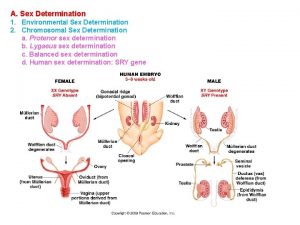
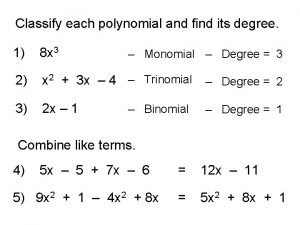Sex Chromosomes X and Y chromosomes that determine





















- Slides: 68


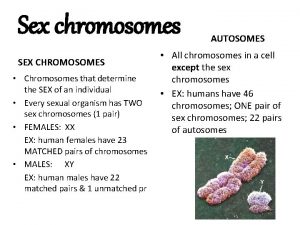
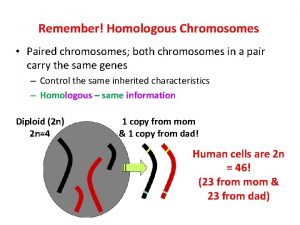
Sex Chromosomes. . . ‘X’ and ‘Y’ chromosomes that determine the sex of an individual in many organisms, Females: XX Males: XY

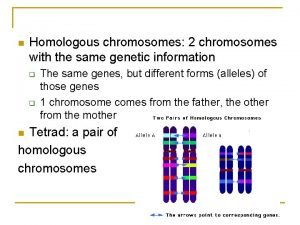
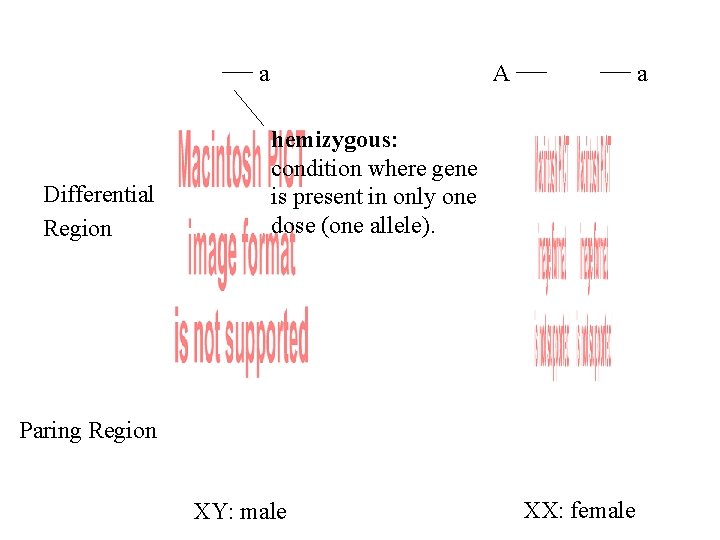
a Differential Region A a hemizygous: condition where gene is present in only one dose (one allele). Differential Region Paring Region XY: male XX: female

X Linkage …the pattern of inheritance resulting from genes located on the X chromosome. X-Linked Genes… …refers specifically to genes on the Xchromosome, with no homologs on the Y chromosome.

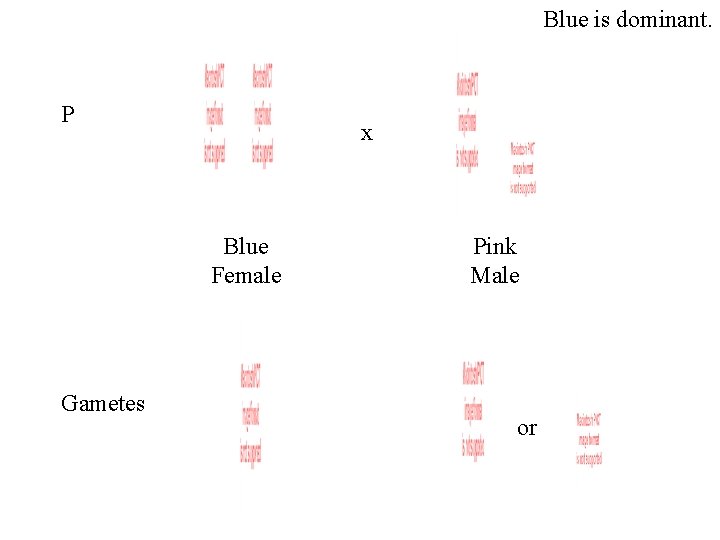
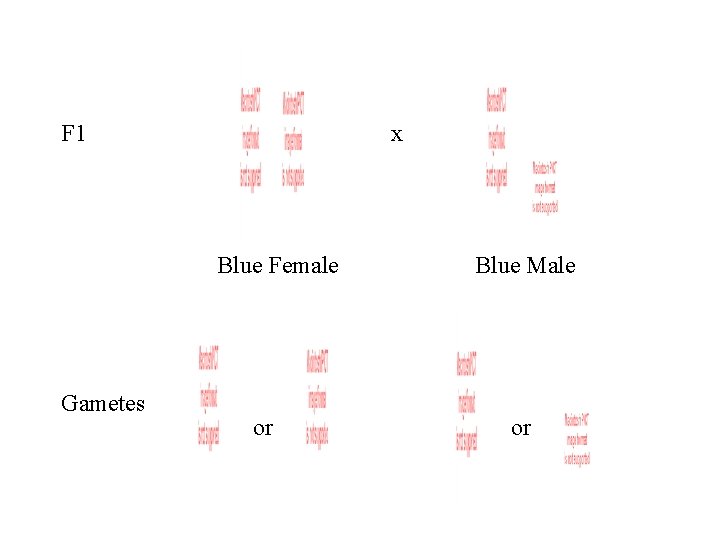
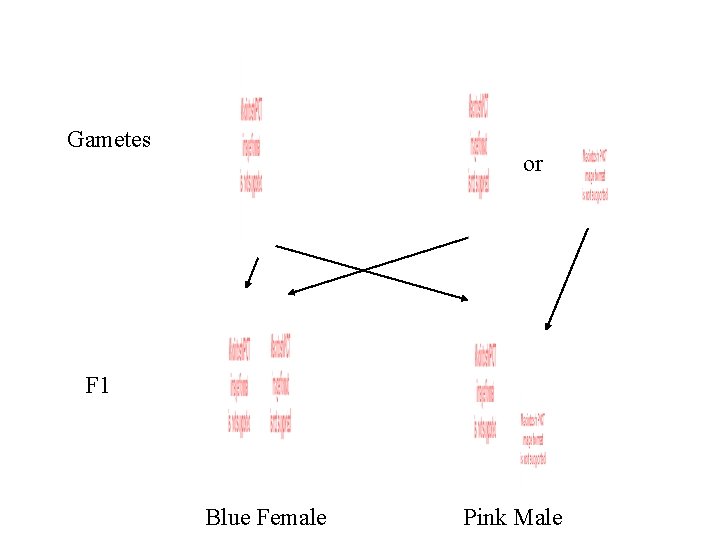
Blue is dominant. P x Blue Female Gametes Pink Male or

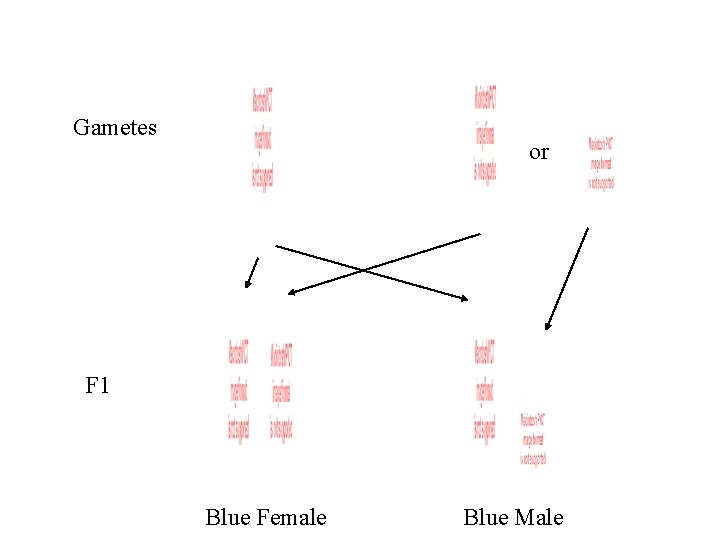
Gametes or F 1 Blue Female Blue Male

F 1 x Blue Female Gametes or Blue Male or

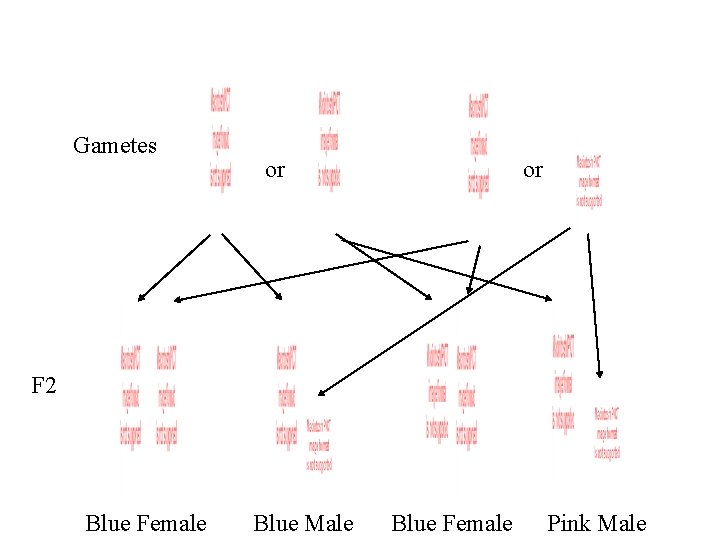
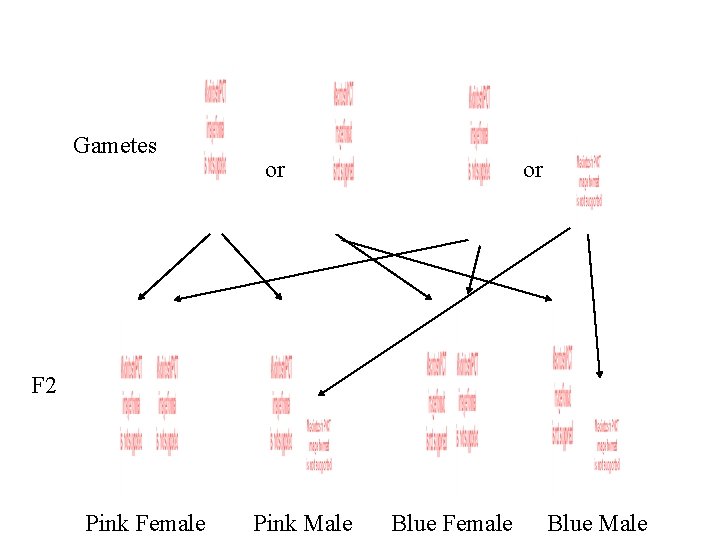
Gametes or or F 2 Blue Female Blue Male Blue Female Pink Male

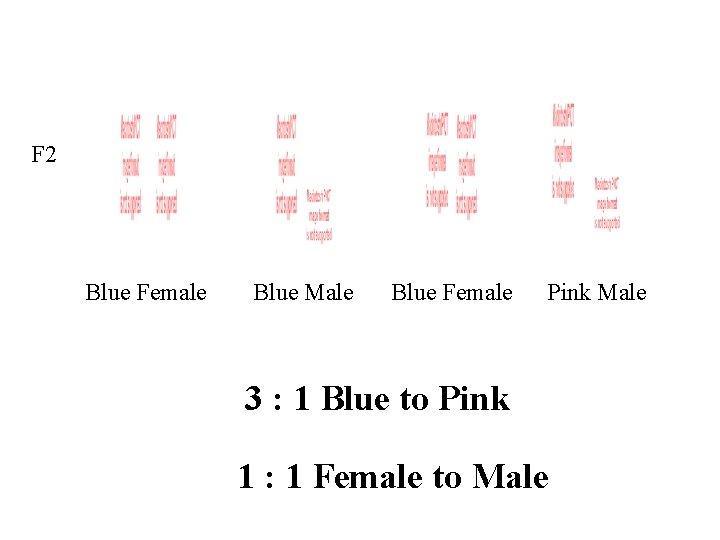
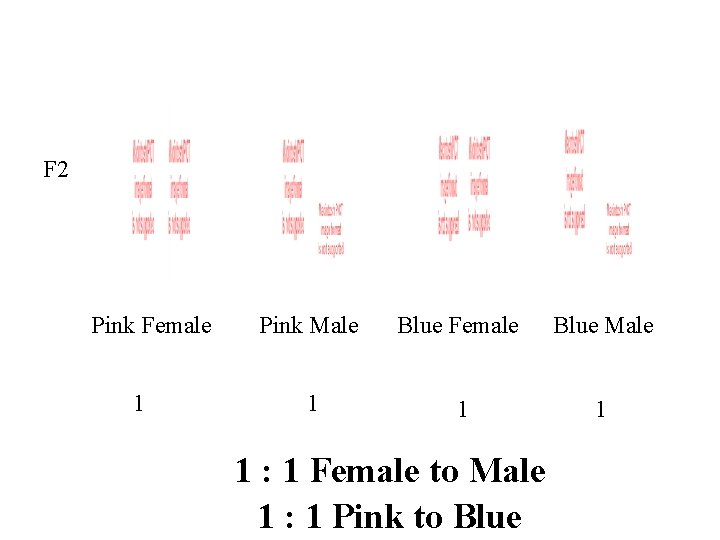
F 2 Blue Female Blue Male Blue Female Pink Male 3 : 1 Blue to Pink 1 : 1 Female to Male

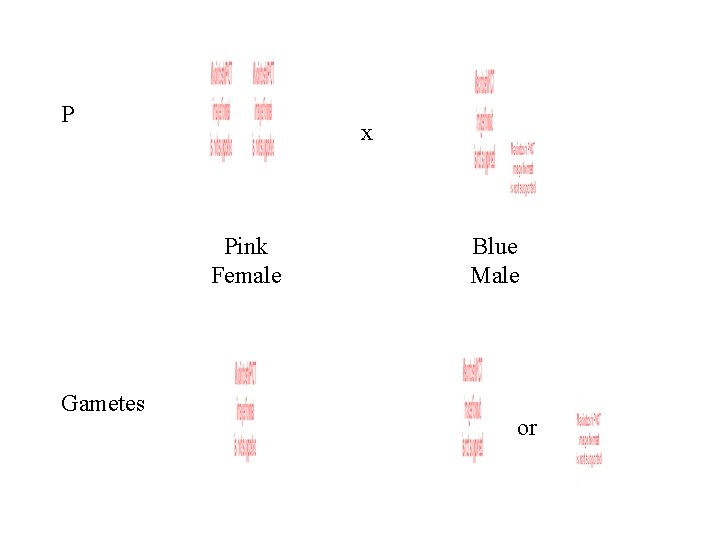
P x Pink Female Gametes Blue Male or

Gametes or F 1 Blue Female Pink Male

Gametes or or F 2 Pink Female Pink Male Blue Female Blue Male

F 2 Pink Female 1 Pink Male Blue Female Blue Male 1 1 : 1 Female to Male 1 : 1 Pink to Blue

Sex Linkage to Ponder • Female is homozygous recessive X-linked gene, – what percentage of male offspring will express? – what percentage of female offspring will express if, • mate is hemizygous for the recessive allele? • mate is hemizygous for the dominant allele? • Repeat at home with female heterozygous Xlinked gene!

Sex-Linked vs. Autosomal • autosomal chromosome: non-sex linked chromosome, • autosomal gene: a gene on an autosomal chromosome, • autosomes segregate identically in reciprocal crosses.

X-Linked Recessive Traits Characteristics • Many more males than females show the phenotype, – female must have both parents carrying the allele, – male only needs a mother with the allele, • Very few (or none) of the offspring of affected males show the disorder, – all of his daughters are carriers, • roughly half of the sons born to these daughters are carriers.


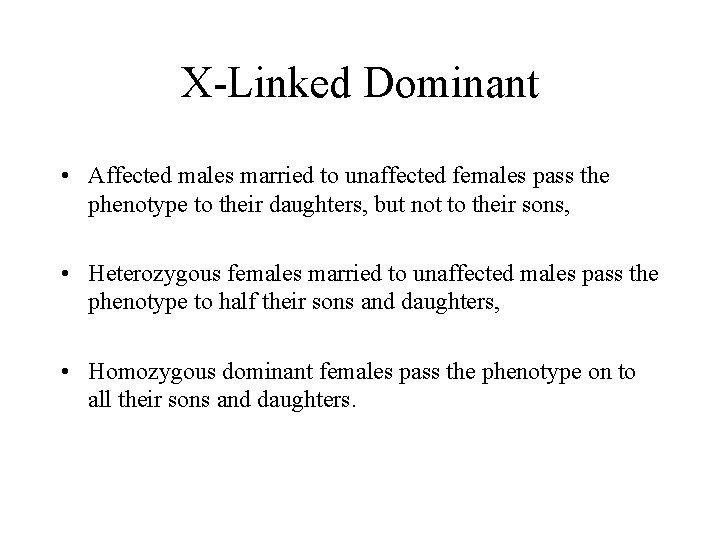
X-Linked Dominant • Affected males married to unaffected females pass the phenotype to their daughters, but not to their sons, • Heterozygous females married to unaffected males pass the phenotype to half their sons and daughters, • Homozygous dominant females pass the phenotype on to all their sons and daughters.


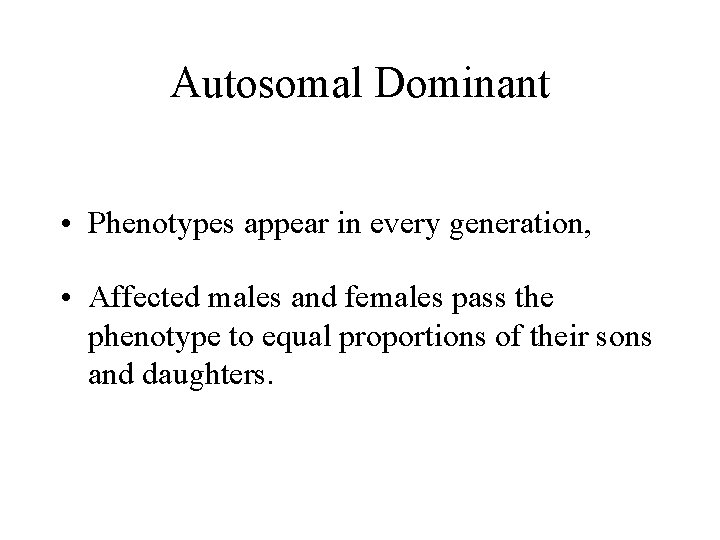
Autosomal Dominant • Phenotypes appear in every generation, • Affected males and females pass the phenotype to equal proportions of their sons and daughters.



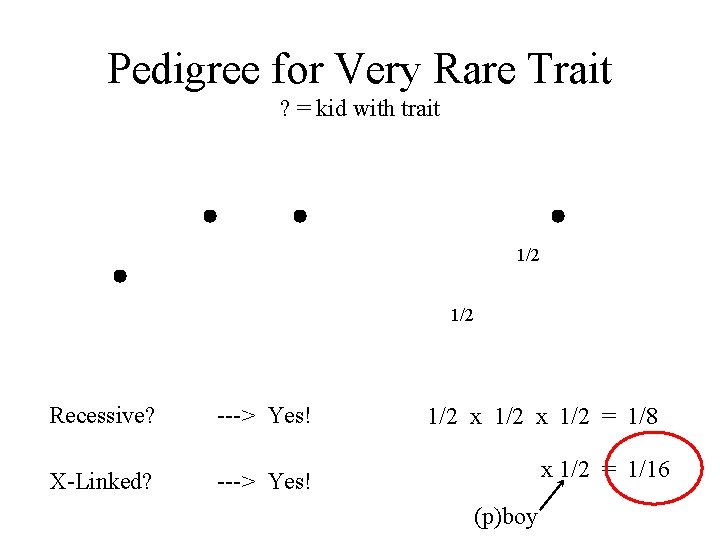
Pedigree for Very Rare Trait ? = kid with trait 1/2 Recessive? ---> Yes! X-Linked? Autosomal? ---> Yes! 1/2 x 1/2 ? = 1/8 x 1/2 = 1/16 (p)boy

X-Linked Dominant examples (OMIM) • HYPOPHOSPHATEMIA: “Vitamin-D resistant Rickett’s”, • LISSENCEPHALY: “smooth brain”, • FRAGILE SITE MENTAL RETARDATION: mild retardation, • RETT Syndrome: neurological disorder, • More on OMIM…

Linkage • Genes linked on the same chromosome may segregate together.

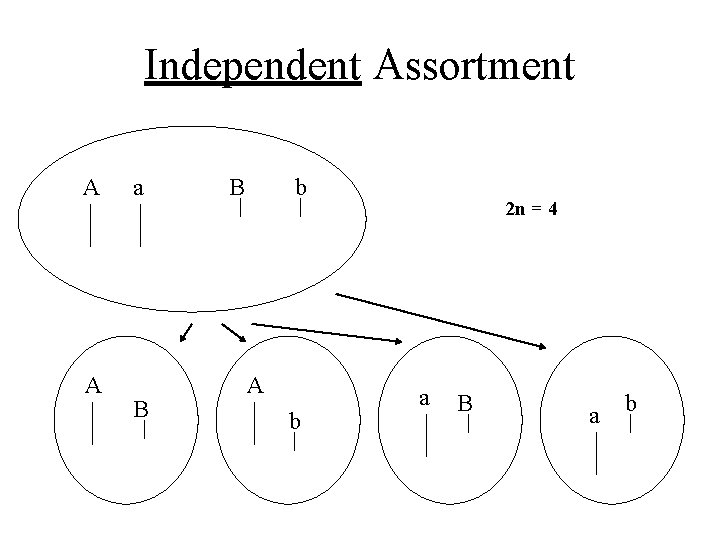
Independent Assortment A A a B B b A b 2 n = 4 a B a b

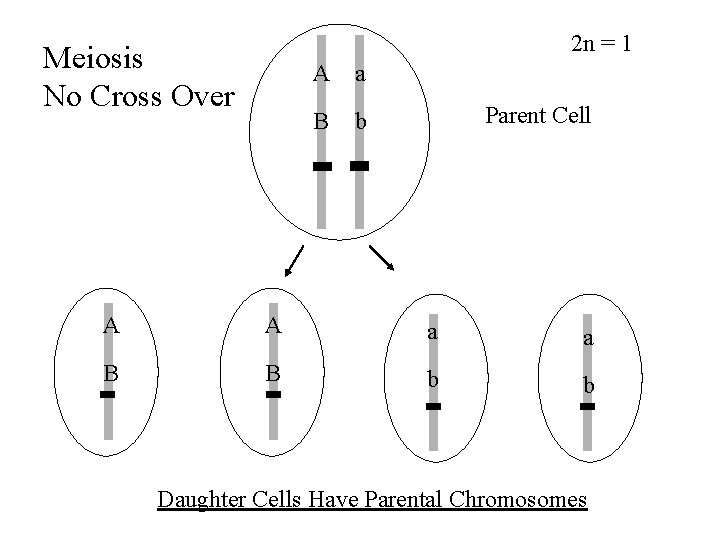
2 n = 1 Meiosis No Cross Over A a B b Parent Cell A A a a B B b b Daughter Cells Have Parental Chromosomes

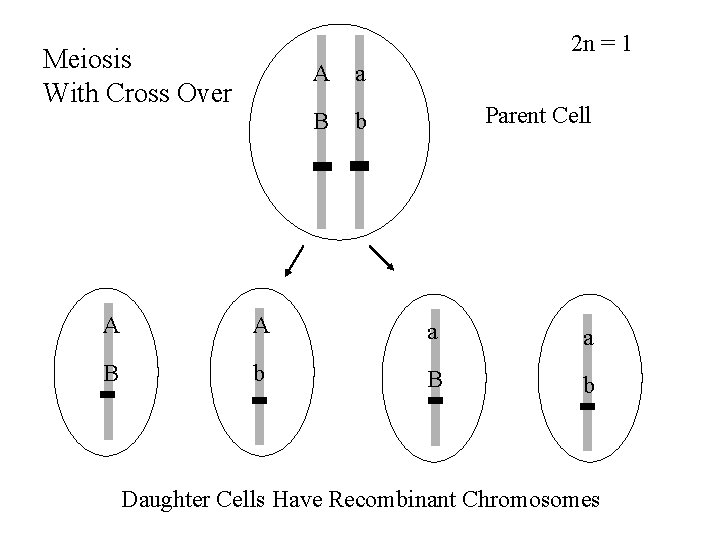
2 n = 1 Meiosis With Cross Over A a B b Parent Cell A A a a B b Daughter Cells Have Recombinant Chromosomes

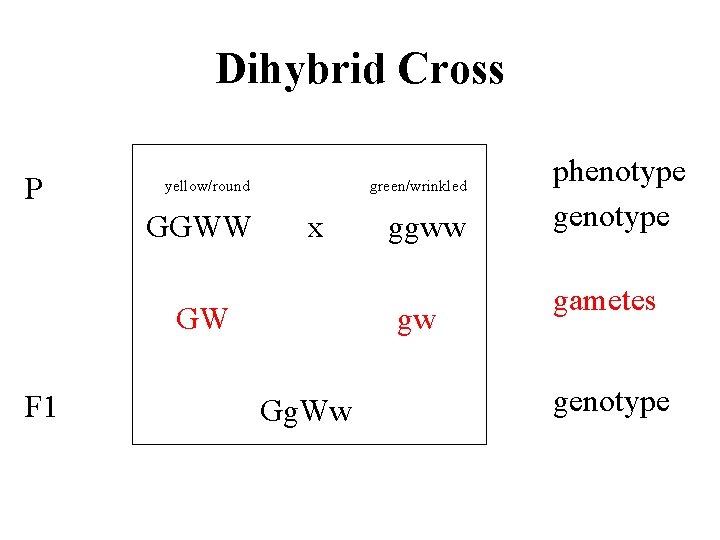
Dihybrid Cross P yellow/round GGWW green/wrinkled x GW F 1 ggww gw Gg. Ww phenotype gametes genotype

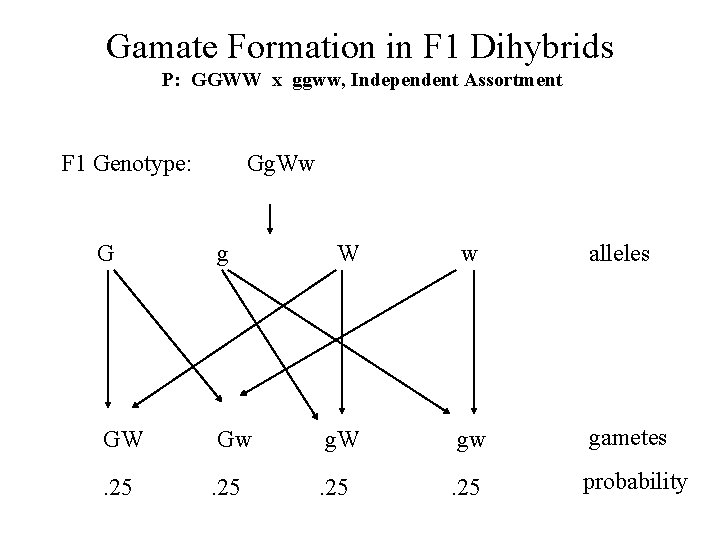
Gamate Formation in F 1 Dihybrids P: GGWW x ggww, Independent Assortment F 1 Genotype: Gg. Ww G g W w alleles GW Gw g. W gw gametes . 25 probability

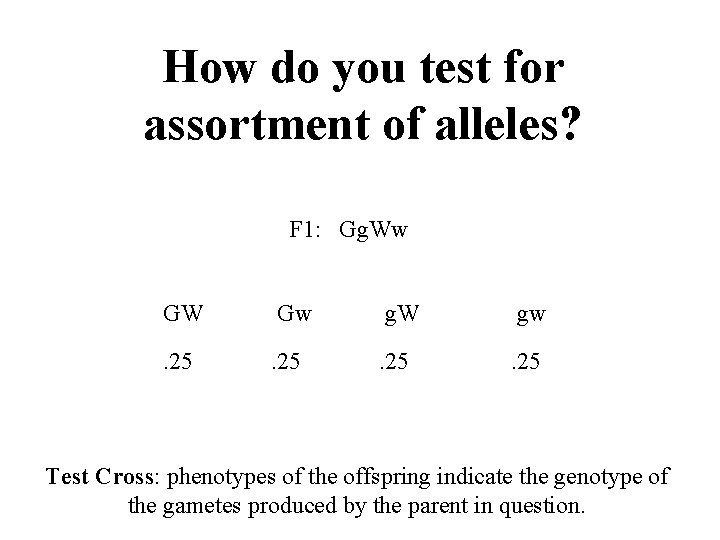
How do you test for assortment of alleles? F 1: Gg. Ww GW Gw g. W gw . 25 Test Cross: phenotypes of the offspring indicate the genotype of the gametes produced by the parent in question.

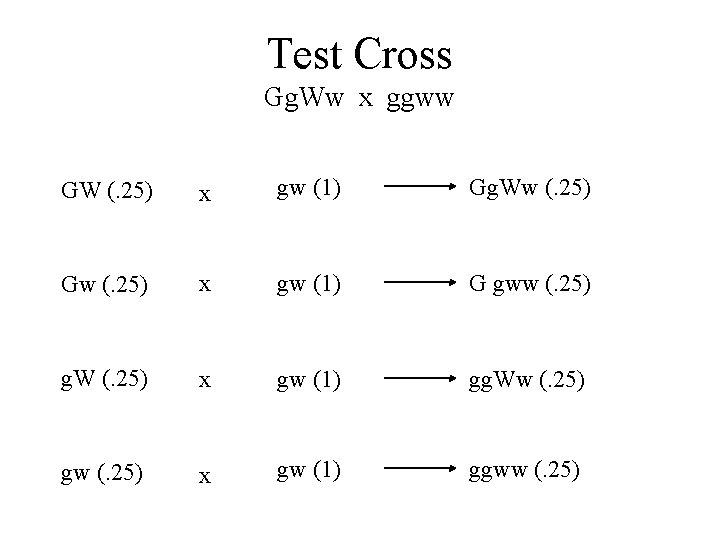
Test Cross Gg. Ww x ggww GW (. 25) x gw (1) Gg. Ww (. 25) Gw (. 25) x gw (1) G gww (. 25) g. W (. 25) x gw (1) gg. Ww (. 25) gw (. 25) x gw (1) ggww (. 25)

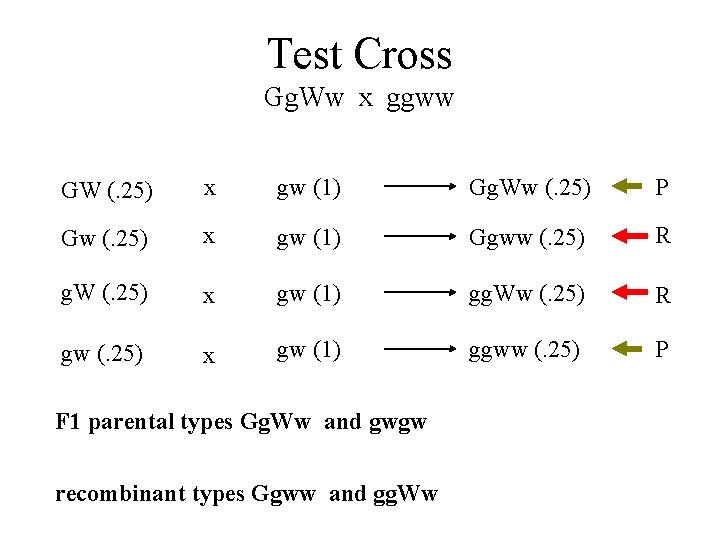
Test Cross Gg. Ww x ggww GW (. 25) x gw (1) Gg. Ww (. 25) P Gw (. 25) x gw (1) Ggww (. 25) R g. W (. 25) x gw (1) gg. Ww (. 25) R gw (. 25) x gw (1) ggww (. 25) P F 1 parental types Gg. Ww and gwgw recombinant types Ggww and gg. Ww

Recombination Frequency …or Linkage Ratio: the percentage of recombinant types, – if 50%, then the genes are not linked, – if less than 50%, then linkage is observed.

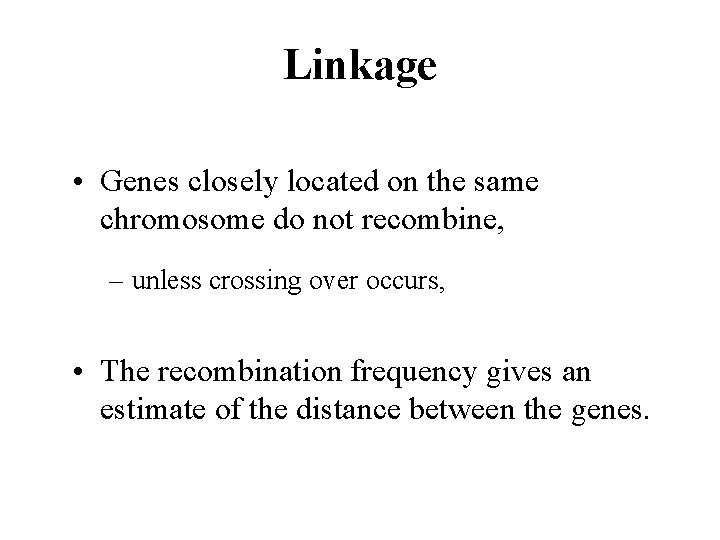
Linkage • Genes closely located on the same chromosome do not recombine, – unless crossing over occurs, • The recombination frequency gives an estimate of the distance between the genes.

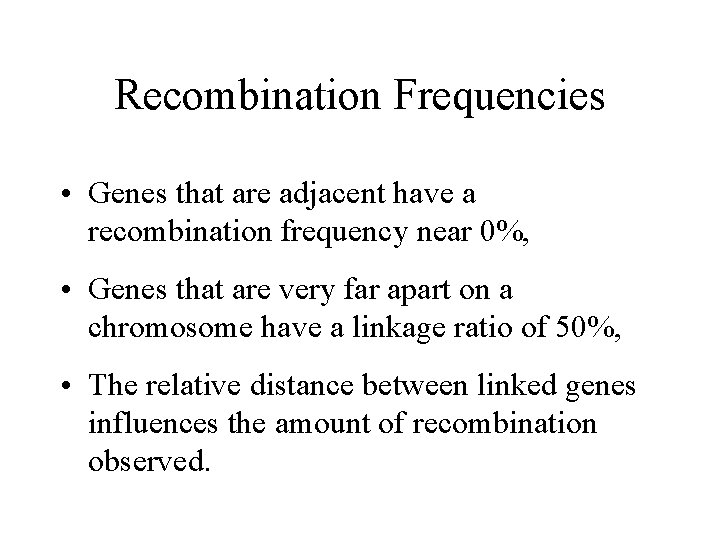
Recombination Frequencies • Genes that are adjacent have a recombination frequency near 0%, • Genes that are very far apart on a chromosome have a linkage ratio of 50%, • The relative distance between linked genes influences the amount of recombination observed.

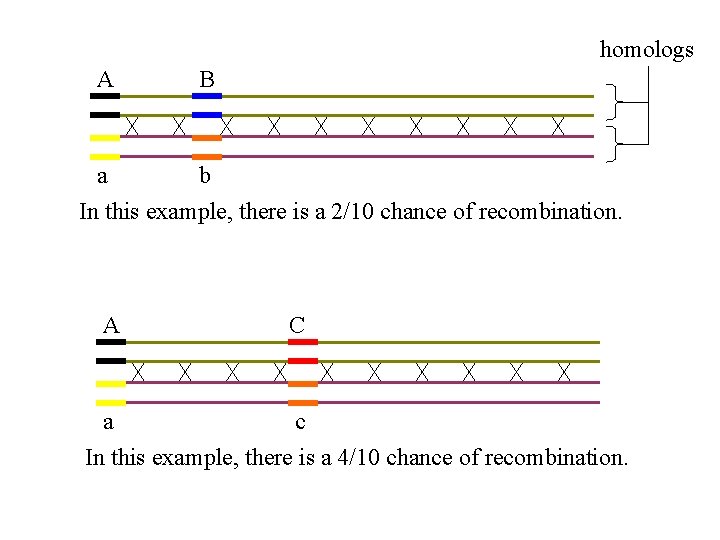
homologs A B a b In this example, there is a 2/10 chance of recombination. A C a c In this example, there is a 4/10 chance of recombination.

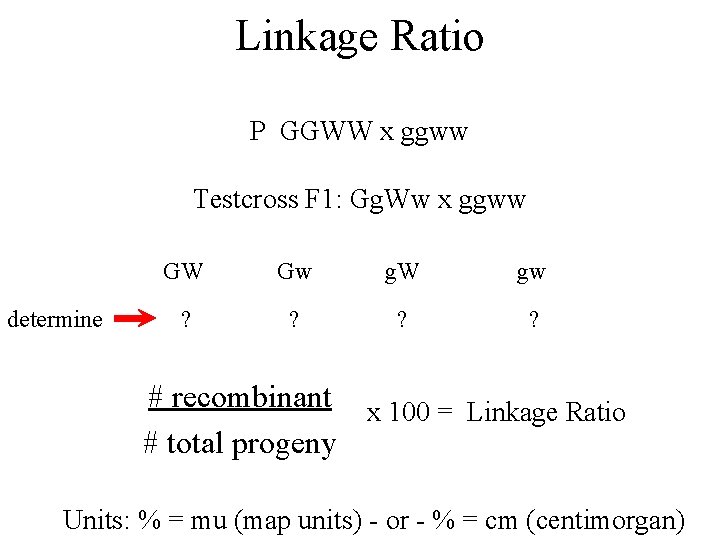
Linkage Ratio P GGWW x ggww Testcross F 1: Gg. Ww x ggww determine GW Gw g. W gw ? ? # recombinant x 100 = Linkage Ratio # total progeny Units: % = mu (map units) - or - % = cm (centimorgan)

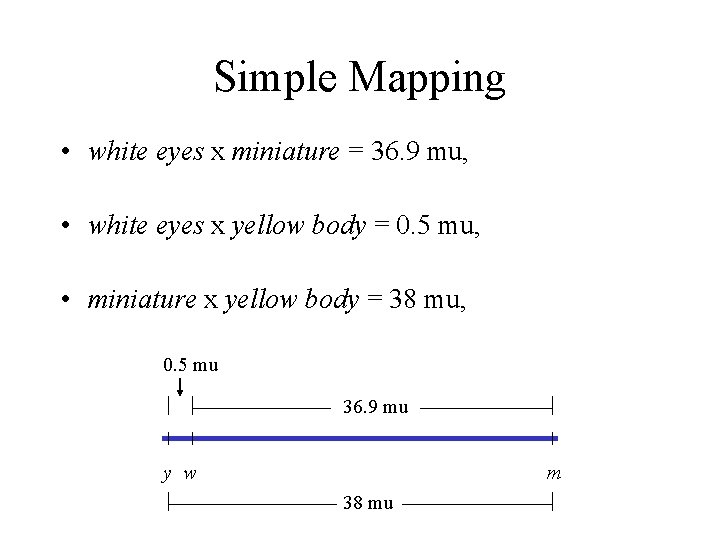
Study Figs 4. 2, 4. 3, and 4. 5 Fly Crosses (simple 3 -point mapping) (white eyes, minature, yellow body) • In a white eyes x miniature cross, 900 of the 2, 441 progeny were recombinant, yielding a map distance of 36. 9 mu, • In a separate white eyes x yellow body cross, 11 of 2, 205 progeny were recombinant, yielding a map distance of 0. 5 mu, • When a miniature x yellow body cross was performed, 650 of 1706 flies were recombinant, yielding a map distance of 38 mu.

Simple Mapping • white eyes x miniature = 36. 9 mu, • white eyes x yellow body = 0. 5 mu, • miniature x yellow body = 38 mu, 0. 5 mu 36. 9 mu y w m 38 mu

Do We have to Learn More Mapping Techniques? • Yes, – three point mapping, • Why, – – Certainty of Gene Order, Double crossovers, To answer Cyril Napp’s questions, and, for example: over 4000 known human diseases have a genetic component, • knowing the protein produced at specific loci facilitates the treatment and testing.

cis “coupling”

trans “repulsion”

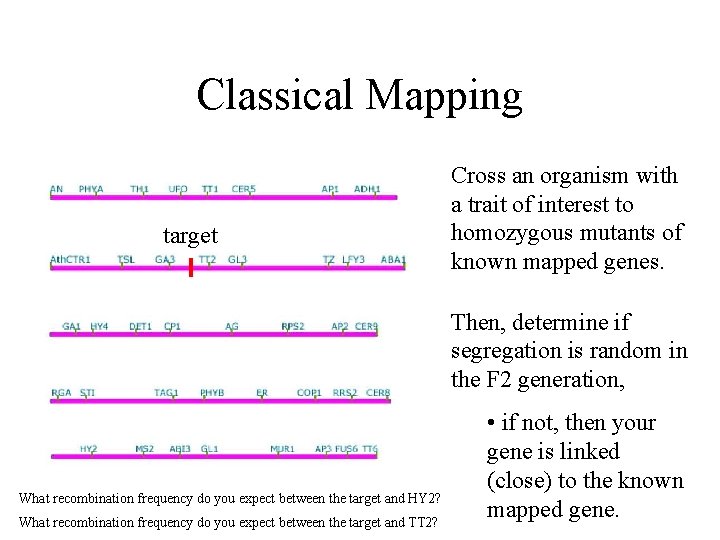
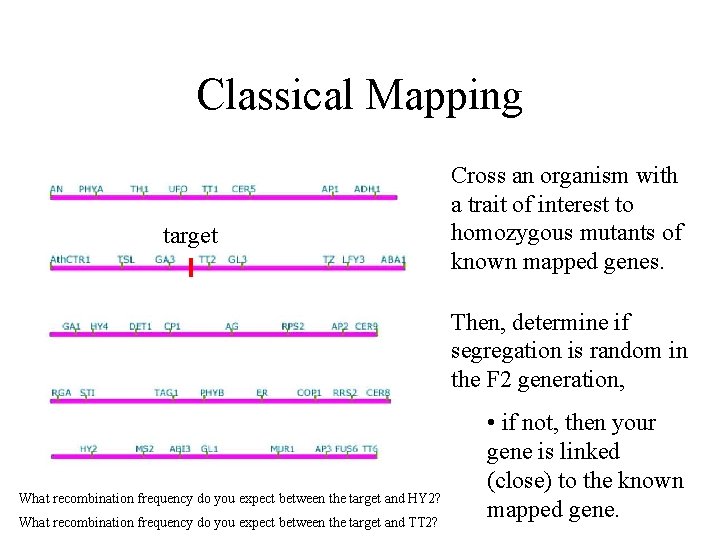
Classical Mapping target Cross an organism with a trait of interest to homozygous mutants of known mapped genes. Then, determine if segregation is random in the F 2 generation, What recombination frequency do you expect between the target and HY 2? What recombination frequency do you expect between the target and TT 2? • if not, then your gene is linked (close) to the known mapped gene.

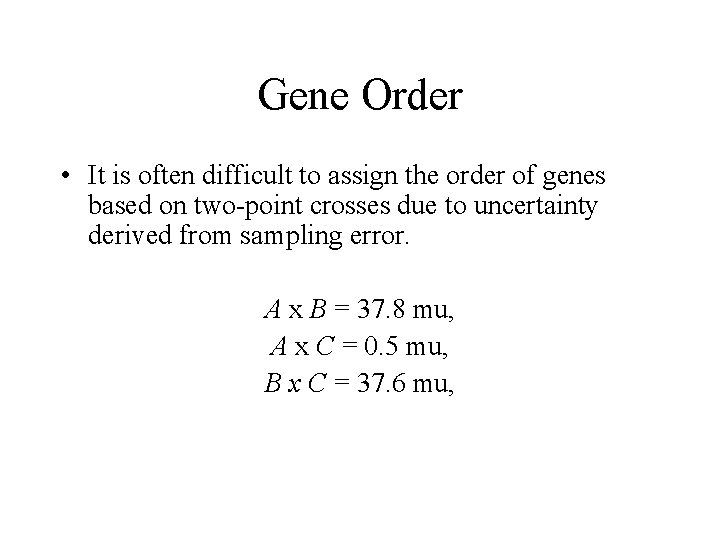
Gene Order • It is often difficult to assign the order of genes based on two-point crosses due to uncertainty derived from sampling error. A x B = 37. 8 mu, A x C = 0. 5 mu, B x C = 37. 6 mu,

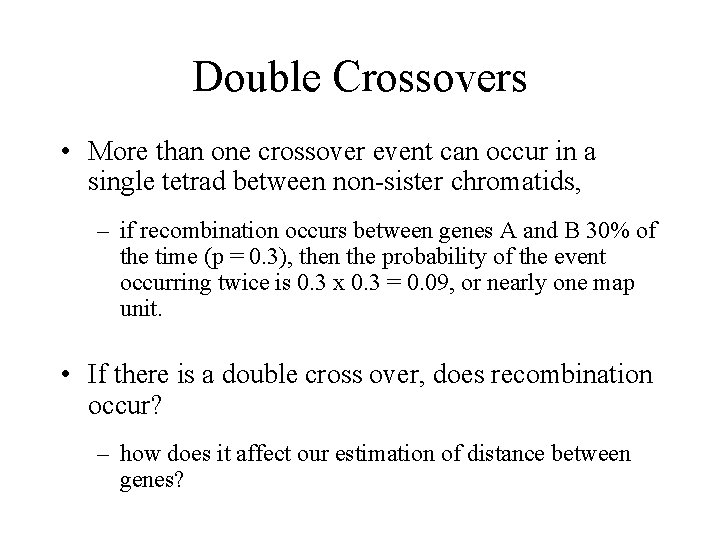
Double Crossovers • More than one crossover event can occur in a single tetrad between non-sister chromatids, – if recombination occurs between genes A and B 30% of the time (p = 0. 3), then the probability of the event occurring twice is 0. 3 x 0. 3 = 0. 09, or nearly one map unit. • If there is a double cross over, does recombination occur? – how does it affect our estimation of distance between genes?

Genetics: …in the News

Classical Mapping target Cross an organism with a trait of interest to homozygous mutants of known mapped genes. Then, determine if segregation is random in the F 2 generation, What recombination frequency do you expect between the target and HY 2? What recombination frequency do you expect between the target and TT 2? • if not, then your gene is linked (close) to the known mapped gene.

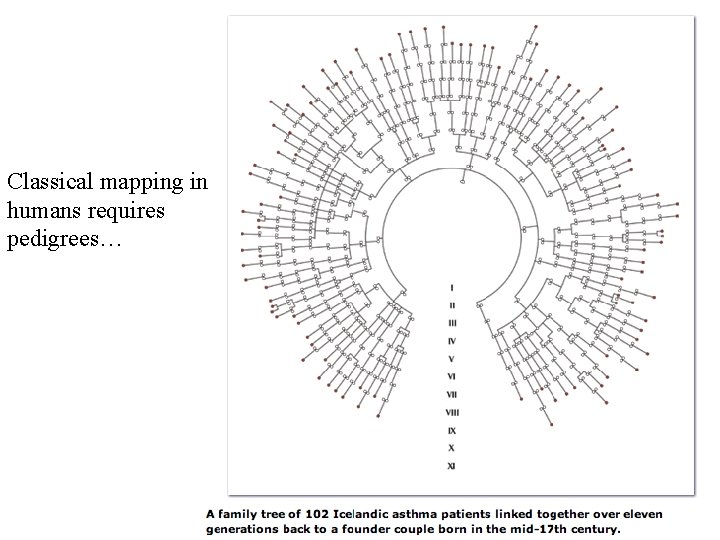
Classical mapping in humans requires pedigrees…

Three Point Testcross Triple Heterozygous (Aa. Bb. Cc ) x Triple Homozygous Recessive (aabbcc)

Three Point Mapping Requirements • The genotype of the organism producing the gametes must be heterozygous at all three loci, • You have to be able to deduce the genotype of the gamete by looking at the phenotype of the offspring, • You must look at enough offspring so that all crossover classes are represented.

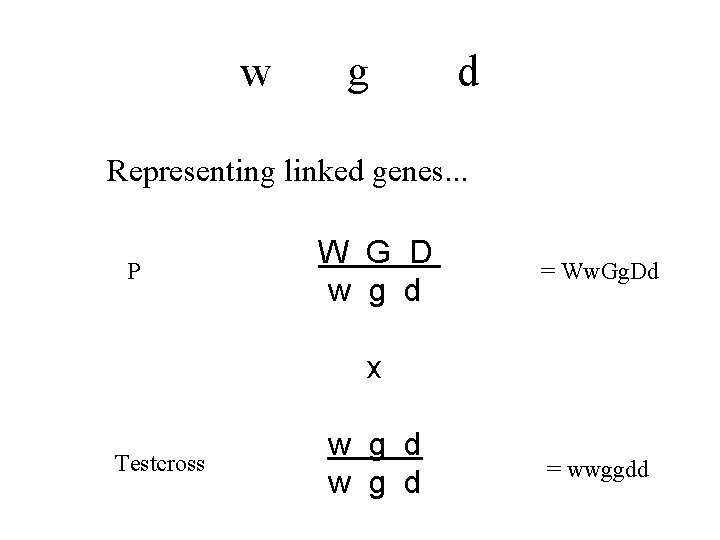
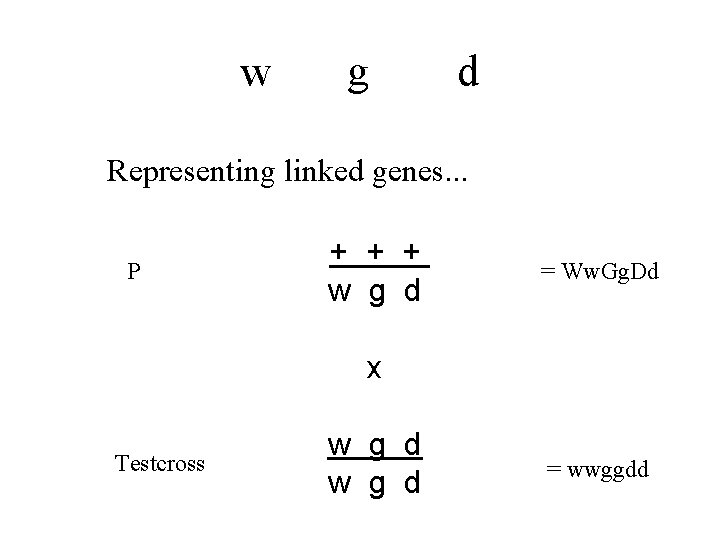
w g d Representing linked genes. . . P W G D w g d = Ww. Gg. Dd x Testcross w g d = wwggdd

w g d Representing linked genes. . . P + + + w g d = Ww. Gg. Dd x Testcross w g d = wwggdd

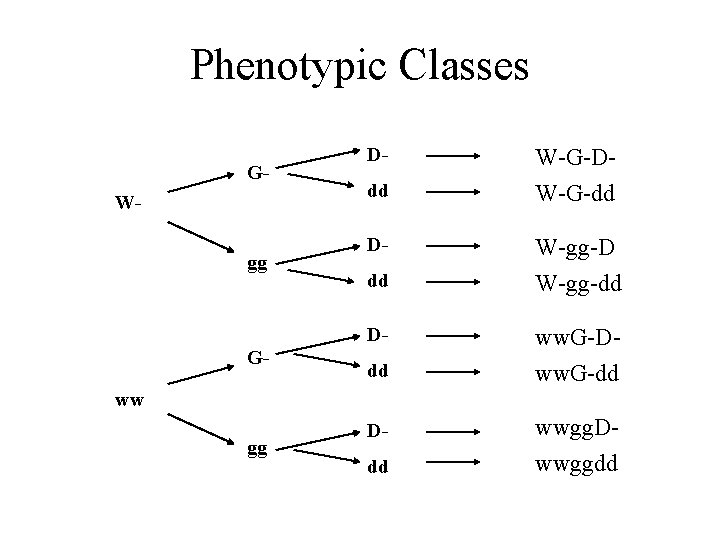
Phenotypic Classes GWgg Ddd D- G- dd W-G-DW-G-dd W-gg-D W-gg-dd ww. G-Dww. G-dd ww gg Ddd wwgg. Dwwggdd

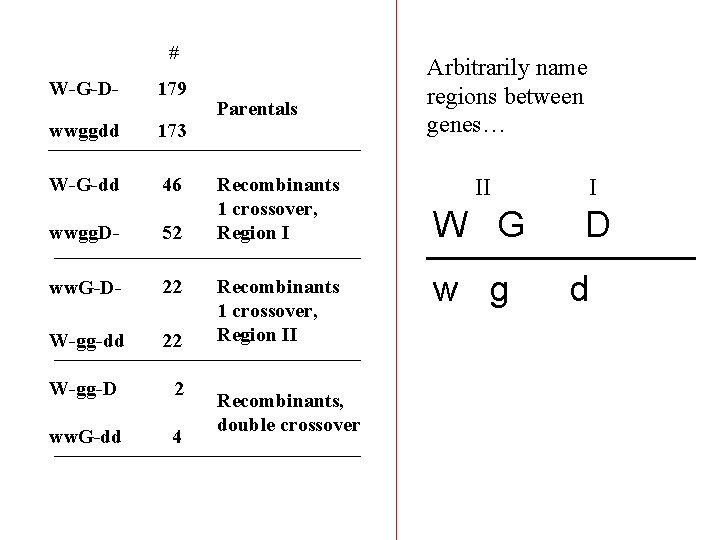
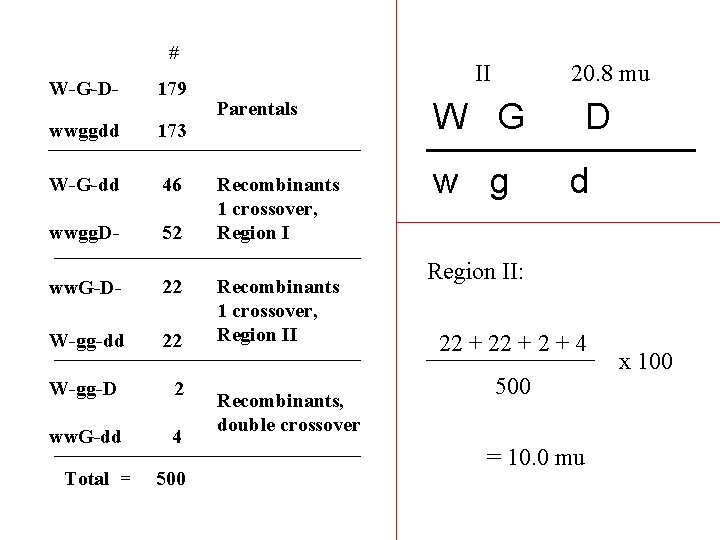
# W-G-D- 179 wwggdd 173 W-G-dd 46 wwgg. D- 52 ww. G-D- 22 W-gg-dd 22 W-gg-D 2 ww. G-dd 4 Parentals Recombinants 1 crossover, Region II Recombinants, double crossover Arbitrarily name regions between genes… II I W G D w g d

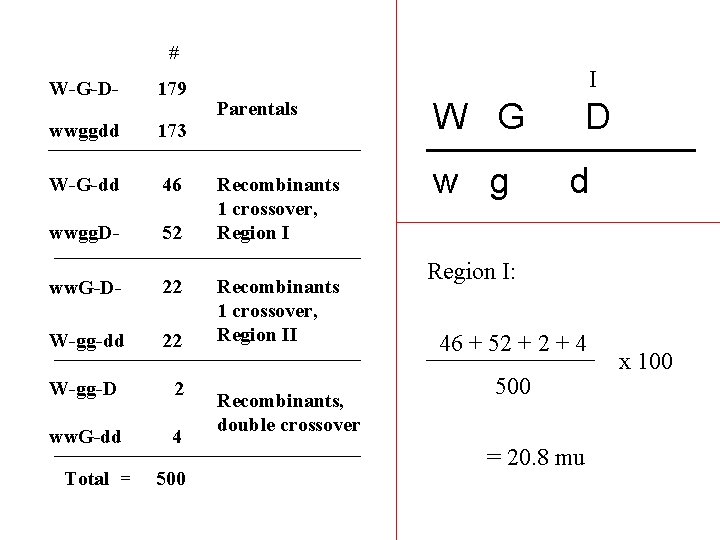
# W-G-D- 179 wwggdd 173 W-G-dd 46 wwgg. D- 52 ww. G-D- 22 W-gg-dd 22 W-gg-D 2 ww. G-dd 4 Total = 500 I Parentals Recombinants 1 crossover, Region II Recombinants, double crossover W G w g D d Region I: 46 + 52 + 4 500 = 20. 8 mu x 100

# W-G-D- 179 wwggdd 173 W-G-dd 46 wwgg. D- 52 ww. G-D- 22 W-gg-dd 22 W-gg-D 2 ww. G-dd 4 Total = 500 II Parentals Recombinants 1 crossover, Region II Recombinants, double crossover 20. 8 mu W G w g D d Region II: 22 + 2 + 4 500 = 10. 0 mu x 100

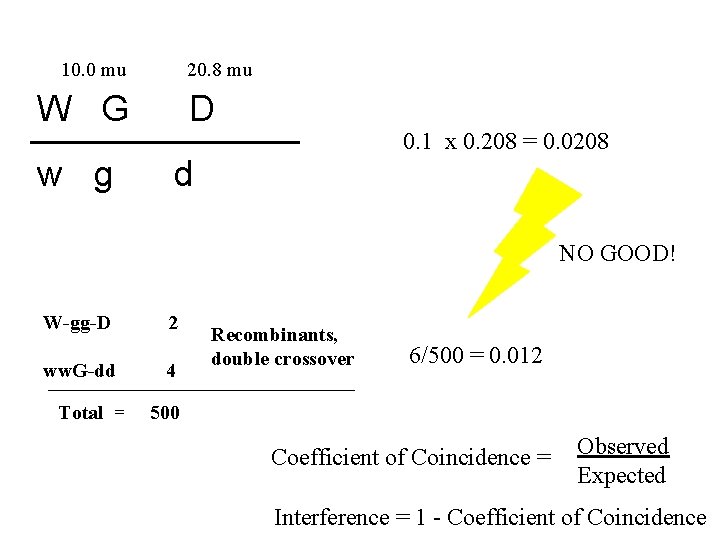
10. 0 mu 20. 8 mu W G w g D 0. 1 x 0. 208 = 0. 0208 d NO GOOD! W-gg-D ww. G-dd Total = 2 4 Recombinants, double crossover 6/500 = 0. 012 500 Coefficient of Coincidence = Observed Expected Interference = 1 - Coefficient of Coincidence

Interference …the effect a crossing over event has on a second crossing over event in an adjacent region of the chromatid, – (positive) interference: decreases the probability of a second crossing over, • most common in eukaryotes, – negative interference: increases the probability of a second crossing over.

Gene Order in Three Point Crosses • Find - either - double cross-over phenotype…based on the recombination frequencies, • Two parental alleles, and one cross over allele will be present, • The cross over allele fits in the middle. . .

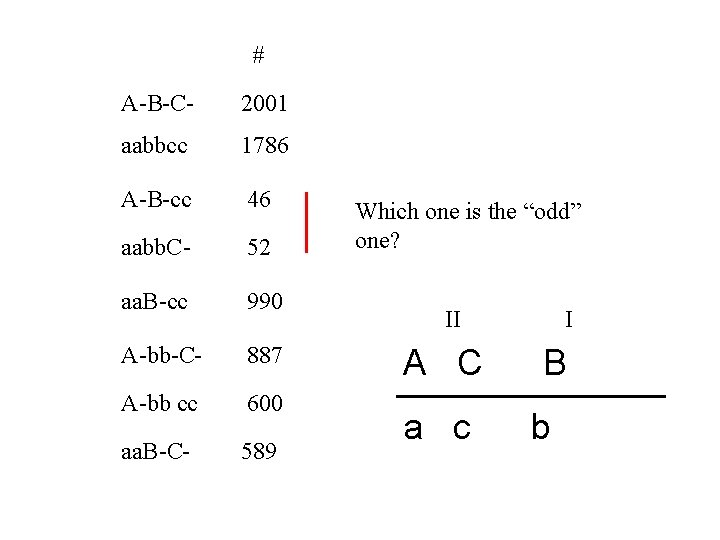
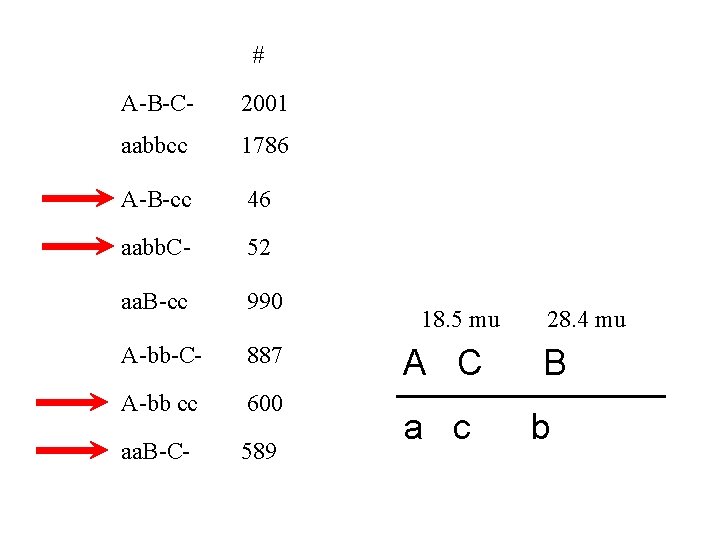
# A-B-C- 2001 aabbcc 1786 A-B-cc 46 aabb. C- 52 aa. B-cc 990 A-bb-C- 887 A-bb cc 600 aa. B-C- 589 Which one is the “odd” one? II A C a c I B b

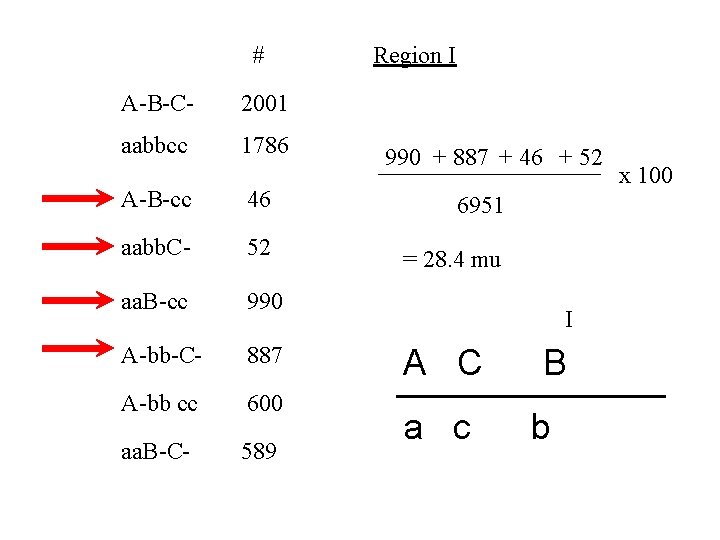
# A-B-C- 2001 aabbcc 1786 Region I 990 + 887 + 46 + 52 A-B-cc 46 6951 aabb. C- 52 = 28. 4 mu aa. B-cc 990 A-bb-C- 887 A-bb cc 600 aa. B-C- 589 I A C a c B b x 100

# A-B-C- 2001 aabbcc 1786 Region II 600 + 589 + 46 + 52 A-B-cc 46 6951 aabb. C- 52 = 18. 5 mu aa. B-cc 990 A-bb-C- 887 A-bb cc 600 aa. B-C- 589 18. 5 II mu A C a c x 100 28. 4 mu B b

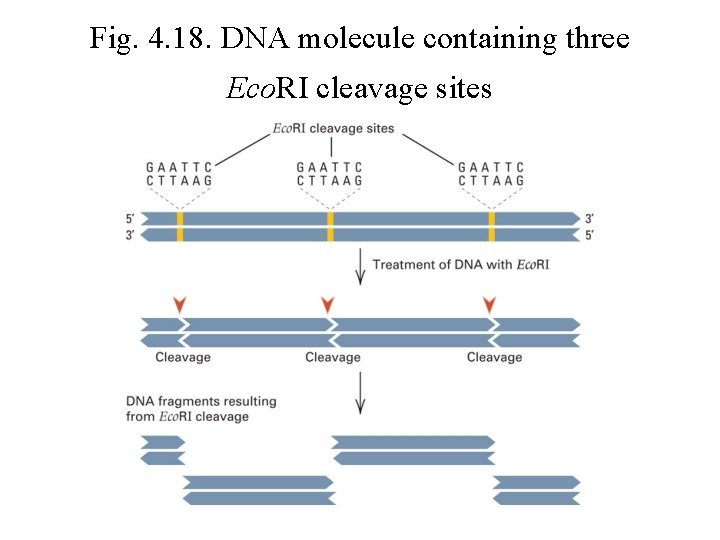
Fig. 4. 18. DNA molecule containing three Eco. RI cleavage sites

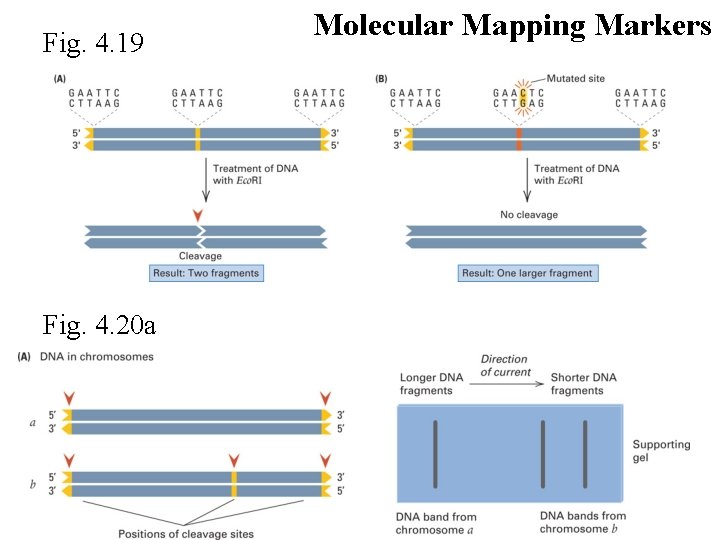
Fig. 4. 19 Fig. 4. 20 a Molecular Mapping Markers

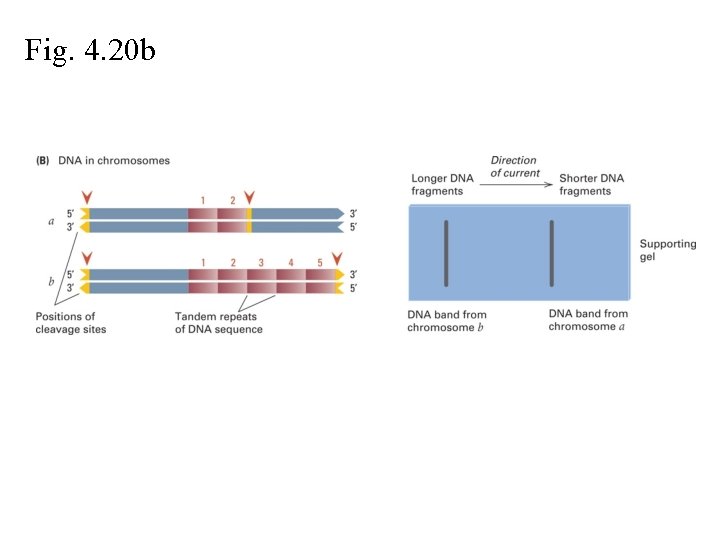
Fig. 4. 20 b

p. 143. Fluorescent dyes are often used to label DNA so that the positions of DAN fragments in a gel can be identified.

Assignments • Read from Chapter 3, 3. 6 (pp. 100 -106), • Master Problems… 3. 12, 3. 15, 3. 20, • Chapter 4, Problems 1, 2, • Questions 4. 1 - 4. 4, 4. 6, 4. 7, 4. 9, 4. 11 -4. 14, 4. 19 4. 20 a, b, c, d. • Exam Wednesday. – One hour (you can use the entire 80 minutes, but no more). One 8” x 11”, one sided crib sheet.
 Sex sex sex
Sex sex sex Sex sex sex
Sex sex sex Xxtesticles
Xxtesticles Sex sex sex
Sex sex sex Secondary sexual characters
Secondary sexual characters Linkage
Linkage Sex linkage
Sex linkage Sex determination and sex linkage
Sex determination and sex linkage Autosomes and sex chromosomes
Autosomes and sex chromosomes Once a sex offender always a sex offender
Once a sex offender always a sex offender Gamete and zygote
Gamete and zygote Genes located
Genes located Autosomes vs sex chromosomes
Autosomes vs sex chromosomes No one likes you
No one likes you Phân độ lown
Phân độ lown Block nhĩ thất độ 3
Block nhĩ thất độ 3 Thơ thất ngôn tứ tuyệt đường luật
Thơ thất ngôn tứ tuyệt đường luật Thơ thất ngôn tứ tuyệt đường luật
Thơ thất ngôn tứ tuyệt đường luật Chiến lược kinh doanh quốc tế của walmart
Chiến lược kinh doanh quốc tế của walmart Tìm vết của đường thẳng
Tìm vết của đường thẳng Hãy nói thật ít để làm được nhiều
Hãy nói thật ít để làm được nhiều Tôn thất thuyết là ai
Tôn thất thuyết là ai Gây tê cơ vuông thắt lưng
Gây tê cơ vuông thắt lưng Sau thất bại ở hồ điển triệt
Sau thất bại ở hồ điển triệt Male vs female fetal pig
Male vs female fetal pig Two copies of each autosomal gene affect
Two copies of each autosomal gene affect Sex male and female
Sex male and female Hard to seem won
Hard to seem won Physiology of coitus
Physiology of coitus Chapter 10 sex gender and sexuality
Chapter 10 sex gender and sexuality Sex and cholesterol
Sex and cholesterol Difference between sex and gender
Difference between sex and gender Noughts and crosses zusammenfassung
Noughts and crosses zusammenfassung Difference between gender and sex
Difference between gender and sex Sex
Sex Harry potter and the inheritance of sex
Harry potter and the inheritance of sex Sex and gender difference
Sex and gender difference Difference between sex and gender
Difference between sex and gender Troy sex and the city
Troy sex and the city Section 1 chromosomes and phenotype
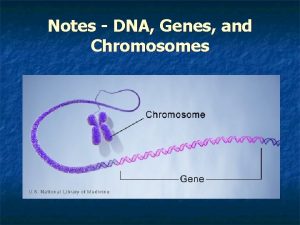
Section 1 chromosomes and phenotype Dna, genes and chromosomes relationship
Dna, genes and chromosomes relationship Difference between maternal and paternal chromosomes
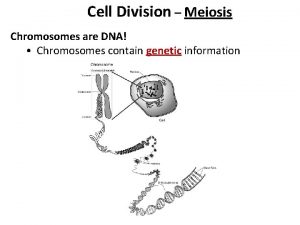
Difference between maternal and paternal chromosomes Meiosis
Meiosis Section 12-1 chromosomes and inheritance
Section 12-1 chromosomes and inheritance Number of chromosomes in meiosis and mitosis
Number of chromosomes in meiosis and mitosis Chromosomes and alleles
Chromosomes and alleles Secondary sex characteristics
Secondary sex characteristics Chromosomes genes and basic genetics foldable answer key
Chromosomes genes and basic genetics foldable answer key Alleles on homologous chromosomes
Alleles on homologous chromosomes Mendel's theory of particulate inheritance __________.
Mendel's theory of particulate inheritance __________. Bill nye genes worksheet
Bill nye genes worksheet Building vocabulary: the nucleus, dna, and chromosomes
Building vocabulary: the nucleus, dna, and chromosomes Dna, genes and chromosomes relationship
Dna, genes and chromosomes relationship Main function of the chromosomes
Main function of the chromosomes Section 1 chromosomes and phenotype
Section 1 chromosomes and phenotype Chromosomes and alleles
Chromosomes and alleles Dna scrunches up and chromosomes are first visible
Dna scrunches up and chromosomes are first visible Chapter 7 extending mendelian genetics vocabulary practice
Chapter 7 extending mendelian genetics vocabulary practice Rr x rr genotype ratio
Rr x rr genotype ratio What is the relationship between dna chromosomes and genes
What is the relationship between dna chromosomes and genes Recombination
Recombination Gamete formation
Gamete formation Section 1 chromosomes and phenotype
Section 1 chromosomes and phenotype Chapter 6 chromosomes and cell reproduction
Chapter 6 chromosomes and cell reproduction Section 1 chromosomes and phenotype
Section 1 chromosomes and phenotype Independent vs dependent variable
Independent vs dependent variable Identify the polynomial written in ascending order.
Identify the polynomial written in ascending order. R and s configuration
R and s configuration Master status
Master status