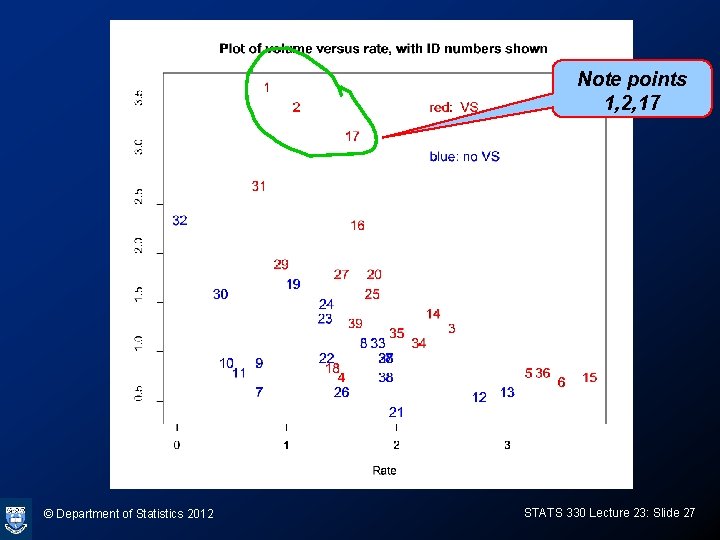
Plot of data > plot(Rate, Volume, type="n", cex=1. 2) > text(Rate, Volume, 1: 39, col=ifelse(Response==1, “red", “blue"), cex=1. 2) > text(2. 3, 3. 5, “blue: no VS", col=“blue", adj=0, cex=1. 2) > text(2. 3, 3. 0, “red: VS", col=“red", adj=0, cex=1. 2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 19

Note points 4 and 18 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 20

Enhanced residual plots > vaso. glm = glm(Response ~ log(Volume) + log(Rate), family=binomial, data=vaso. df) > pear. r<-residuals(vaso. glm, type="pearson") > dev. r<-residuals(vaso. glm, type="deviance") > par(mfrow=c(1, 2)) > plot(pear. r, ylab="residuals", main="Pearson", type="n") > text(pear. r, cex=0. 7) > abline(h=0, lty=2) > abline(h=2, lty=2, lwd=2) > abline(h=-2, lty=2, lwd=2) > plot(dev. r, ylab="residuals", main="Deviance", type="h") > text(dev. r, cex=0. 7) > abline(h=0, lty=2) > abline(h=2, lty=2, lwd=2) > abline(h=-2, lty=2, lwd=2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 21

© Department of Statistics 2012 STATS 330 Lecture 23: Slide 22

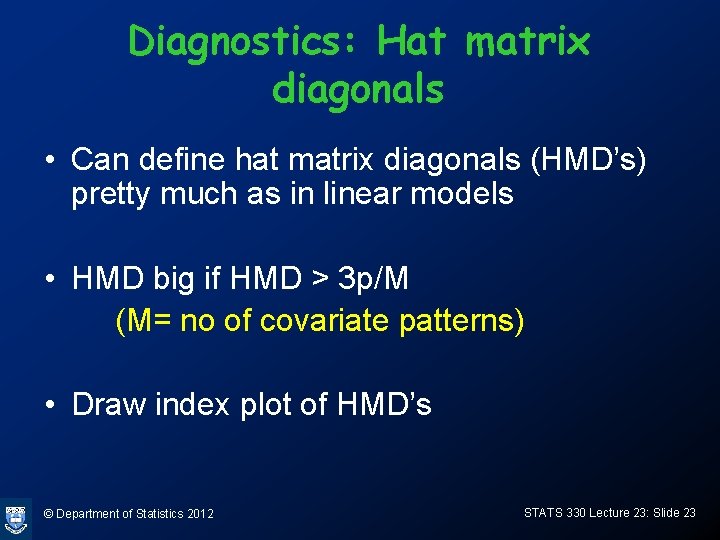
Diagnostics: Hat matrix diagonals • Can define hat matrix diagonals (HMD’s) pretty much as in linear models • HMD big if HMD > 3 p/M (M= no of covariate patterns) • Draw index plot of HMD’s © Department of Statistics 2012 STATS 330 Lecture 23: Slide 23

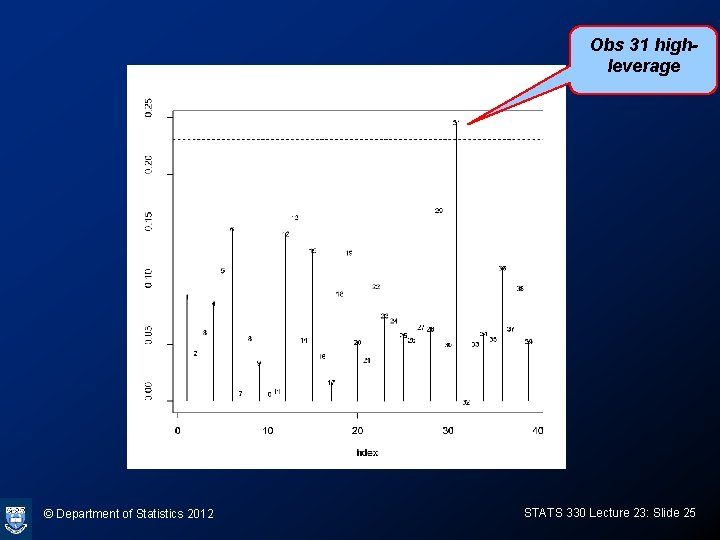
Plotting HMD’s > > HMD<-hatvalues(vaso. glm) plot(HMD, ylab="HMD's", type="h") text(HMD, cex=0. 7) abline(h=3*3/39, lty=2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 24

Obs 31 highleverage © Department of Statistics 2012 STATS 330 Lecture 23: Slide 25

Hat matrix diagonals • In ordinary regression, the hat matrix diagonals measure how “outlying” the covariates for an observation are • In logistic regression, the HMD’s measure the same thing, but are down-weighted according to the estimated probability for the observation. The weights gets small if the probability is close to 0 or 1. • In the vaso-constriction data, points 1, 2, 17 had very small weights, since the probabilities are close to 1 for these points. © Department of Statistics 2012 STATS 330 Lecture 23: Slide 26

Note points 1, 2, 17 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 27

Diagnostics: Cooks distance • Can define an analogue of Cook’s distance for each point CD = (Pearson resid )2 x HMD/(p*(1 -HMD)2) p = number of coeficients • CD big if more than about 10% quantile of the chisquared distribution on k+1 df, divided by k+1 • Calculate with qchisq(0. 1, k+1)/(k+1) • But not that reliable as a measure © Department of Statistics 2012 STATS 330 Lecture 23: Slide 28

Cooks D: calculating and plotting p<-3 CD<-cooks. distance(vaso. glm) plot(CD, ylab="Cook's D", type="h", main="index plot of Cook's distances") text(CD, cex=0. 7) bigcook<-qchisq(0. 1, p)/p abline(h=bigcook, lty=2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 29

Points 4 and 18 influential © Department of Statistics 2012 STATS 330 Lecture 23: Slide 30

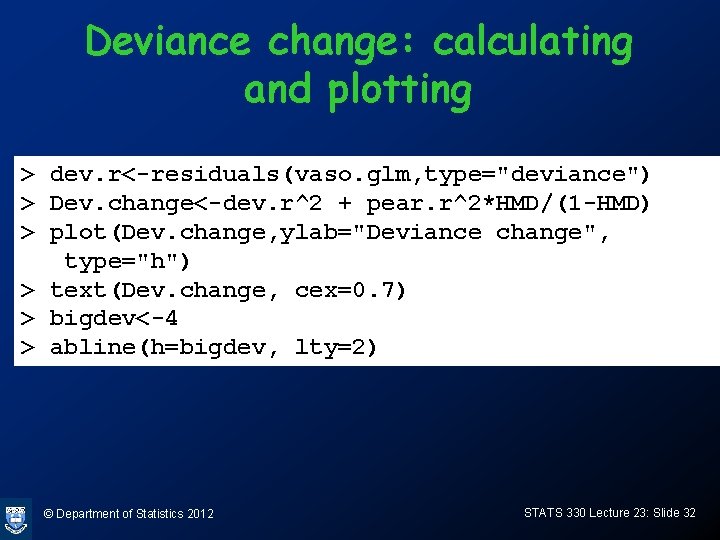
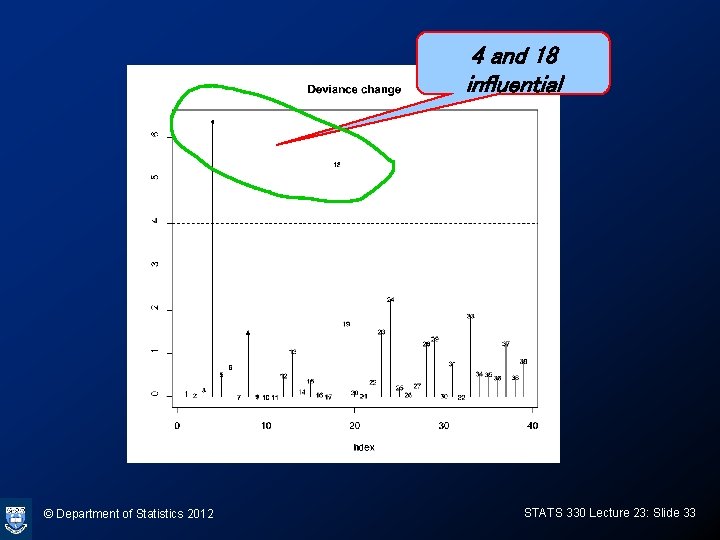
Diagnostics: leave-one-out deviance change • If the ith covariate pattern is left out, the change in the deviance is approximately (Dev. Res) 2 + (Pearson. Res)2 HMD/(1 -HMD) Big if more than about 4 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 31

Deviance change: calculating and plotting > dev. r<-residuals(vaso. glm, type="deviance") > Dev. change<-dev. r^2 + pear. r^2*HMD/(1 -HMD) > plot(Dev. change, ylab="Deviance change", type="h") > text(Dev. change, cex=0. 7) > bigdev<-4 > abline(h=bigdev, lty=2) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 32

4 and 18 influential © Department of Statistics 2012 STATS 330 Lecture 23: Slide 33

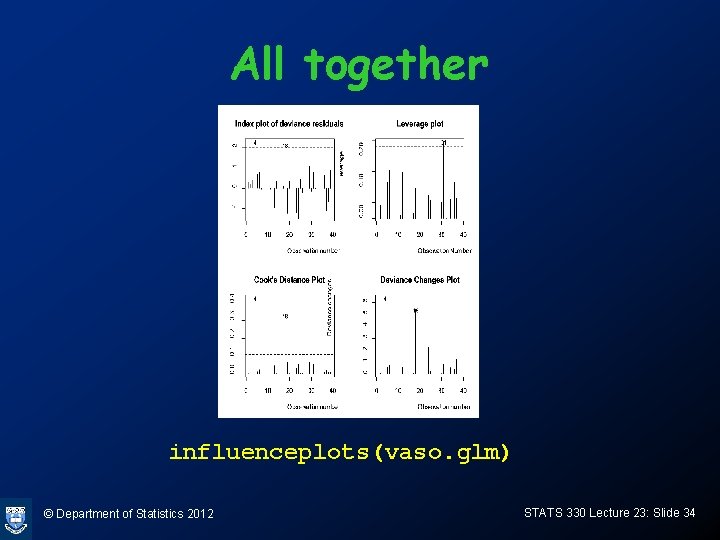
All together influenceplots(vaso. glm) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 34

Should we delete points? • How influential are the 3 points? • Can delete each in turn and examine changes in coefficients, predicted probabilities • First, coefficients: Deleting: None 31 4 18 All 3 (Intercept) -2. 875 -3. 041 -5. 206 -4. 758 -24. 348 log(Volume) 5. 179 4. 966 8. 468 7. 671 39. 142 log(Rate) 4. 562 4. 765 7. 455 6. 880 31. 642 © Department of Statistics 2012 STATS 330 Lecture 23: Slide 35

Should we delete points (2)? • Next, fitted probabilities: Fitted at None 31 point 31 0. 722 0. 627 point 4 0. 075 0. 073 point 18 0. 106 0. 100 delete points 4 18 4 and 18 0. 743 0. 707 0. 996 0. 010 0. 015 0. 000 0. 018 0. 026 0. 000 All 3 0. 996 0. 000 • Conclusion: points 4 and 18 have a big effect. © Department of Statistics 2012 STATS 330 Lecture 23: Slide 36

Should we delete points (3)? • Should we delete? • They could be genuine – no real evidence they are wrong • If we delete them, we increase the regression coefficients, make fitted probabilities more extreme • Overstate the predictive ability of the model © Department of Statistics 2012 STATS 330 Lecture 23: Slide 37

Residuals for ungrouped data • If all cases have distinct covariate patterns, then the residuals lie along two curves (corresponding to success and failure) and have little or no diagnostic value. • Thus, there is a pattern even if everything is OK. © Department of Statistics 2012 STATS 330 Lecture 23: Slide 38

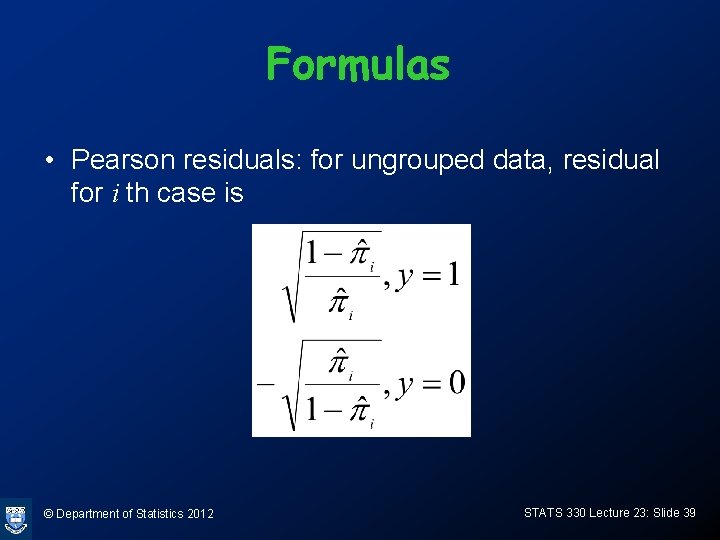
Formulas • Pearson residuals: for ungrouped data, residual for i th case is © Department of Statistics 2012 STATS 330 Lecture 23: Slide 39

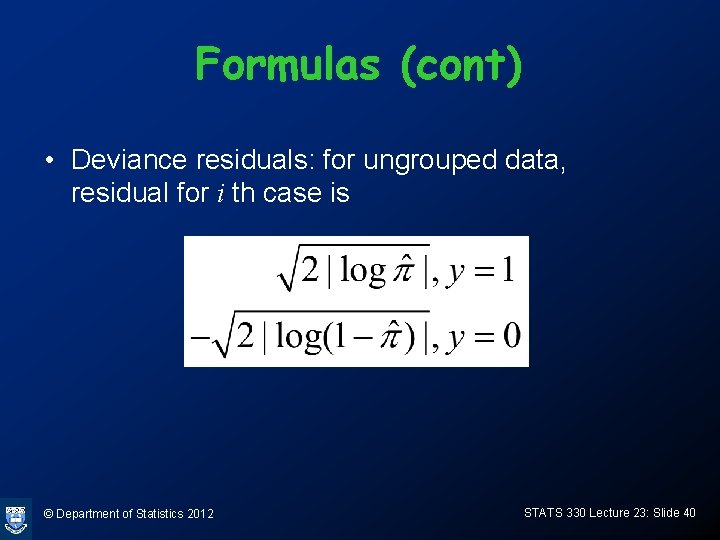
Formulas (cont) • Deviance residuals: for ungrouped data, residual for i th case is © Department of Statistics 2012 STATS 330 Lecture 23: Slide 40

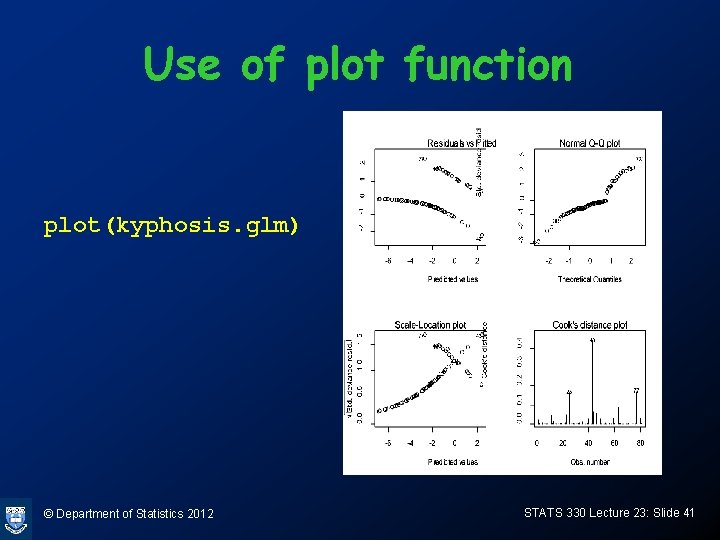
Use of plot function plot(kyphosis. glm) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 41

Analogue of R 2? • There is no satisfactory analogue of R 2 for logistic regression. • For the “small m big n” situation we can use the residual deviance, since we can obtain an approximate p-value. • For other situations we can use the Hosmer –Lemeshow statistic (next slide) © Department of Statistics 2012 STATS 330 Lecture 23: Slide 42

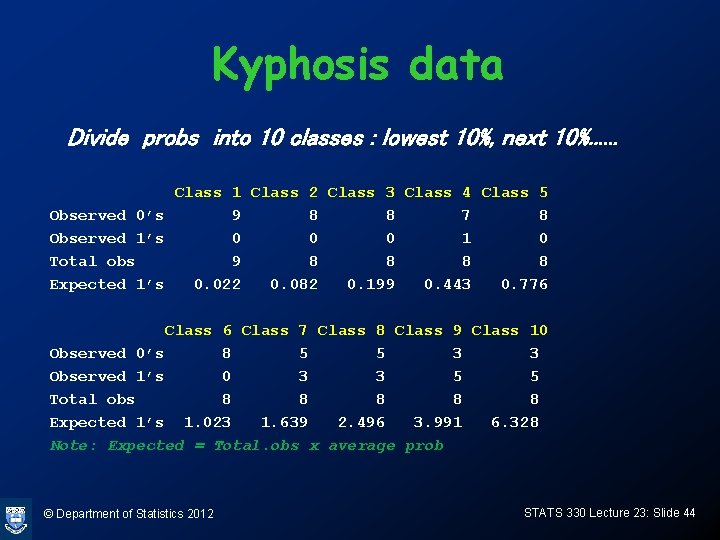
Hosmer-Lemeshow statistic • How can we judge goodness of fit for ungrouped data? • Can use the Hosmer-Lemeshow statistic, which groups the data into cases having similar fitted probabilities – Sort the cases in increasing order of fitted probabilities – Divide into 10 (almost) equal groups – Do a chi-square test to see if the number of successes in each group matches the estimated probability © Department of Statistics 2012 STATS 330 Lecture 23: Slide 43

Kyphosis data Divide probs into 10 classes : lowest 10%, next 10%. . . Class 1 Class 2 Class 3 Class 4 Class 5 Observed 0’s 9 8 8 7 8 Observed 1’s 0 0 0 1 0 Total obs 9 8 8 Expected 1’s 0. 022 0. 082 0. 199 0. 443 0. 776 Class 7 Class 8 Class 9 Class 10 Observed 0’s 8 5 5 3 3 Observed 1’s 0 3 3 5 5 Total obs 8 8 8 Expected 1’s 1. 023 1. 639 2. 496 3. 991 6. 328 Note: Expected = Total. obs x average prob © Department of Statistics 2012 STATS 330 Lecture 23: Slide 44

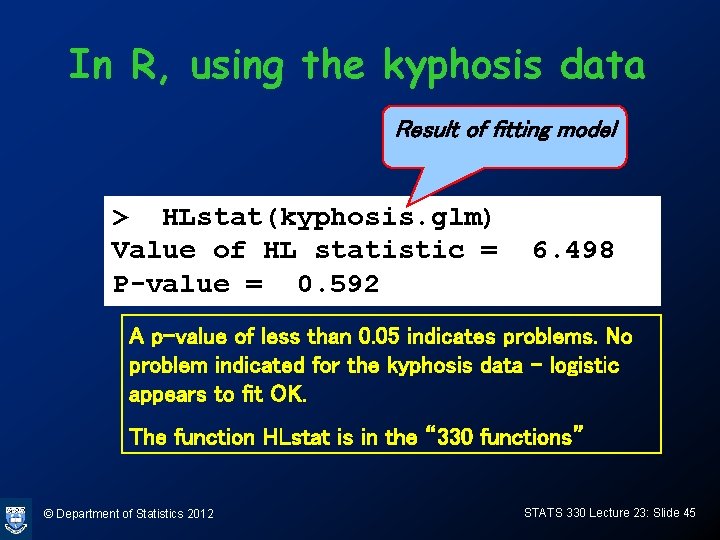
In R, using the kyphosis data Result of fitting model > HLstat(kyphosis. glm) Value of HL statistic = P-value = 0. 592 6. 498 A p-value of less than 0. 05 indicates problems. No problem indicated for the kyphosis data – logistic appears to fit OK. The function HLstat is in the “ 330 functions” © Department of Statistics 2012 STATS 330 Lecture 23: Slide 45
 Stats 330
Stats 330 Stats 330
Stats 330 Stats 330
Stats 330 01:640:244 lecture notes - lecture 15: plat, idah, farad
01:640:244 lecture notes - lecture 15: plat, idah, farad Medical statistics lecture
Medical statistics lecture Introduction to statistics what is statistics
Introduction to statistics what is statistics Astm c 330
Astm c 330 Indica la forma correcta de cada verbo en el imperfecto.
Indica la forma correcta de cada verbo en el imperfecto. Art. 317 do código penal
Art. 317 do código penal The byzantine empire reached its greatest size under
The byzantine empire reached its greatest size under Ordenanza 330 de publicidad exterior
Ordenanza 330 de publicidad exterior Isa 330 bahasa indonesia
Isa 330 bahasa indonesia Kapitalstorlek
Kapitalstorlek 550+330
550+330 The roman principate (31 bce- 330 ce) was installed by
The roman principate (31 bce- 330 ce) was installed by Isa 330
Isa 330 1453-330
1453-330 330 ml di latte quanti bicchieri sono
330 ml di latte quanti bicchieri sono Astm c 330
Astm c 330 Nuremberg code
Nuremberg code 649+330
649+330 S-330 test answers
S-330 test answers Bts 330
Bts 330 Room 330
Room 330 1453-330
1453-330 Garmin mode s transponder
Garmin mode s transponder Definition of byzantium
Definition of byzantium Reva freedman
Reva freedman Ssis 330
Ssis 330 Nwcg s-330
Nwcg s-330 1453-330
1453-330 Nep 530
Nep 530 395-1453
395-1453 1453-330
1453-330 Chapter 16 ap stats
Chapter 16 ap stats Webmetrics statistics
Webmetrics statistics Mobile commerce stats
Mobile commerce stats Stats symbols
Stats symbols Stats refund
Stats refund Freesurfer stats
Freesurfer stats It&m stats
It&m stats What is absolute language in reading
What is absolute language in reading In cc stats
In cc stats Statistics for beginners
Statistics for beginners A botanist is running an experiment on two fertilizers
A botanist is running an experiment on two fertilizers Chapter 21 comparing two proportions
Chapter 21 comparing two proportions