SOME TRAINING ON NUCLEOTIDE SEQUENCES EDITION REGISTRATION ALIGNMENT















- Slides: 33

SOME TRAINING ON NUCLEOTIDE SEQUENCES: EDITION, REGISTRATION, ALIGNMENT AND TREE BUILDING Y. Ph. Kartavtsev A. V. Zhirmunsky Institute of Marine Biology of Far Eastern Branch of Russian Academy of Sciences, Vladivostok 690041, Russia, e-mail: yuri. kartavtsev 48@hotmail. com

ГЛАВНЫЕ ВОПРОСЫ 1. Sequence edition and their registration in Gen. Bank. 2. Data format and gene banks available. 3. Sequence alignment. 4. Finding an optimal model of nucleotide substitution. 5. Tree building with software package MEGA-3 (MEGA-4). 6. Annotation on PAUP, Mr. Bayes and some other programs.

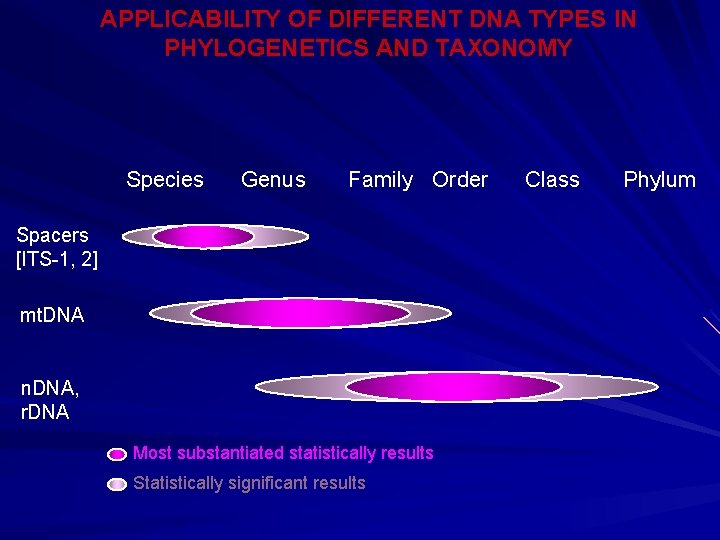
APPLICABILITY OF DIFFERENT DNA TYPES IN PHYLOGENETICS AND TAXONOMY Species Genus Family Order Class Phylum Spacers [ITS-1, 2] mt. DNA n. DNA, r. DNA Most substantiated statistically results Statistically significant results

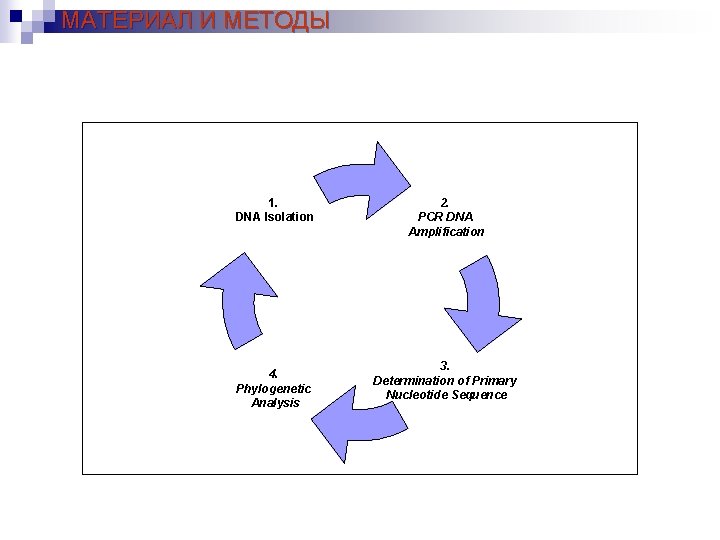
МАТЕРИАЛ И МЕТОДЫ 1. DNA Isolation 4. Phylogenetic Analysis 2. PCR DNA Amplification 3. Determination of Primary Nucleotide Sequence

1. SEQUENCE EDITION AND THEIR REGISTRATION IN THE GENBANK, NCBI (1) Original sequence that obtained from a sequencing machine requires an edition. Many requirement for the edition meet such program packages (PP) as MEGA-3 or MEGA-4 (http: //www. megasoftware. net/ ), Gene. DOC (http: //www. nrbsc. org/) etc. Most suitable PP tool for the primary edition is Chromas (Chromas-pro, that is available at http: //www. flu. org. cn/en or http: //www. technelysium. com. au/chromas. html ). Currently realized version (Chromas-pro 2. 31) let to perform a number of edition options. Opens chromatogram files from Applied Biosystems and Amersham Mega. Bace DNA sequencers. Opens SCF format chromatogram files created by ALF, Li-Cor, Visible Genetics Open. Gene, Beckman CEQ 2000 XL and CEQ 8000, and other sequencers. View Genescan genotype files. Save in SCF or Applied Biosystems format. Prints chromatogram with options to zoom or fit to one page. Exports sequences in plaint text, formatted with base numbering, FASTA, EMBL, Gen. Bank or GCG formats. Copy the sequence to the clipboard in plain text or FASTA format for pasting into other applications. Export sequences from batches of chromatogram files, with automatic removal of vector sequence. Reverse & complement the sequence and chromatogram. Search for sequences by exact matching or optimal alignment. Display translations in 3 frames along with the sequence. Copy an image of a chromatogram section for pasting into documents or presentations.

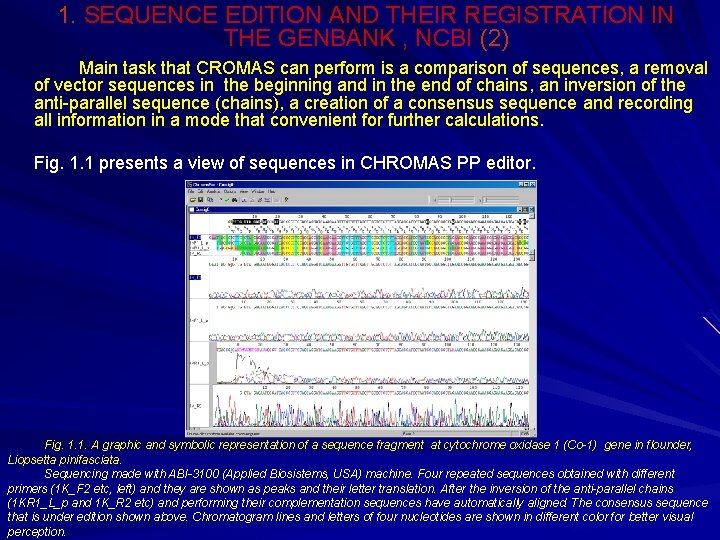
1. SEQUENCE EDITION AND THEIR REGISTRATION IN THE GENBANK , NCBI (2) Main task that CROMAS can perform is a comparison of sequences, a removal of vector sequences in the beginning and in the end of chains, an inversion of the anti-parallel sequence (chains), a creation of a consensus sequence and recording all information in a mode that convenient for further calculations. Fig. 1. 1 presents a view of sequences in CHROMAS PP editor. Fig. 1. 1. A graphic and symbolic representation of a sequence fragment at cytochrome oxidase 1 (Со-1) gene in flounder, Liopsetta pinifasciata. Sequencing made with АBI-3100 (Applied Biosistems, USA) machine. Four repeated sequences obtained with different primers (1 K_F 2 etc, left) and they are shown as peaks and their letter translation. After the inversion of the anti-parallel chains (1 KR 1_L_p and 1 K_R 2 etc) and performing their complementation sequences have automatically aligned. The consensus sequence that is under edition shown above. Chromatogram lines and letters of four nucleotides are shown in different color for better visual perception.

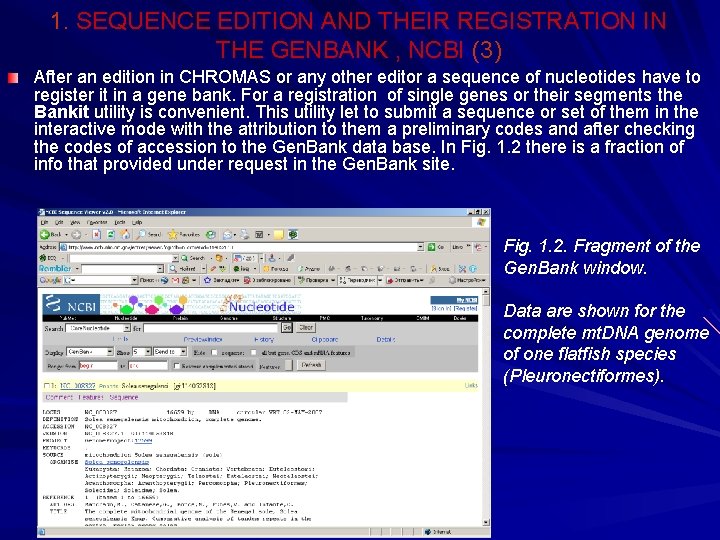
1. SEQUENCE EDITION AND THEIR REGISTRATION IN THE GENBANK , NCBI (3) After an edition in CHROMAS or any other editor a sequence of nucleotides have to register it in a gene bank. For a registration of single genes or their segments the Bankit utility is convenient. This utility let to submit a sequence or set of them in the interactive mode with the attribution to them a preliminary codes and after checking the codes of accession to the Gen. Bank data base. In Fig. 1. 2 there is a fraction of info that provided under request in the Gen. Bank site. Fig. 1. 2. Fragment of the Gen. Bank window. Data are shown for the complete mt. DNA genome of one flatfish species (Pleuronectiformes).

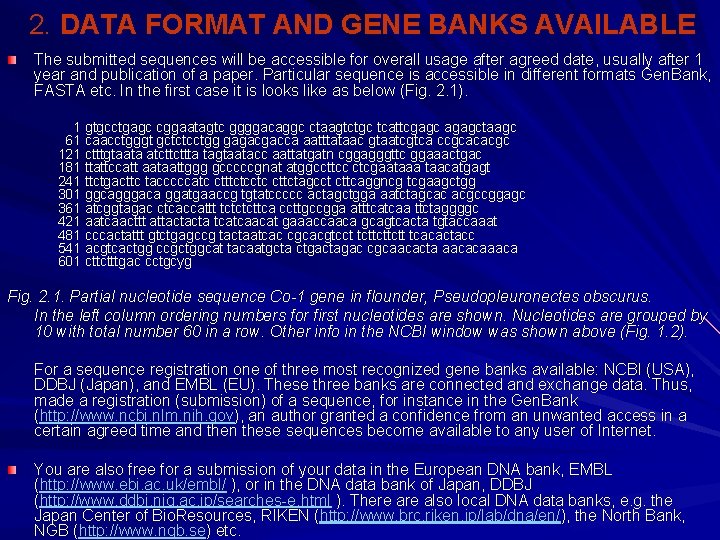
2. DATA FORMAT AND GENE BANKS AVAILABLE The submitted sequences will be accessible for overall usage after agreed date, usually after 1 year and publication of a paper. Particular sequence is accessible in different formats Gen. Bank, FASTA etc. In the first case it is looks like as below (Fig. 2. 1). 1 gtgcctgagc cggaatagtc ggggacaggc ctaagtctgc tcattcgagc agagctaagc 61 caacctgggt gctctcctgg gagacgacca aatttataac gtaatcgtca ccgcacacgc 121 ctttgtaata atcttcttta tagtaatacc aattatgatn cggagggttc ggaaactgac 181 ttattccatt aataattggg gcccccgnat atggccttcc ctcgaataaa taacatgagt 241 ttctgacttc tacccccatc ctttctcctc cttctagcct cttcaggncg tcgaagctgg 301 ggcagggaca ggatgaaccg tgtatccccc actagctgga aatctagcac acgccggagc 361 atcggtagac ctcaccattt tctctcttca ccttgccgga atttcatcaa ttctaggggc 421 aatcaacttt attactacta tcatcaacat gaaaccaaca gcagtcacta tgtaccaaat 481 cccactattt gtctgagccg tactaatcac cgcacgtcct tcttcttctt tcacactacc 541 acgtcactgg ccgctggcat tacaatgcta ctgactagac cgcaacacta aacacaaaca 601 cttctttgac cctgcyg Fig. 2. 1. Partial nucleotide sequence Со-1 gene in flounder, Pseudopleuronectes obscurus. In the left column ordering numbers for first nucleotides are shown. Nucleotides are grouped by 10 with total number 60 in a row. Other info in the NCBI window was shown above (Fig. 1. 2). For a sequence registration one of three most recognized gene banks available: NCBI (USA), DDBJ (Japan), and EMBL (EU). These three banks are connected and exchange data. Thus, made a registration (submission) of a sequence, for instance in the Gen. Bank (http: //www. ncbi. nlm. nih. gov), an author granted a confidence from an unwanted access in a certain agreed time and then these sequences become available to any user of Internet. You are also free for a submission of your data in the European DNA bank, EMBL (http: //www. ebi. ac. uk/embl/ ), or in the DNA data bank of Japan, DDBJ (http: //www. ddbj. nig. ac. jp/searches-e. html ). There also local DNA data banks, e. g. the Japan Center of Bio. Resources, RIKEN (http: //www. brc. riken. jp/lab/dna/en/), the North Bank, NGB (http: //www. ngb. se) etc.

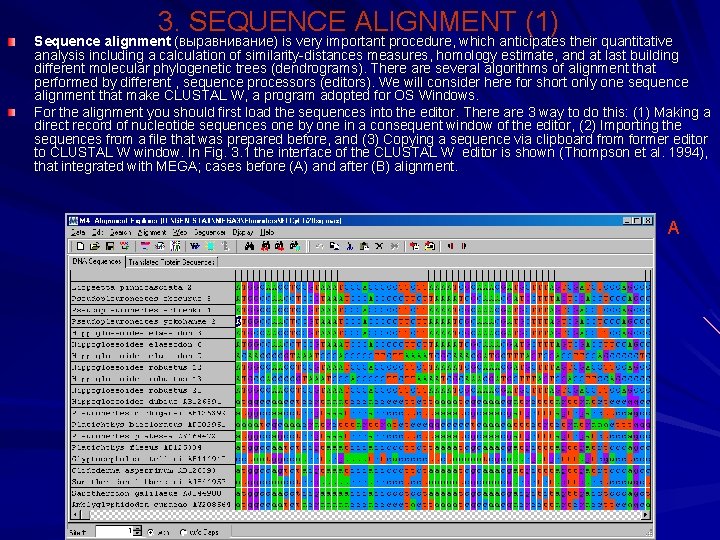
3. SEQUENCE ALIGNMENT (1) Sequence alignment (выравнивание) is very important procedure, which anticipates their quantitative analysis including a calculation of similarity-distances measures, homology estimate, and at last building different molecular phylogenetic trees (dendrograms). There are several algorithms of alignment that performed by different , sequence processors (editors). We will consider here for short only one sequence alignment that make CLUSTAL W, a program adopted for OS Windows. For the alignment you should first load the sequences into the editor. There are 3 way to do this: (1) Making a direct record of nucleotide sequences one by one in a consequent window of the editor, (2) Importing the sequences from a file that was prepared before, and (3) Copying a sequence via clipboard from former editor to CLUSTAL W window. In Fig. 3. 1 the interface of the CLUSTAL W editor is shown (Thompson et al. 1994), that integrated with MEGA; cases before (А) and after (В) alignment. А

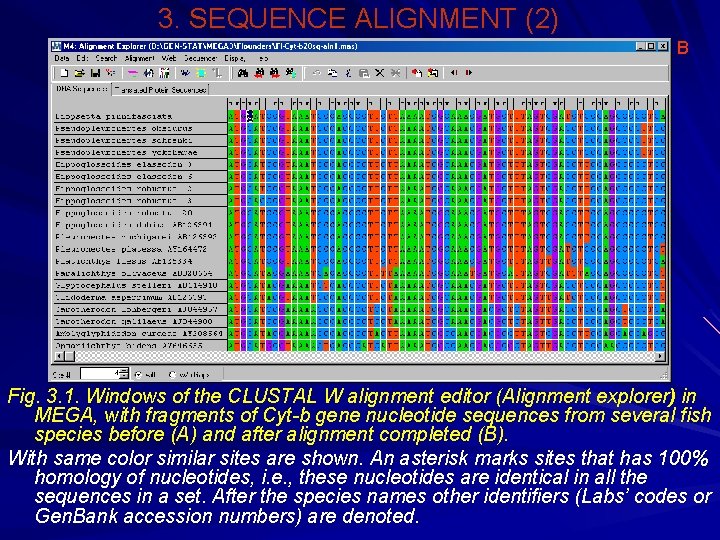
3. SEQUENCE ALIGNMENT (2) В Fig. 3. 1. Windows of the CLUSTAL W alignment editor (Alignment explorer) in MEGA, with fragments of Сyt-b gene nucleotide sequences from several fish species before (А) and after alignment completed (В). With same color similar sites are shown. An asterisk marks sites that has 100% homology of nucleotides, i. e. , these nucleotides are identical in all the sequences in a set. After the species names other identifiers (Labs’ codes or Gen. Bank accession numbers) are denoted.

3. SEQUENCE ALIGNMENT (3) In the above case the sequences were loaded via clipboard (Fig. 3. 1). Make run of MEGA-3 (MEGA-4), we can chouse in the main menu: Alignment explorer/Clustal Create a new alignment ( «выравнивание» «редактор выравнивания/Clustal» «создать новое выравнивание» ). In the last options there actually 3 possibilities: Create a new alignment ( «создать новое выравнивание» ), Open a saved alignment session ( «открыть сохраненную сессию выравнивания» ), Retrieve sequence from a file ( «вывести последовательность из файла» ). When sequences are loaded, an author meets, as a rule, with a dimension problem: sequences length is unequal and their starts & ends are not complemented; more over, many sequences have deletions/insertions (Gaps), which are not coincide in different individuals and species. Alignment allows to solve all these problems.

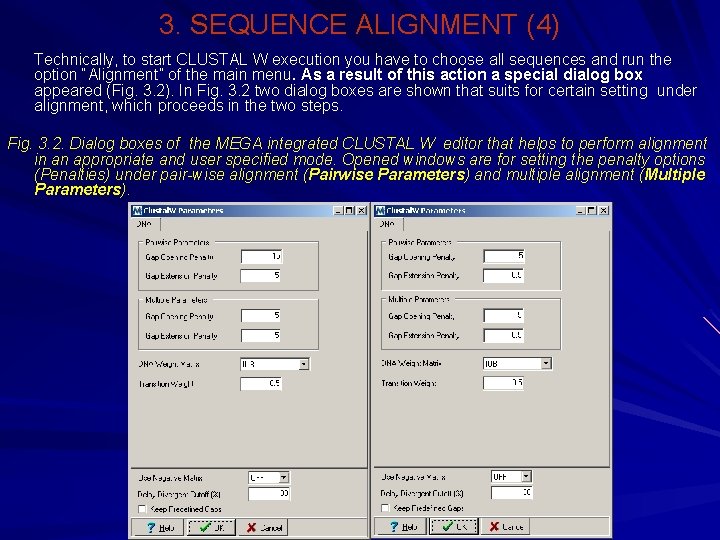
3. SEQUENCE ALIGNMENT (4) Technically, to start CLUSTAL W execution you have to choose all sequences and run the option “Alignment” of the main menu. As a result of this action a special dialog box appeared (Fig. 3. 2). In Fig. 3. 2 two dialog boxes are shown that suits for certain setting under alignment, which proceeds in the two steps. Fig. 3. 2. Dialog boxes of the MEGA integrated CLUSTAL W editor that helps to perform alignment in an appropriate and user specified mode. Opened windows are for setting the penalty options (Penalties) under pair-wise alignment (Pairwise Parameters) and multiple alignment (Multiple Parameters).

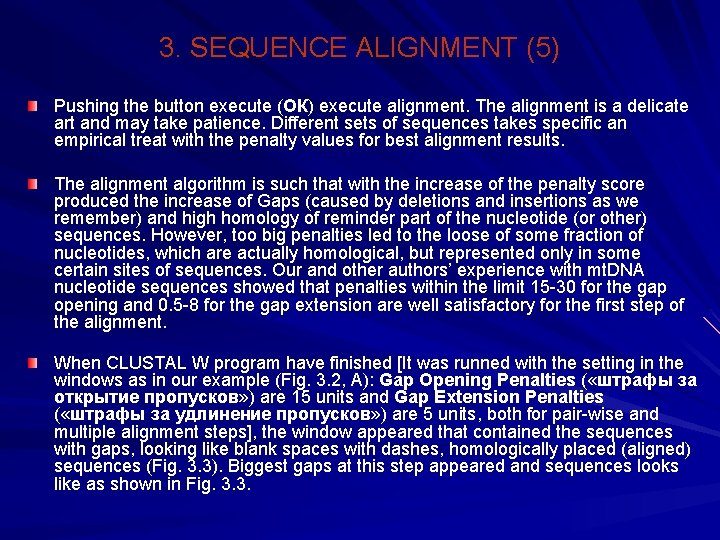
3. SEQUENCE ALIGNMENT (5) Pushing the button execute (ОК) execute alignment. The alignment is a delicate art and may take patience. Different sets of sequences takes specific an empirical treat with the penalty values for best alignment results. The alignment algorithm is such that with the increase of the penalty score produced the increase of Gaps (caused by deletions and insertions as we remember) and high homology of reminder part of the nucleotide (or other) sequences. However, too big penalties led to the loose of some fraction of nucleotides, which are actually homological, but represented only in some certain sites of sequences. Our and other authors’ experience with mt. DNA nucleotide sequences showed that penalties within the limit 15 -30 for the gap opening and 0. 5 -8 for the gap extension are well satisfactory for the first step of the alignment. When CLUSTAL W program have finished [It was runned with the setting in the windows as in our example (Fig. 3. 2, А): Gap Opening Penalties ( «штрафы за открытие пропусков» ) are 15 units and Gap Extension Penalties ( «штрафы за удлинение пропусков» ) are 5 units, both for pair-wise and multiple alignment steps], the window appeared that contained the sequences with gaps, looking like blank spaces with dashes, homologically placed (aligned) sequences (Fig. 3. 3). Biggest gaps at this step appeared and sequences looks like as shown in Fig. 3. 3.

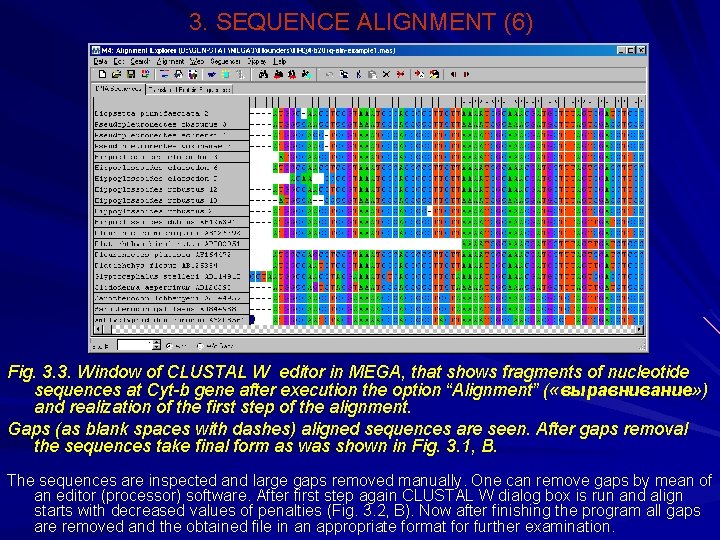
3. SEQUENCE ALIGNMENT (6) Fig. 3. 3. Window of CLUSTAL W editor in MEGA, that shows fragments of nucleotide sequences at Сyt-b gene after execution the option “Alignment” ( «выравнивание» ) and realization of the first step of the alignment. Gaps (as blank spaces with dashes) aligned sequences are seen. After gaps removal the sequences take final form as was shown in Fig. 3. 1, В. The sequences are inspected and large gaps removed manually. One can remove gaps by mean of an editor (processor) software. After first step again CLUSTAL W dialog box is run and align starts with decreased values of penalties (Fig. 3. 2, В). Now after finishing the program all gaps are removed and the obtained file in an appropriate format for further examination.

4. FINDING AN OPTIMAL MODEL OF NUCLEOTIDE SUBSTITUTION (1) For choosing a model that is most suitable for particular empirical data sets you need some tool. The MODELTEST 3. 06 (Posada, Grandal, 1998) program and later versions 3. 6 - 3. 7 are very convenient for that. I could not present here info about models but you can easily know on model properties in the program manual and in the literature (Nei, Kumar, 2000; Hall, 2001; Sanderson, Shaffer, 2003; Felsenstein, 2004); there is also a brief info in my book (Kartavtsev, 2005). To use MODELTEST you have to learn firstly the PAUP PP, because this program uses some of PAUP modules. The work with the program is basically simple and includes 5 steps.

4. FINDING AN OPTIMAL MODEL OF NUCLEOTIDE SUBSTITUTION (2) 1. First you must make a working file in the Nexus (. nex) format with the nucleotide sequences and necessary identifiers of the program parameters, in acordance with the PAUP demands; 2. Next you should reach the MODELTEST website and load all recommended modules and copy in the nexus-file made before the file “modelblock. PAUPb 10. txt”, which is distributed with the MODELTEST (it suits for PAUP 4 b 10 version for Windows); 3. Run then PAUP 4 b 10 installed before (better to rename original data file) and start the execution of the working file; 4. When program stops normally, in the same directory (folder), from which working file have been executed, the new file will appeared with the name “model. scores”; 5. Now it is necessary to run the program, MODELTEST 3. 7 is best, from an OS DOS window; better to do this from the directory that contain executable file “modeltest 3. 7. win. exe”. Consequent identifiers in the command line will be as follows: “modeltest 3. 7. exe <model. scores> test. out” (last output file may have an arbitrary name). In the output file all necessary information will be presented and the parameters of one or two best fit models of 57 estimated model types will be given as well; testing is performed by the Maximum Likelihood (ML) algorithm and by the Acaike Information Criteria.

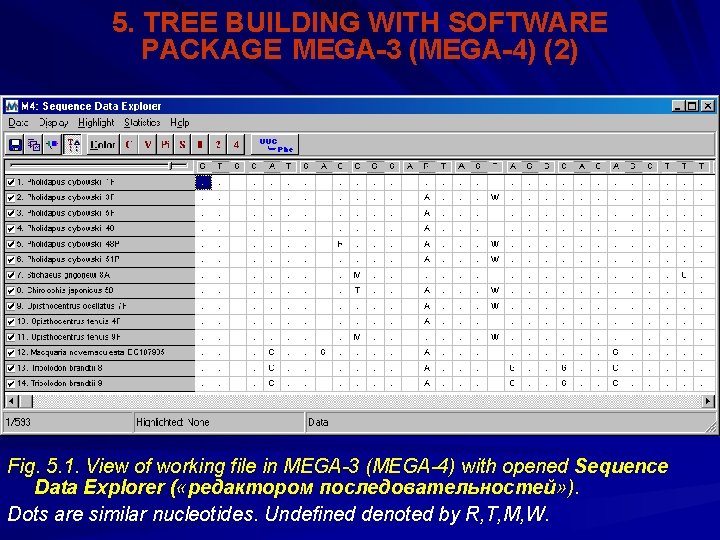
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (1) Options and model parameters as well model themselves for calculation of molecular phylogenetic tress are provided by different programs: PAUP* (Swofford, 2000), MEGA-3 (MEGA-4) (Kumar et al. , 1993; 2000) etc. Book by Hall (2001) is very good manual for a molecular phylogenetic analysis. This manual is focused mainly on PAUP*. However, in the book the exact examples available and recommendations are given on PP CLUSTAL X, Mr. Bayes etc. Beginning an analytical job in MEGA-3 and MEGA-4 may be accomplished right after alignment completed. Closing saved file in the Alignment Explorer (редактора выравнивания; it has the extension. mas). Under this action a window appear with a notice: “Save data to MEGA file: Yes, No, Cancel’ ( «сохранить файл для MEGA» , с опциями: «да» , «нет» , «сброс» ). Choosing the option “YES” opens the next window with the file name ready to be saved on the hard disk. By default the file name is supposed same as the alignment file, but with different extension: “. meg”. By choosing the option save ( «сохранить» ), we run the MEGA PP itself. Before openning the meg-file for the execution, it is necessary to note in the opened window, what sequence is processed: “Protein-coding nucleotide sequence data” ( «данные с белок-кодирующей нуклеотидной последовательностью» ), with the alternative YES or NO. At last the dialog box appeared with the question: “ Open Data File in MEGA ( «открыть файл с данными в MEGA» ), YES, NO. In a choose YES we get MEGA working file, following by opening a special editor “Sequence Data Explorer” ( «редактора последовательностей» ) (Fig. 5. 1).

5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (2) Fig. 5. 1. View of working file in MEGA-3 (MEGA-4) with opened Sequence Data Explorer ( «редактором последовательностей» ). Dots are similar nucleotides. Undefined denoted by R, T, M, W.

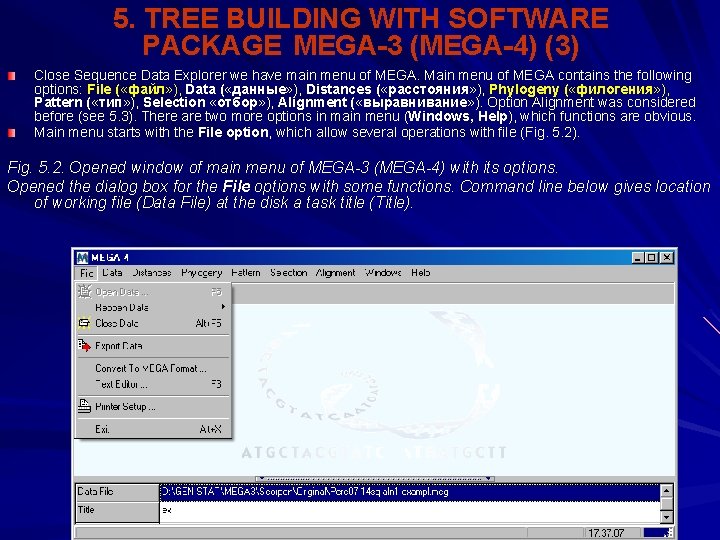
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (3) Close Sequence Data Explorer we have main menu of MEGA. Main menu of MEGA contains the following options: File ( «файл» ), Data ( «данные» ), Distances ( «расстояния» ), Phylogeny ( «филогения» ), Pattern ( «тип» ), Selection «отбор» ), Alignment ( «выравнивание» ). Option Alignment was considered before (see 5. 3). There are two more options in main menu (Windows, Help), which functions are obvious. Main menu starts with the File option, which allow several operations with file (Fig. 5. 2). Fig. 5. 2. Opened window of main menu of MEGA-3 (MEGA-4) with its options. Opened the dialog box for the File options with some functions. Command line below gives location of working file (Data File) at the disk a task title (Title).

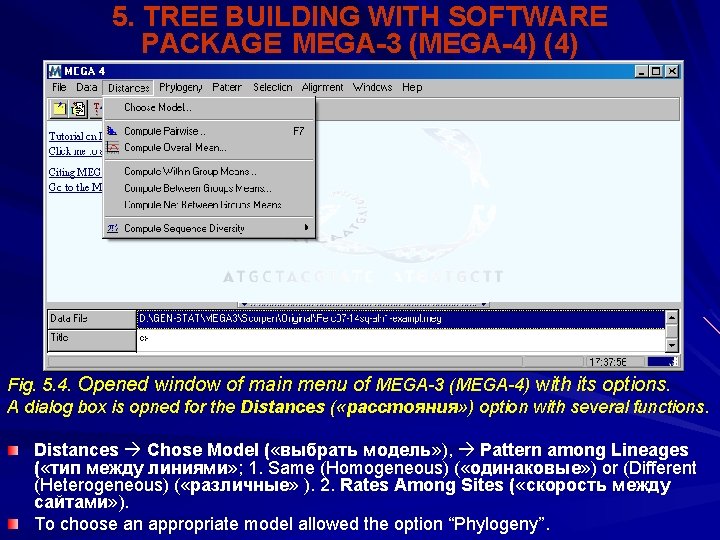
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (4) Fig. 5. 4. Opened window of main menu of MEGA-3 (MEGA-4) with its options. A dialog box is opned for the Distances ( «расстояния» ) option with several functions. Distances Chose Model ( «выбрать модель» ), Pattern among Lineages ( «тип между линиями» ; 1. Same (Homogeneous) ( «одинаковые» ) or (Different (Heterogeneous) ( «различные» ). 2. Rates Among Sites ( «скорость между сайтами» ). To choose an appropriate model allowed the option “Phylogeny”.

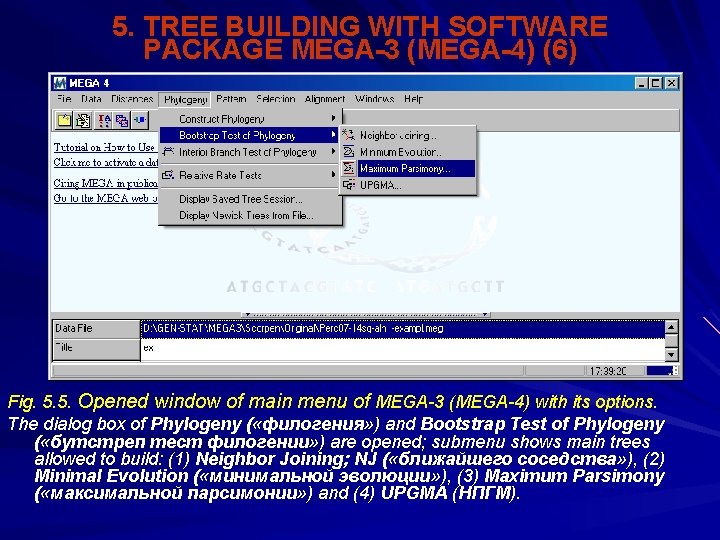
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (5) Next option in main menu is Phylogeny ( «филогения» ) (Fig. 5. 5). Actions: Construct Phylogeny ( «построить филогению» ), or Bootstrap Test of Phylogeny ( «бутстреп тест филогении» ); give the access to 4 different programs of tree building. From up to bottom that are: (1) Neighbor Joining; NJ ( «ближайшего соседства» ), (2) Minimal Evolution ( «минимальной эволюции» ), (3) Maximum Parsimony ( «максимальной парсимонии» ) and (4) UPGMA (НПГМА). Comments.

5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (6) Fig. 5. 5. Opened window of main menu of MEGA-3 (MEGA-4) with its options. The dialog box of Phylogeny ( «филогения» ) and Bootstrap Test of Phylogeny ( «бутстреп тест филогении» ) are opened; submenu shows main trees allowed to build: (1) Neighbor Joining; NJ ( «ближайшего соседства» ), (2) Minimal Evolution ( «минимальной эволюции» ), (3) Maximum Parsimony ( «максимальной парсимонии» ) and (4) UPGMA (НПГМ).

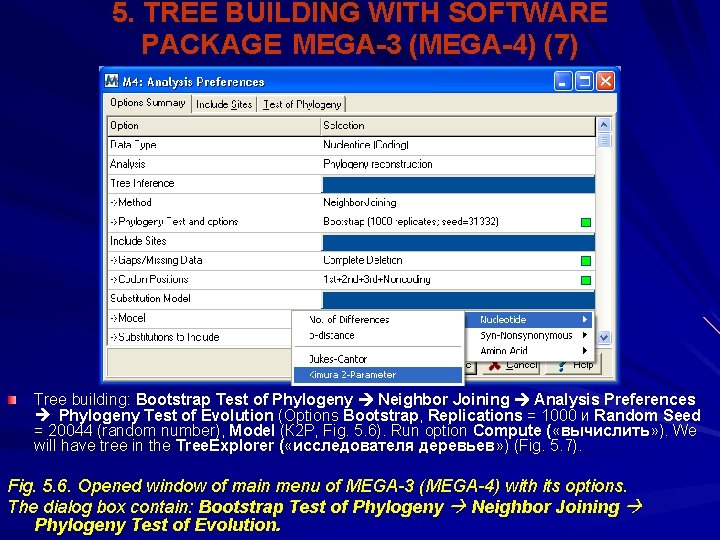
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (7) Tree building: Bootstrap Test of Phylogeny Neighbor Joining Analysis Preferences Phylogeny Test of Evolution (Options Bootstrap, Replications = 1000 и Random Seed = 20044 (random number), Model (К 2 Р, Fig. 5. 6). Run option Compute ( «вычислить» ). We will have tree in the Tree. Explorer ( «исследователя деревьев» ) (Fig. 5. 7). Fig. 5. 6. Opened window of main menu of MEGA-3 (MEGA-4) with its options. The dialog box contain: Bootstrap Test of Phylogeny Neighbor Joining Phylogeny Test of Evolution.

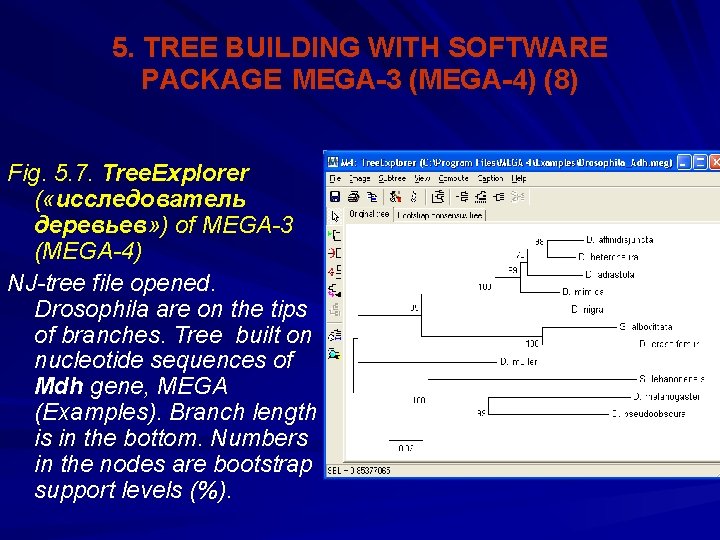
5. TREE BUILDING WITH SOFTWARE PACKAGE MEGA-3 (MEGA-4) (8) Fig. 5. 7. Tree. Explorer ( «исследователь деревьев» ) of MEGA-3 (MEGA-4) NJ-tree file opened. Drosophila are on the tips of branches. Tree built on nucleotide sequences of Mdh gene, MEGA (Examples). Branch length is in the bottom. Numbers in the nodes are bootstrap support levels (%).

6. ANNOTATION ON PAUP, MRBAYES AND SOME OTHER PROGRAMS Other widely used PP are PAUP 4. 0, Mr. Bayes, PHYLIP etc. PAUP 4. 0 (Swofford, 2002): Macintosh ( «Макинтош» ). This PAUP 4. 0 version explained in Hall (2001; 2003). For OS Windows there is PAUP 4. 0 10 b. PAUP 4. 0 is very important tool (MODELTEST!). Main its PP: Maximum Likelihood, ML, NJ- and MP Trees. Sustainability of tree quality is fine in PAUP. Time in ML is bad property of PAUP; 67 seq at Cyt-b (Kartavtsev et al. , 2007 a), took 3 weeks. There is PAUP for Linux/Unix. Mr. Bayes (Hulsenbeck, Rondquist, 2001; Ronquist, Huelsenbeck, 2003) is relatively small PP. Very effective. Set of 67 seq was processed during 2 days. Bayesian trees are MCMC based trees. Mr. Bayes provides other opportunities, say phylogenetic trees based on morphology. Mr. Bayes is not able to drow a tree. PP Tree. View (Page, 1996) is necessary to view a tree and build a consensus tree. PP PHYLIP (Felsenstein, 1995) is very good tool too. Theoretic background is fine for it (Felsenstein, 2004). PHYLIP gives opportunity to build main trees. Interface is for OS DOS not very convenient.

THANKS!




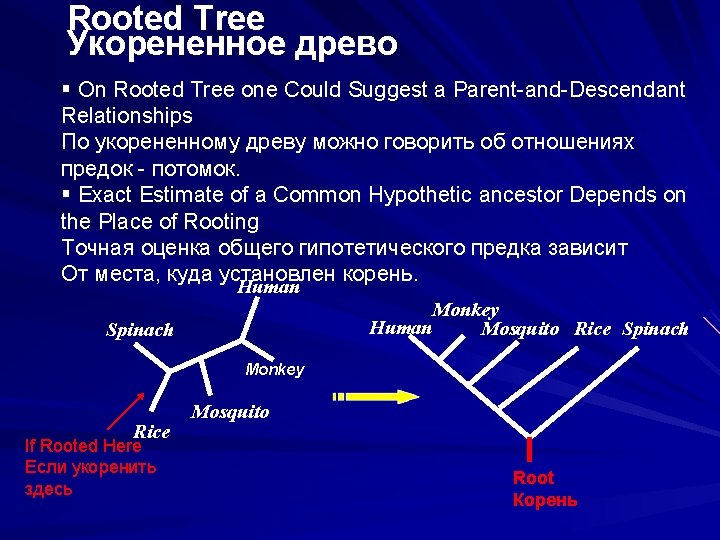
Rooted Tree Укорененное древо § On Rooted Tree one Could Suggest a Parent-and-Descendant Relationships По укорененному древу можно говорить об отношениях предок - потомок. § Exact Estimate of a Common Hypothetic ancestor Depends on the Place of Rooting Точная оценка общего гипотетического предка зависит От места, куда установлен корень. Human Spinach Monkey Human Mosquito Rice Spinach Monkey Rice If Rooted Here Если укоренить здесь Mosquito Root Корень

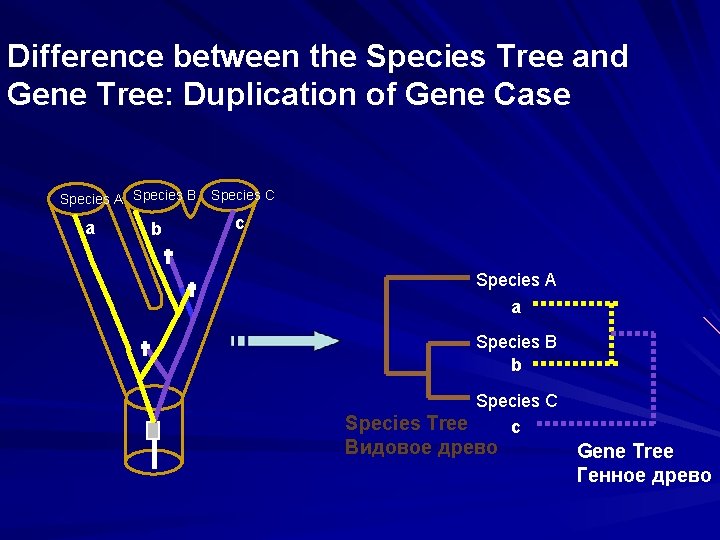
Difference between the Species Tree and Gene Tree: Duplication of Gene Case Species A Species B a b Species C c Species A a Species B b Species C Species Tree c Видовое древо Gene Tree Генное древо


Sequence Submission to the Gen. Bank Подписка последовательностей в Gen. Bank (NCBI)