Specialpurpose computers for scientific simulations Makoto Taiji Processor

- Slides: 34

Special-purpose computers for scientific simulations Makoto Taiji Processor Research Team RIKEN Advanced Institute for Computational Science Computational Biology Research Core RIKEN Quantitative Biology Center taiji@riken. jp

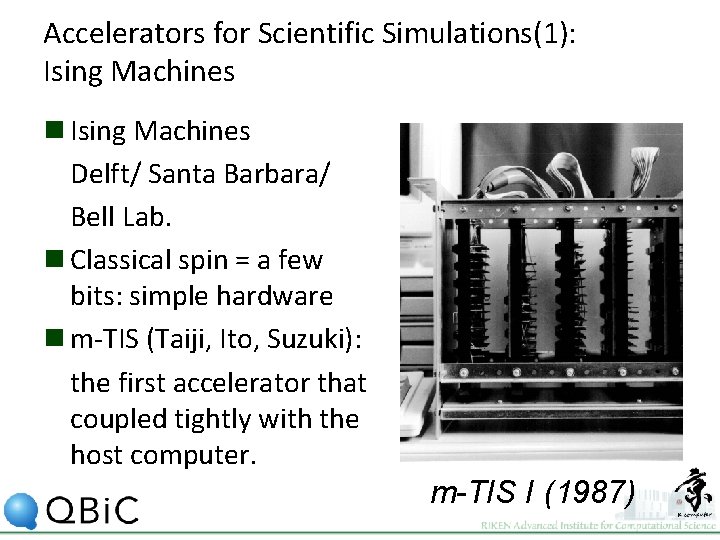
Accelerators for Scientific Simulations(1): Ising Machines n Ising Machines Delft/ Santa Barbara/ Bell Lab. n Classical spin = a few bits: simple hardware n m-TIS (Taiji, Ito, Suzuki): the first accelerator that coupled tightly with the host computer. m-TIS I (1987)

Accelerators for Scientific Simulations(2): FPGA-based Systems n Splash n m-TIS II n Edinburgh n Heidelberg

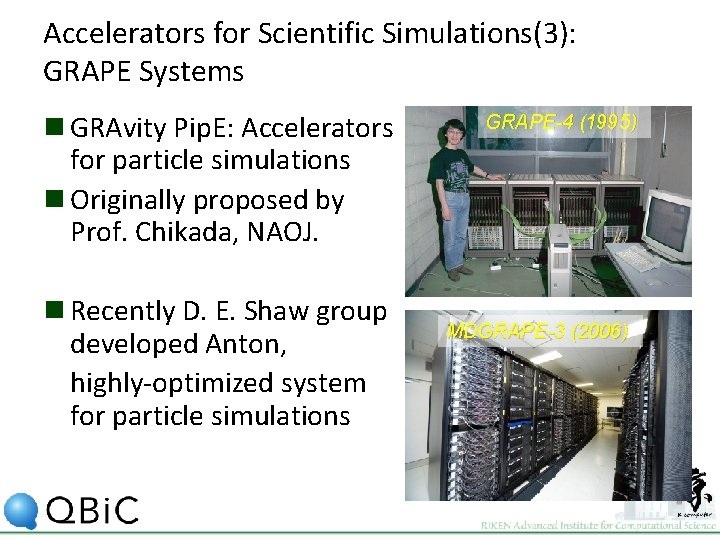
Accelerators for Scientific Simulations(3): GRAPE Systems n GRAvity Pip. E: Accelerators for particle simulations n Originally proposed by Prof. Chikada, NAOJ. n Recently D. E. Shaw group developed Anton, highly-optimized system for particle simulations GRAPE-4 (1995) MDGRAPE-3 (2006)

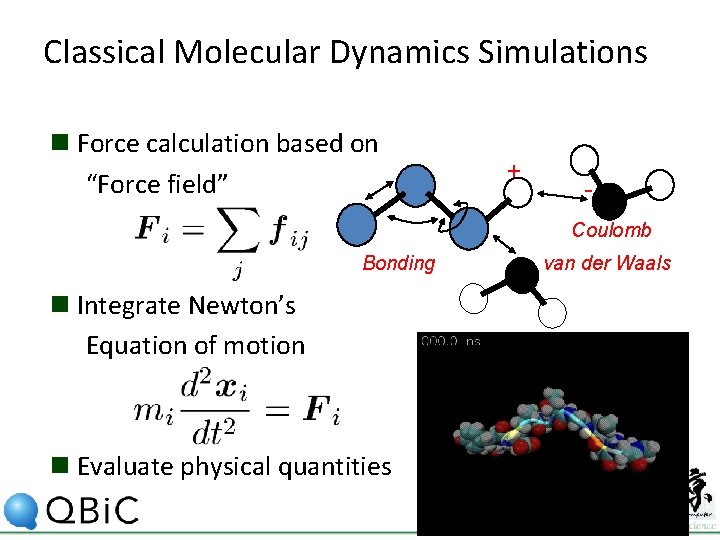
Classical Molecular Dynamics Simulations n Force calculation based on “Force field” + Coulomb Bonding n Integrate Newton’s Equation of motion n Evaluate physical quantities van der Waals

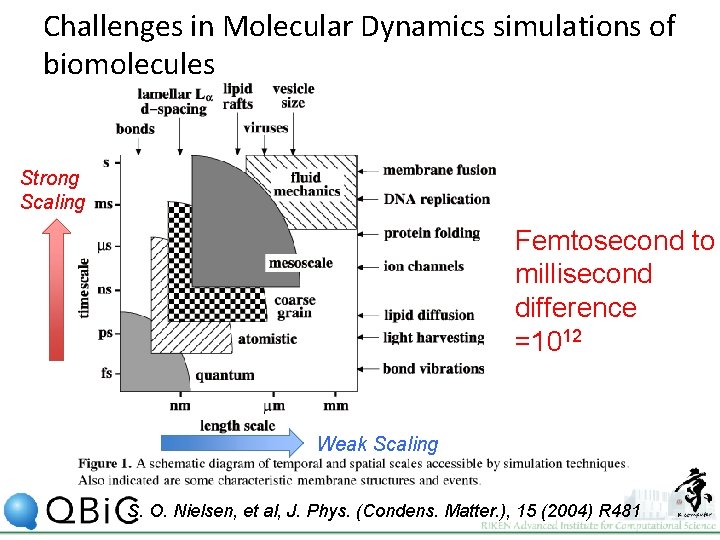
Challenges in Molecular Dynamics simulations of biomolecules Strong Scaling Femtosecond to millisecond difference =1012 Weak Scaling S. O. Nielsen, et al, J. Phys. (Condens. Matter. ), 15 (2004) R 481

Scaling challenges in MD n 〜 50, 000 FLOP/particle/step n Typical system size : N=105 n 5 GFLOP/step n 5 TFLOPS effective performance 1 msec/step = 170 nsec/day Rather Easy n 5 PFLOPS effective performance 1μsec/step = 200μsec/day? ? ? Difficult, but important

What is GRAPE? n GRAvity Pip. E n Special-purpose accelerator for classical particle simulations ▷ Astrophysical N-body simulations ▷ Molecular Dynamics Simulations J. Makino & M. Taiji, Scientific Simulations with Special-Purpose Computers, John Wiley & Sons, 1997.

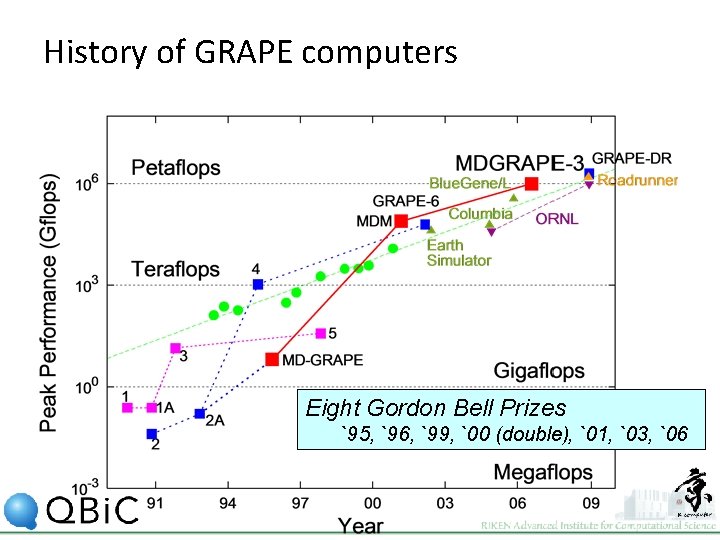
History of GRAPE computers Eight Gordon Bell Prizes `95, `96, `99, `00 (double), `01, `03, `06

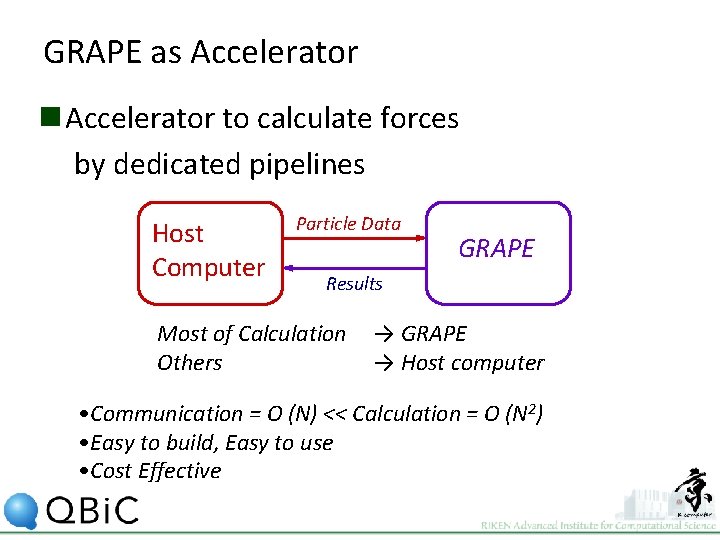
GRAPE as Accelerator n Accelerator to calculate forces by dedicated pipelines Host Computer Particle Data GRAPE Results Most of Calculation Others → GRAPE → Host computer • Communication = O (N) << Calculation = O (N 2) • Easy to build, Easy to use • Cost Effective

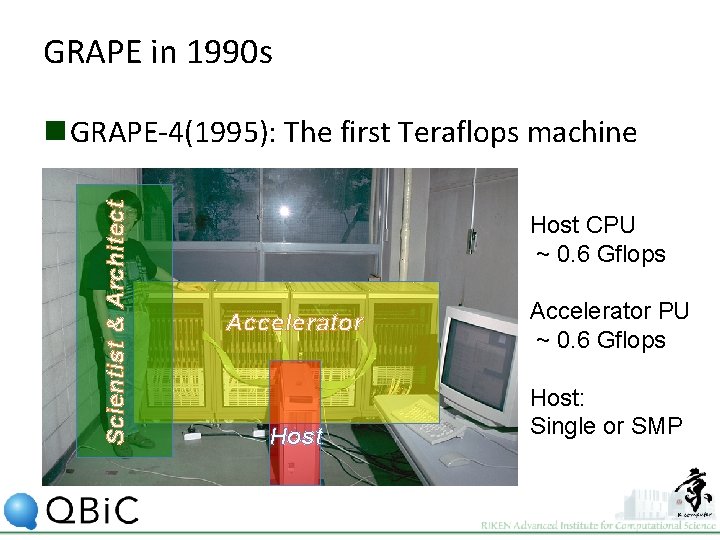
GRAPE in 1990 s Scientist & Architect n GRAPE-4(1995): The first Teraflops machine Host CPU ~ 0. 6 Gflops Accelerator Host Accelerator PU ~ 0. 6 Gflops Host: Single or SMP

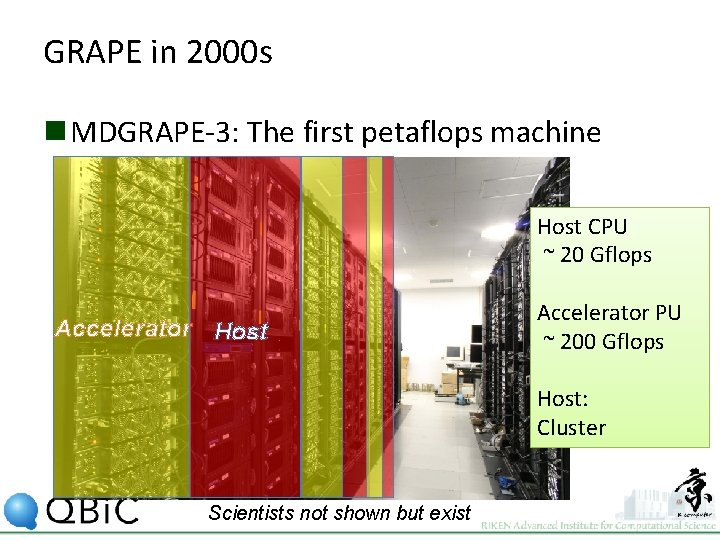
GRAPE in 2000 s n MDGRAPE-3: The first petaflops machine Host CPU ~ 20 Gflops Accelerator Host Accelerator PU ~ 200 Gflops Host: Cluster Scientists not shown but exist

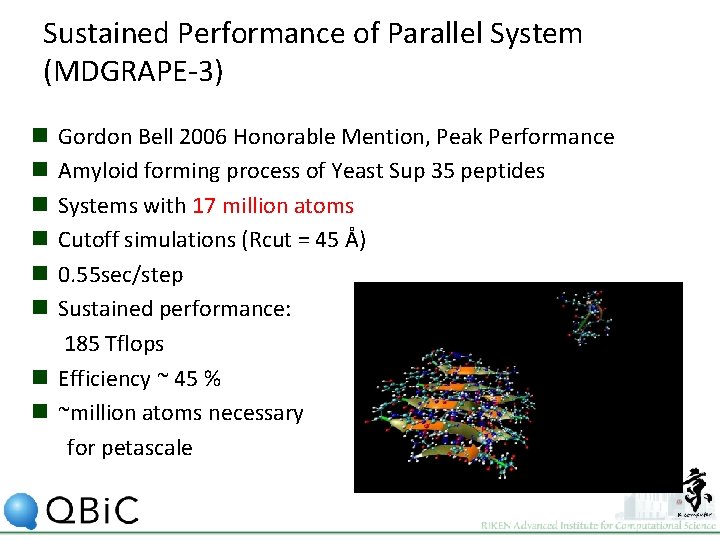
Sustained Performance of Parallel System (MDGRAPE-3) Gordon Bell 2006 Honorable Mention, Peak Performance Amyloid forming process of Yeast Sup 35 peptides Systems with 17 million atoms Cutoff simulations (Rcut = 45 Å) 0. 55 sec/step Sustained performance: 185 Tflops n Efficiency ~ 45 % n ~million atoms necessary for petascale n n n

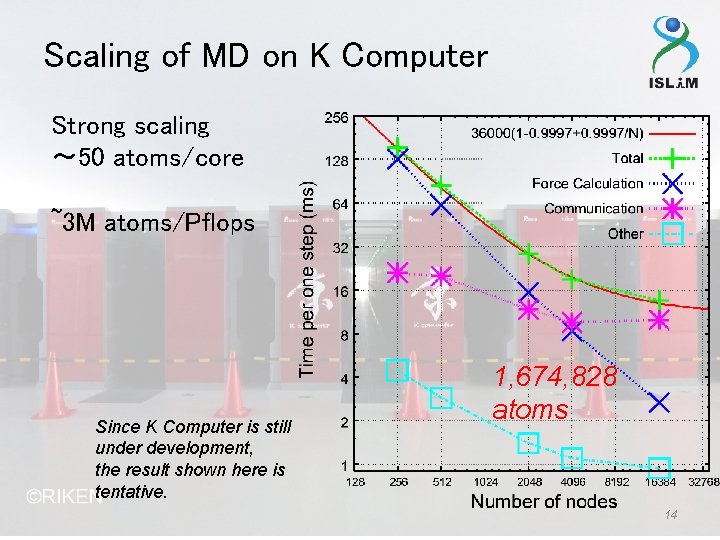
Scaling of MD on K Computer Strong scaling 〜 50 atoms/core ~3 M atoms/Pflops Since K Computer is still under development, the result shown here is tentative. 1, 674, 828 atoms 14

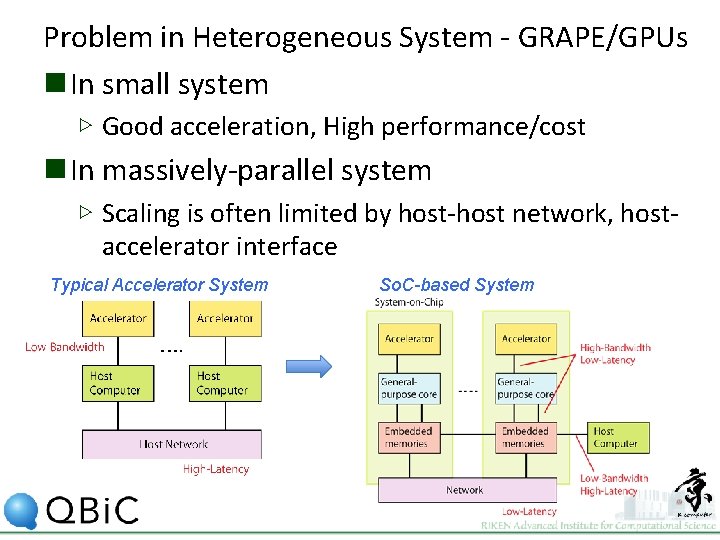
Problem in Heterogeneous System - GRAPE/GPUs n In small system ▷ Good acceleration, High performance/cost n In massively-parallel system ▷ Scaling is often limited by host-host network, hostaccelerator interface Typical Accelerator System So. C-based System

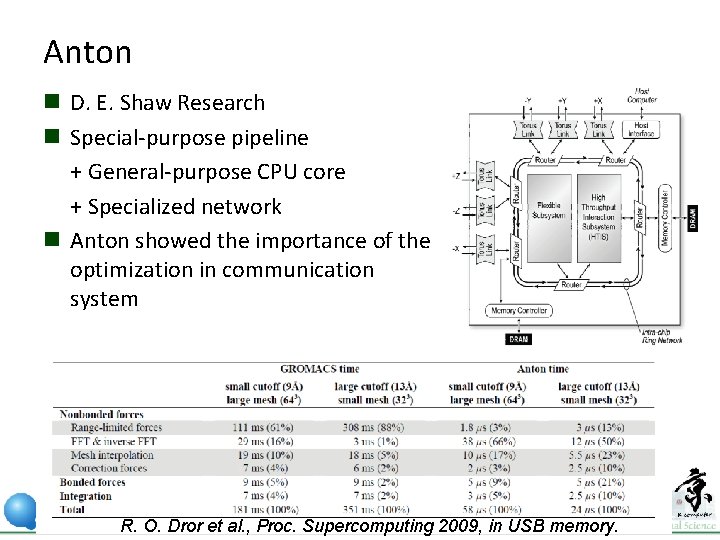
Anton n D. E. Shaw Research n Special-purpose pipeline + General-purpose CPU core + Specialized network n Anton showed the importance of the optimization in communication system R. O. Dror et al. , Proc. Supercomputing 2009, in USB memory.

MDGRAPE-4 n Special-purpose computer for MD simulation n Test platform for special-purpose machine n Target performance ▷ 10μsec/step for 100 K atom system – At this moment it seems difficult to achieve… ▷ 8. 6μsec/day (1 fsec/step) n Target application : GROMACS n Completion: ~2013 n Enhancement from MDGRAPE-3 ▷ 130 nm → 40 nm process ▷ Integration of Network / CPU n The design of the So. C is not finished yet, so we cannot report performance estimation.

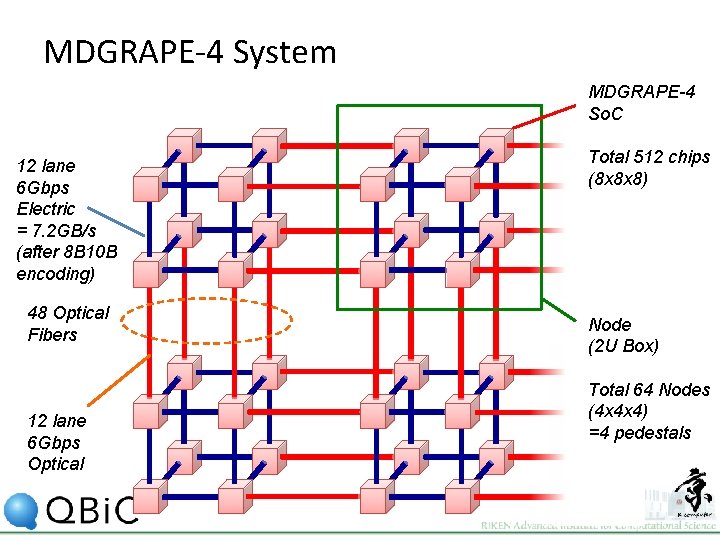
MDGRAPE-4 System MDGRAPE-4 So. C 12 lane 6 Gbps Electric = 7. 2 GB/s (after 8 B 10 B encoding) 48 Optical Fibers 12 lane 6 Gbps Optical Total 512 chips (8 x 8 x 8) Node (2 U Box) Total 64 Nodes (4 x 4 x 4) =4 pedestals

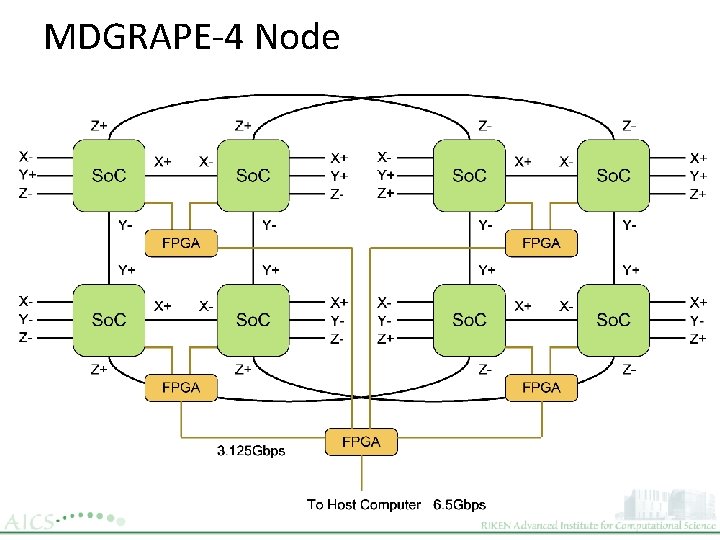
MDGRAPE-4 Node

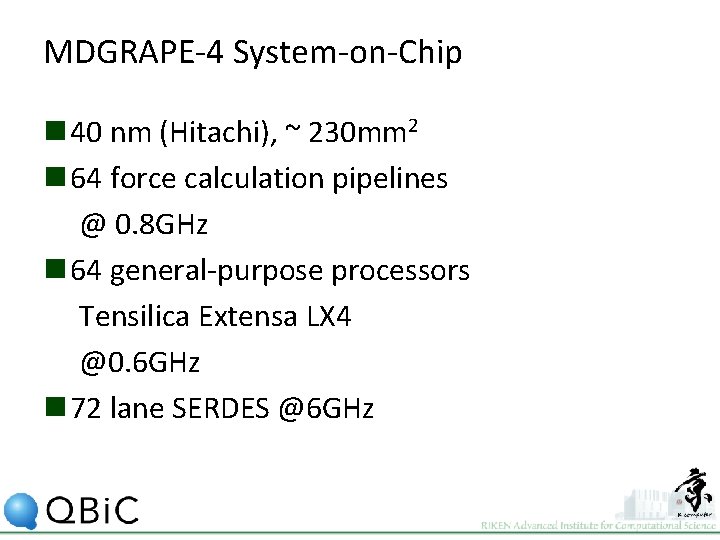
MDGRAPE-4 System-on-Chip n 40 nm (Hitachi), ~ 230 mm 2 n 64 force calculation pipelines @ 0. 8 GHz n 64 general-purpose processors Tensilica Extensa LX 4 @0. 6 GHz n 72 lane SERDES @6 GHz

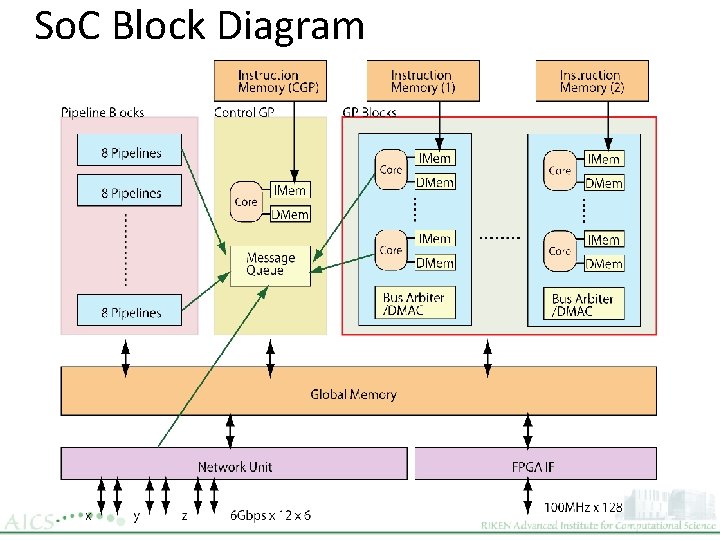
So. C Block Diagram

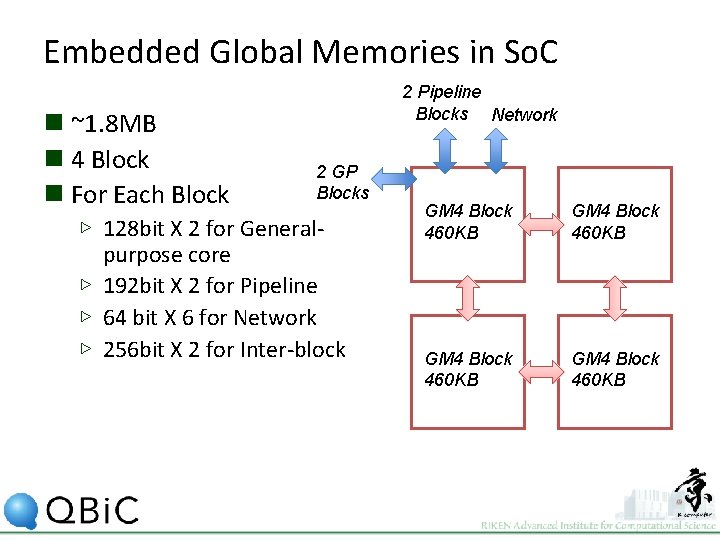
Embedded Global Memories in So. C n ~1. 8 MB n 4 Block n For Each Block 2 Pipeline Blocks Network 2 GP Blocks ▷ 128 bit X 2 for Generalpurpose core ▷ 192 bit X 2 for Pipeline ▷ 64 bit X 6 for Network ▷ 256 bit X 2 for Inter-block GM 4 Block 460 KB

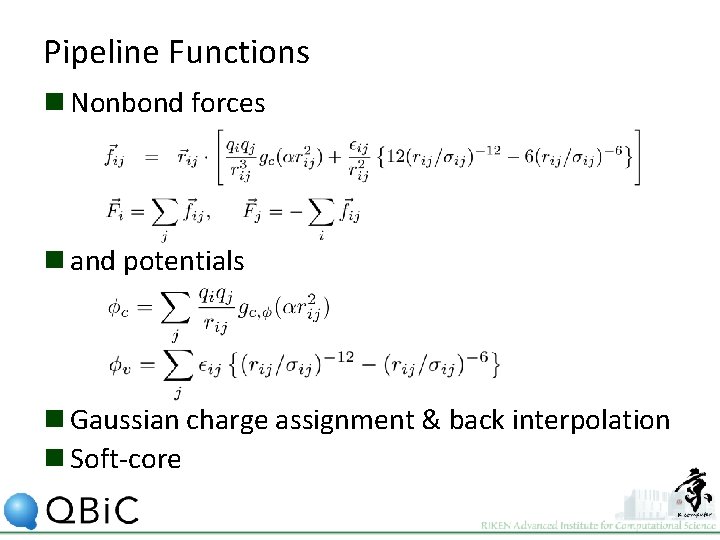
Pipeline Functions n Nonbond forces n and potentials n Gaussian charge assignment & back interpolation n Soft-core

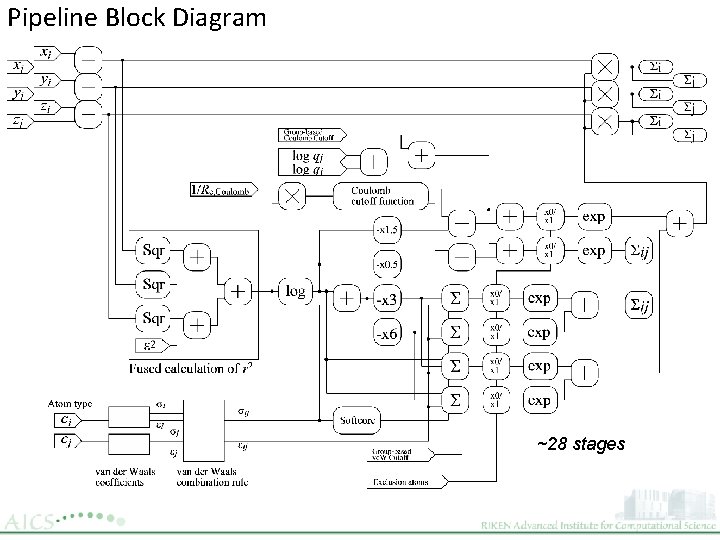
Pipeline Block Diagram ~28 stages

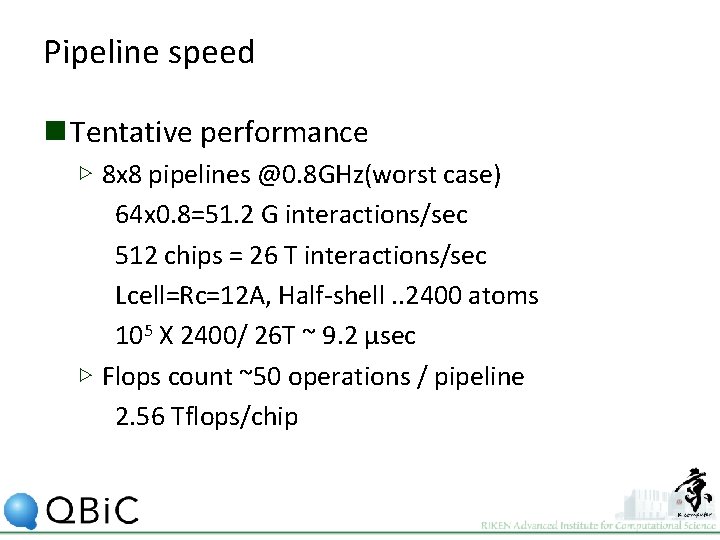
Pipeline speed n Tentative performance ▷ 8 x 8 pipelines @0. 8 GHz(worst case) 64 x 0. 8=51. 2 G interactions/sec 512 chips = 26 T interactions/sec Lcell=Rc=12 A, Half-shell. . 2400 atoms 105 X 2400/ 26 T ~ 9. 2 μsec ▷ Flops count ~50 operations / pipeline 2. 56 Tflops/chip

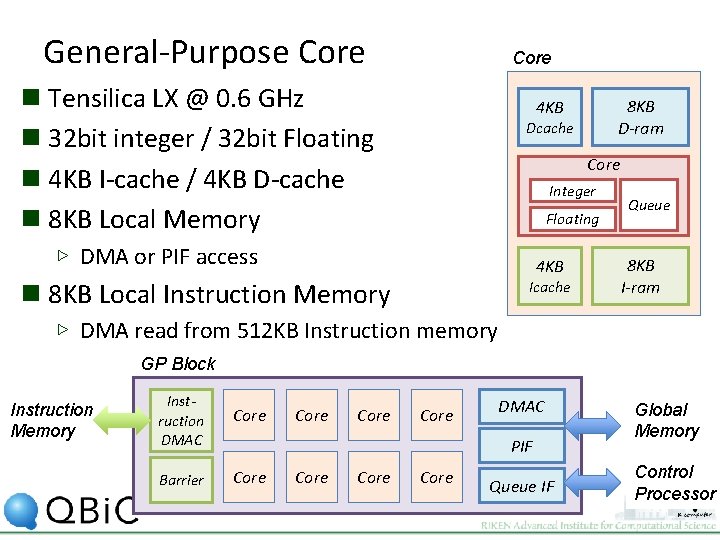
General-Purpose Core n Tensilica LX @ 0. 6 GHz n 32 bit integer / 32 bit Floating n 4 KB I-cache / 4 KB D-cache n 8 KB Local Memory 8 KB D-ram 4 KB Dcache Core Integer Floating ▷ DMA or PIF access 4 KB n 8 KB Local Instruction Memory Icache Queue 8 KB I-ram ▷ DMA read from 512 KB Instruction memory GP Block Instruction Memory Instruction DMAC Core Barrier Core DMAC PIF Core Queue IF Global Memory Control Processor

Synchronization n 8 -core synchronization unit n Tensilica Queue-based synchronization send messages ▷ Pipeline → Control GP ▷ Network IF → Control GP ▷ GP Block → Control GP n Synchronization at memory – accumulation at memory

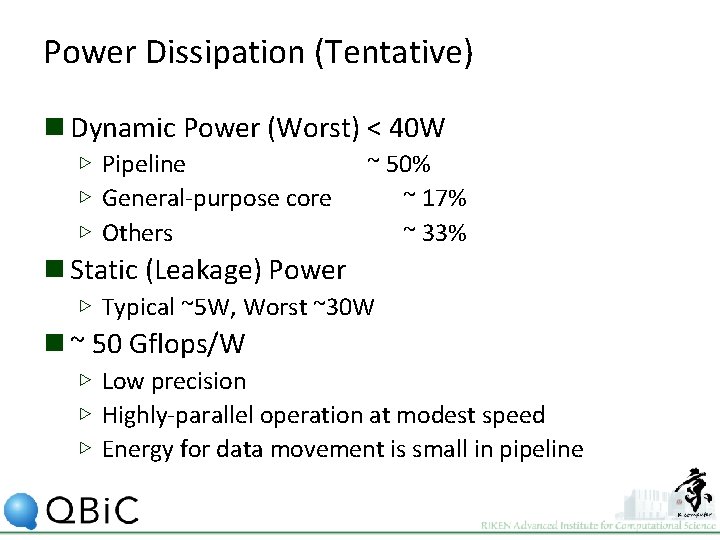
Power Dissipation (Tentative) n Dynamic Power (Worst) < 40 W ▷ Pipeline ▷ General-purpose core ▷ Others ~ 50% ~ 17% ~ 33% n Static (Leakage) Power ▷ Typical ~5 W, Worst ~30 W n ~ 50 Gflops/W ▷ Low precision ▷ Highly-parallel operation at modest speed ▷ Energy for data movement is small in pipeline

Reflection Though the design is not finished yet… n Latency in Memory Subsystem ▷ More distribution inside So. C n Latency in Network ▷ More intelligent Network controller n Pipeline / General-purpose balance ▷ Shift for general-purpose? ▷ # of Control GP

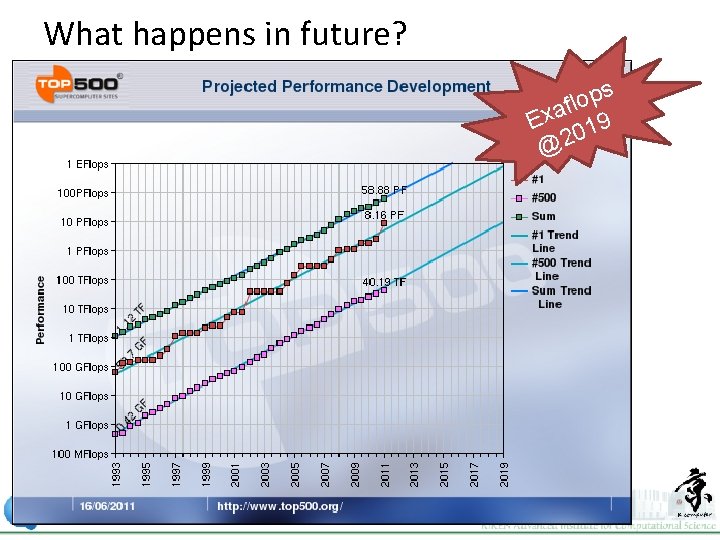
What happens in future? s p o l f a x E 019 @2

Merit of specialization(Repeat) n Low frequency / Highly parallel n Low cost for data movement ▷ Dedicated pipelines, for example n Dark silicon problem ▷ All transistors cannot be operated simultaneously due to power limitation ▷ Advantages of dedicated approach: – High power-efficiency – Little damage to general-purpose performance

Future Perspectives (1) n In life science fields ▷ High computing demand, but ▷ we do not have so many applications that scale to exaflops (even petascale is difficult) n Requirement for strong scaling ▷ Molecular Dynamics ▷ Quantum Chemistry (QM-MM) ▷ There remain problems to be solved in petascale

Future Perspectives (2) For Molecular Dynamics n Single-chip system ▷ >1/10 of the MDGRAPE-4 system can be embedded with 11 nm process ▷ For typical simulation system it will be the most convenient ▷ Still network is necessary inside So. C n For further strong scaling ▷ # of operations / step / 20 Katom ~ 109 ▷ # of arithmetic units in system ~ 106 /Pflops Exascale means “Flash” (one-path) calculation ▷ More specialization is required

Acknowledgements n RIKEN Mr. Itta Ohmura Dr. Gentaro Morimoto Dr. Yousuke Ohno n Japan IBM Service Mr. Ken Namura Mr. Mitsuru Sugimoto Mr. Masaya Mori Mr. Tsubasa Saitoh n Hitachi Co. Ltd. Mr. Iwao Yamazaki Mr. Tetsuya Fukuoka Mr. Makio Uchida Mr. Toru Kobayashi and many other staffs n Hitachi JTE Co. Ltd. Mr. Satoru Inazawa Mr. Takeshi Ohminato