Rietveld texture analysis of SKAT diffractometer data R



- Slides: 64

Rietveld texture analysis of SKAT diffractometer data R. N. Vasin STI-2011, 6 -9 June, 2011, Dubna, Russia

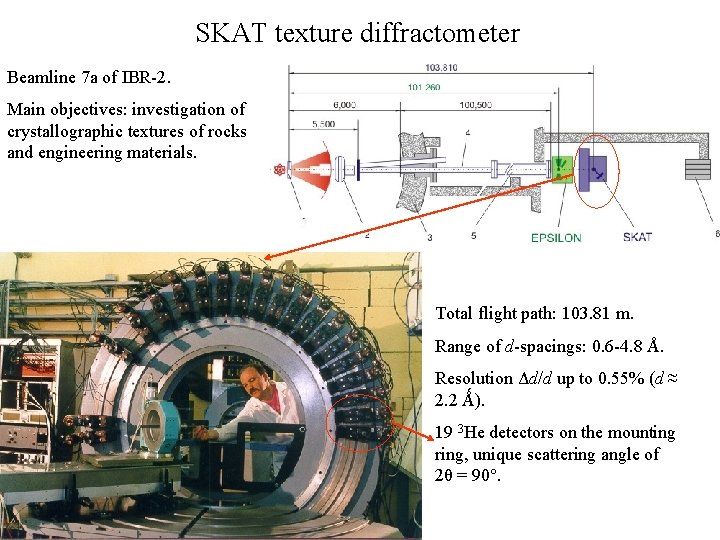
SKAT texture diffractometer Beamline 7 a of IBR-2. Main objectives: investigation of crystallographic textures of rocks and engineering materials. Total flight path: 103. 81 m. Range of d-spacings: 0. 6 -4. 8 Å. Resolution Δd/d up to 0. 55% (d ≈ 2. 2 Ǻ). 19 3 He detectors on the mounting ring, unique scattering angle of 2θ = 90.

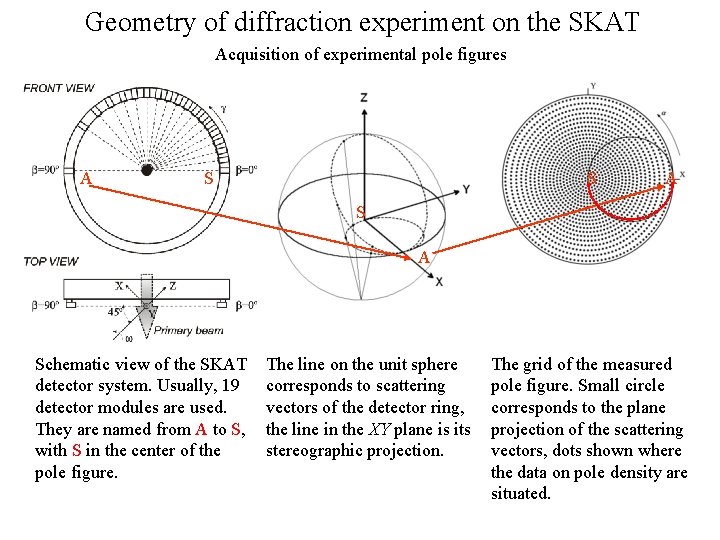
Geometry of diffraction experiment on the SKAT Acquisition of experimental pole figures A S S A Schematic view of the SKAT detector system. Usually, 19 detector modules are used. They are named from A to S, with S in the center of the pole figure. The line on the unit sphere corresponds to scattering vectors of the detector ring, the line in the XY plane is its stereographic projection. The grid of the measured pole figure. Small circle corresponds to the plane projection of the scattering vectors, dots shown where the data on pole density are situated.

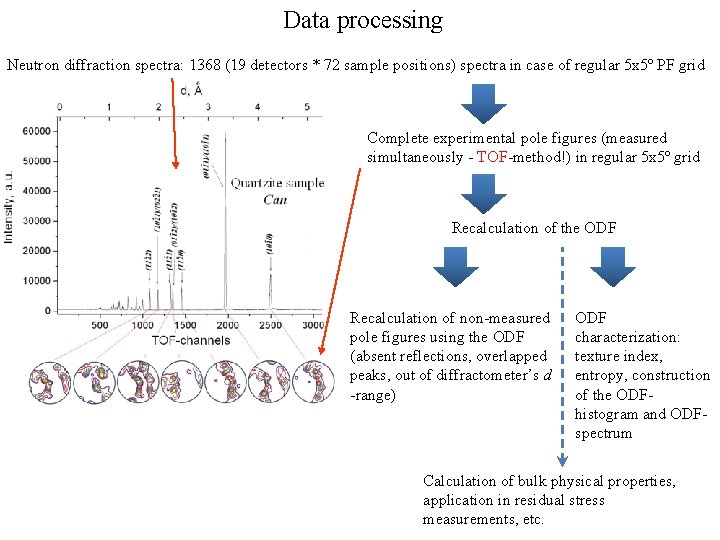
Data processing Neutron diffraction spectra: 1368 (19 detectors * 72 sample positions) spectra in case of regular 5 х5º PF grid Complete experimental pole figures (measured simultaneously - TOF-method!) in regular 5 х5º grid Recalculation of the ODF Recalculation of non-measured pole figures using the ODF (absent reflections, overlapped peaks, out of diffractometer’s d -range) ODF characterization: texture index, entropy, construction of the ODFhistogram and ODFspectrum Calculation of bulk physical properties, application in residual stress measurements, etc.

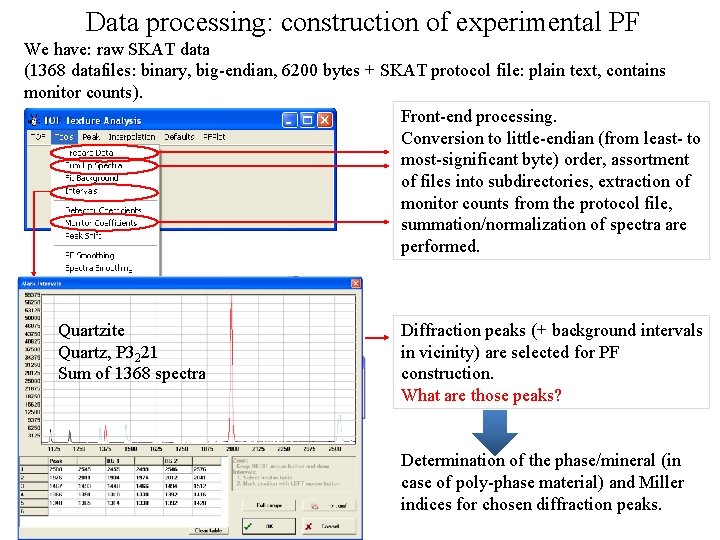
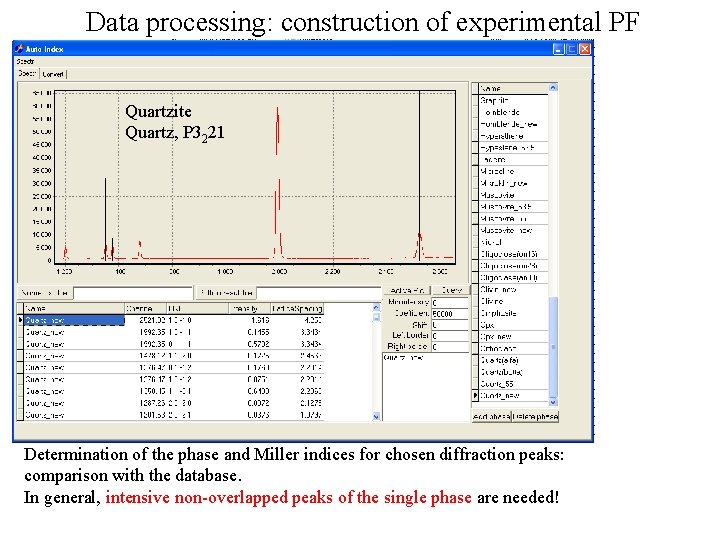
Data processing: construction of experimental PF We have: raw SKAT data (1368 datafiles: binary, big-endian, 6200 bytes + SKAT protocol file: plain text, contains monitor counts). Front-end processing. Conversion to little-endian (from least- to most-significant byte) order, assortment of files into subdirectories, extraction of monitor counts from the protocol file, summation/normalization of spectra are performed. Quartzite Quartz, P 3221 Sum of 1368 spectra Diffraction peaks (+ background intervals in vicinity) are selected for PF construction. What are those peaks? Determination of the phase/mineral (in case of poly-phase material) and Miller indices for chosen diffraction peaks.

Data processing: construction of experimental PF Quartzite Quartz, P 3221 Determination of the phase and Miller indices for chosen diffraction peaks: comparison with the database. In general, intensive non-overlapped peaks of the single phase are needed!

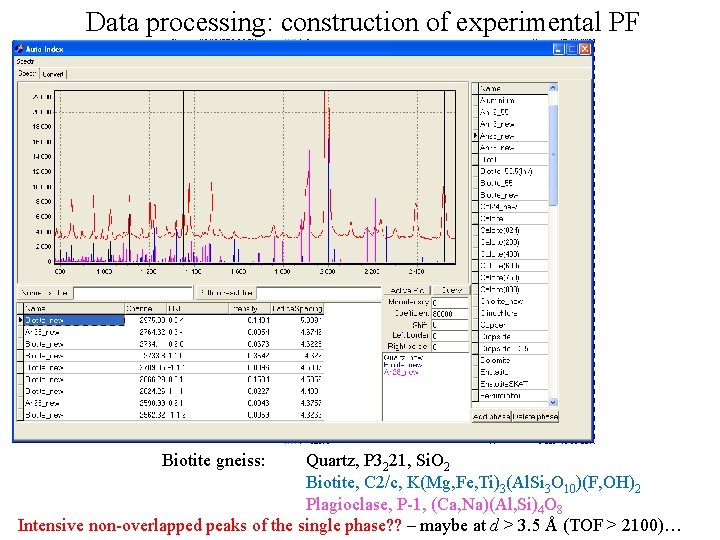
Data processing: construction of experimental PF Biotite gneiss: Quartz, P 3221, Si. O 2 Biotite, С 2/c, K(Mg, Fe, Ti)3(Al. Si 3 O 10)(F, OH)2 Plagioclase, P-1, (Ca, Na)(Al, Si)4 O 8 Intensive non-overlapped peaks of the single phase? ? – maybe at d > 3. 5 Å (TOF > 2100)…

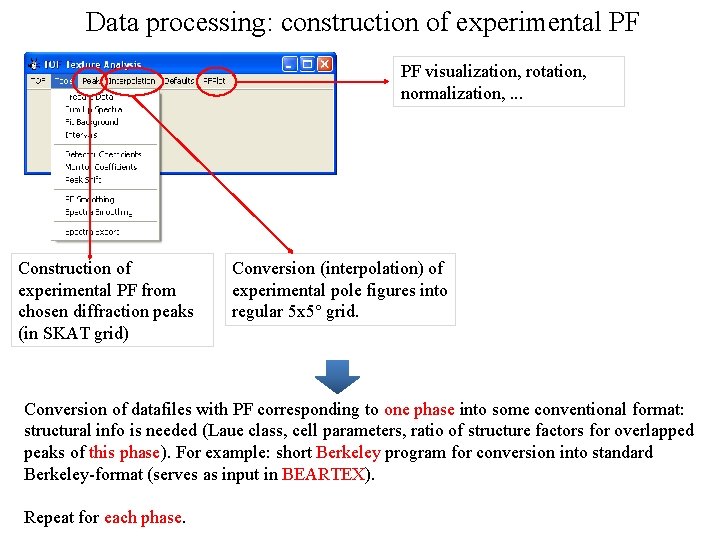
Data processing: construction of experimental PF PF visualization, rotation, normalization, . . . Construction of experimental PF from chosen diffraction peaks (in SKAT grid) Conversion (interpolation) of experimental pole figures into regular 5 х5° grid. Conversion of datafiles with PF corresponding to one phase into some conventional format: structural info is needed (Laue class, cell parameters, ratio of structure factors for overlapped peaks of this phase). For example: short Berkeley program for conversion into standard Berkeley-format (serves as input in BEARTEX). Repeat for each phase.

Orientation distribution function calculation from experimental PF + some other options BEARTEX (WIMV method) H. -R. Wenk & S. Matthies http: //eps. berkeley. edu/~wenk/Texture. Page/beartex. htm Operations with ODF, PF, inverse PF, PF modeling, tensor averaging (calculation of physical properties), … Single license – 2000$ (academic – 1000$) Some routines do not function in 64 -bit OS! LABOTEX (ADC method) K. Pawlik http: //www. labosoft. com. pl/index. htm Operations with ODF, PF, inverse PF, … Single license – 6000$ (academic – 3000$)

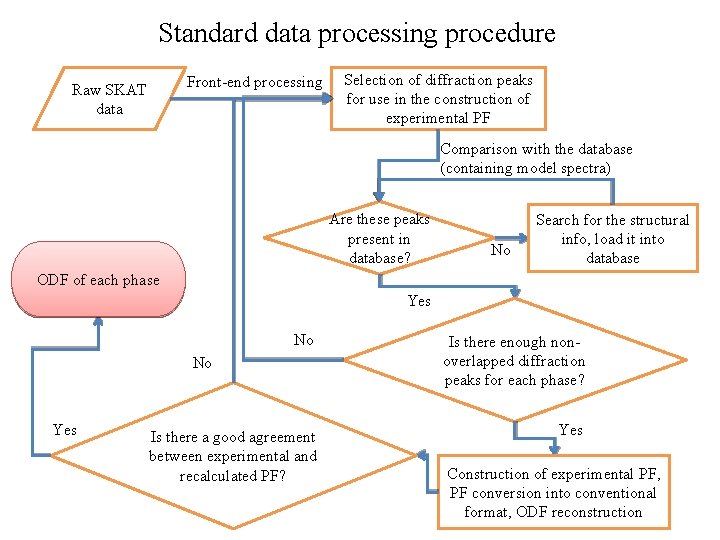
Standard data processing procedure Front-end processing Raw SKAT data Selection of diffraction peaks for use in the construction of experimental PF Comparison with the database (containing model spectra) Are these peaks present in database? No Search for the structural info, load it into database ODF of each phase Yes No Is there enough nonoverlapped diffraction peaks for each phase? Is there a good agreement between experimental and recalculated PF? Yes No Yes Construction of experimental PF, PF conversion into conventional format, ODF reconstruction

Standard data processing procedure: drawbacks • Evident problems with the selection of non-overlapped intensive peaks from diffraction patterns in case of multiphase sample (especially if several low-symmetry phases are present). Usually in this case diffraction peaks are selected at high-d region, where counting statistics are not so good. • Only a small part of acquired diffraction patterns is used (≤ 6 peaks for each phase). And, for example, for Ni (space group Fm-3 m) in SKAT d-range at least 12 peaks are easily available, for oligoclase An 16 (space group P-1) in interval d > 1. 5 Å – more than 400 peaks. • The result of the standard data processing is the ODF, no additional information is retrieved (like cell parameters, phase volume fractions, etc. ) • New detector rings for SKAT (at different scattering angles) may add some complexities. Solution: it is possible to use Rietveld method for simultaneous processing of all (e. g. , 1368) SKAT spectra with account for crystallographic texture (this is requirement!). No need for manual peak selection, most part of available data is used, additional info about crystal structure of the sample is received.

MAUD http: //www. ing. unitn. it/~maud/ L. Lutterotti, "Total pattern fitting for the combined sizestrain-stress-texture determination in thin film diffraction“, Nuclear Inst. and Methods in Physics Research, B, 268, 334 -340, 2010. Freeware! Java-based, needs JVM to work, but this exists for very much every system. Versions for Windows, Mac OS, Linux, Unix (x 86 и x 64) are available. User-friendly (fine GUI). It’s possible to work with X-ray /synchrotron, electron, neutron diffraction patterns (TOF is included). Great options for texture evaluation: ODF calculation from the set of diffraction spectra (several methods are available).

MAUD And what about TOF spectra?

SKAT data analysis by MAUD: what do we need? • SKAT instrument parameter file (TOF channel to d conversion: scattering angle, delay, DIFC constant ~ L·sinθ; effective spectrum; peak shape) in GSAS format (. prm). Calibrations have been made for each of 19 detectors of the SKAT (different calibrations for different reactor cycles!); vanadium spectrum have been fitted by Fit. Spec routine of GSAS, function № 4 has been used (MAUD uses functions № 0… 5): Maxvellian term + first 10 Chebyshev polynomials of the first kind; peak shape have been approximated by function № 1 TOF (the only one available in MAUD? ? ? ): Gaussian convoluted with two exponentials, accounts only for the instrument-dependent peak broadening in MAUD. • SKAT datafiles in standard GSAS format (. gda) It’s not a problem to convert binary file to the formatted ASCII with the proper header. But it’s too complicated and time consuming to perform manually for each of 1368 spectra. • Load all the datafiles into MAUD taking into account the position on the pole figure (two angles!) and monitor counts. It’s too complicated and time consuming to perform manually for each of 1368 spectra. The fit is not perfect due to some peculiarities of the vanadium spectrum, its better to use “point-to-point” normalization.

SKAT 2 MAUD «Mass transformation» of spectra and construction of the special script file to automatically load all the data into MAUD. C++ based. Ease to use GUI. Tested in Win. XP Pro x 86 & Win 7 HP x 64. File access is made through Win API functions → it is fast. Conversion + normalization + script creation for 1368 files takes ≈ 15 sec. A possibility to easily add some other IBR-2 diffractometers.

SKAT 2 MAUD Selection of parameters for the data conversion Spectra selection Monitor counts extraction from the SKAT protocol file Construction of the script file for MAUD and of the protocol. Options for point Data type -to-point data selection normalization

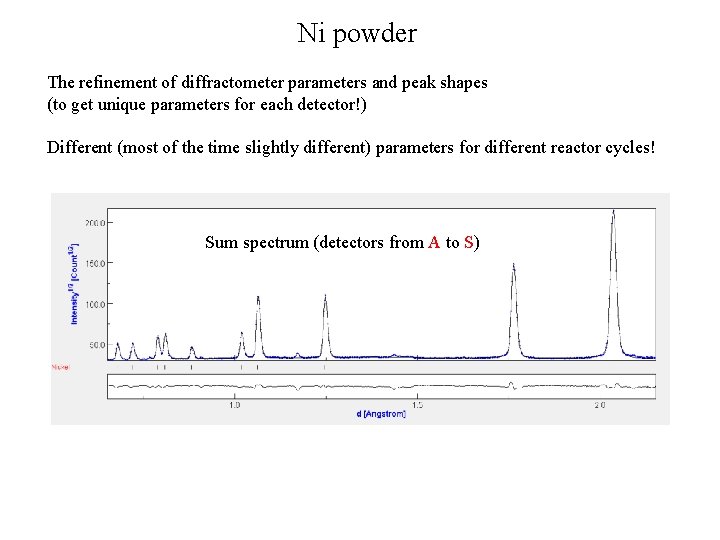
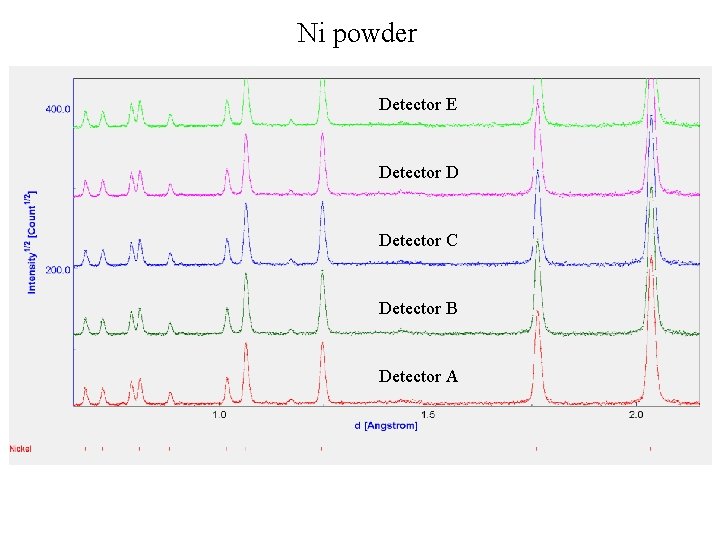
Ni powder The refinement of diffractometer parameters and peak shapes (to get unique parameters for each detector!) Different (most of the time slightly different) parameters for different reactor cycles! Sum spectrum (detectors from A to S)

Ni powder Detector E Detector D Detector C Detector B Detector А

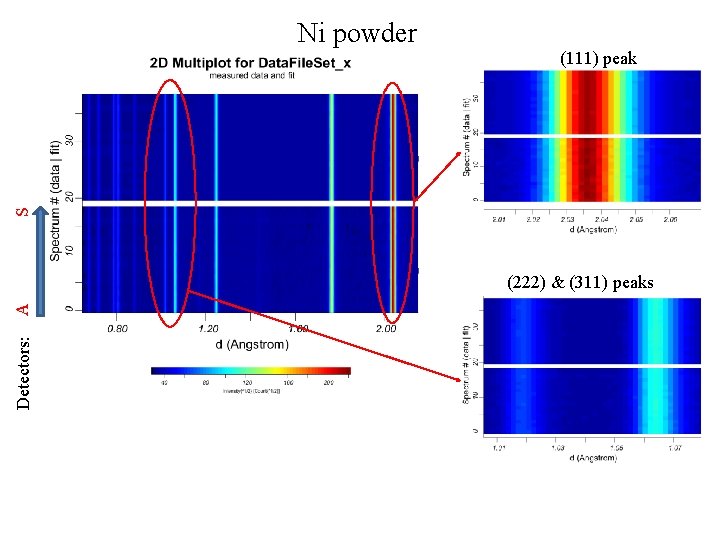
Ni powder S (111) peak Detectors: A (222) & (311) peaks

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Sample: Outokumpu borehole (Finland), 676 m depth. Mineral composition: quartz – 42. 6 vol. %, plagioclase – 37. 6 vol. %, biotite – 19. 8 vol. % (thin sections analysis, Kern, Mengel, Strauss et al. // PEPI, 175, 2009. 151 -166). Volume of the sample is > 100 cm 3. Lattice spacing range: d = 1. 51… 4. 31 Å → about 700 diffraction peaks/pole figures. Getting prepared for intensive calculations: MAUD command file is altered – now MAUD exclusively allocates 3 Gb of RAM (should be working on x 64 OS or x 86 with enabled PAE). Free parameters: phase volume fractions, individual background for each spectrum (3· 1386 parameters), cell parameters of each phase, 1 thermal factor for all atoms (isotropic approximation), crystallite size and microdeformation for each phase (isotropic approximation) – in total 4145 parameters. + ODF calculation for each phase, E-WIMV method, 5° ODF resolution, peaks with intensity less than 1% of maximum for the given phase were not used for the ODF calculation. 1 iteration took ≈ 3 h. 40 min. (Intel Core i 5 -430 M, 4 Gb DDR 3 -1066(CL 7), Win 7 HP x 64, x 64 Java Virtual Machine).

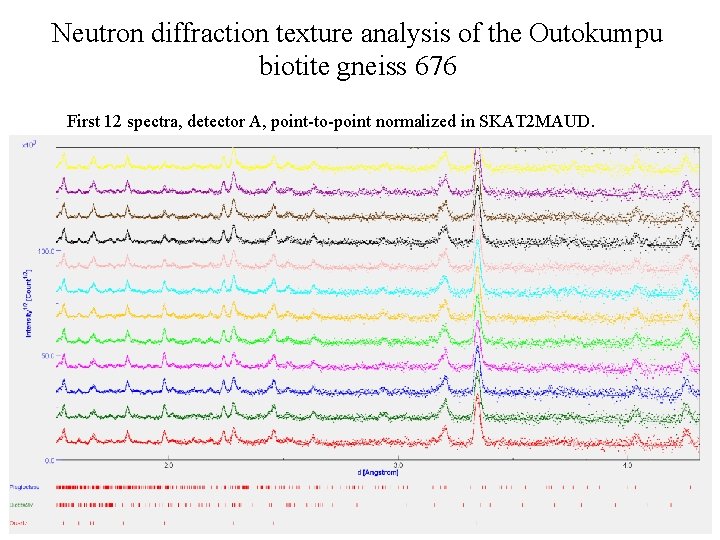
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 First 12 spectra, detector A, point-to-point normalized in SKAT 2 MAUD.

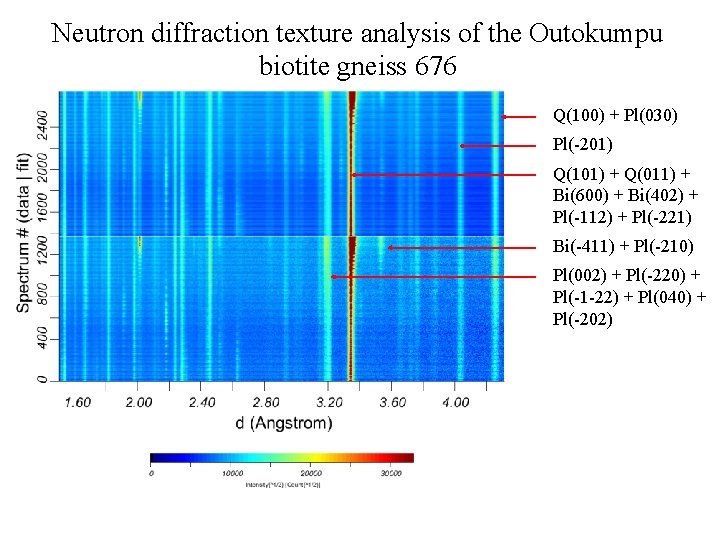
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Q(100) + Pl(030) Pl(-201) Q(101) + Q(011) + Bi(600) + Bi(402) + Pl(-112) + Pl(-221) Bi(-411) + Pl(-210) Pl(002) + Pl(-220) + Pl(-1 -22) + Pl(040) + Pl(-202)

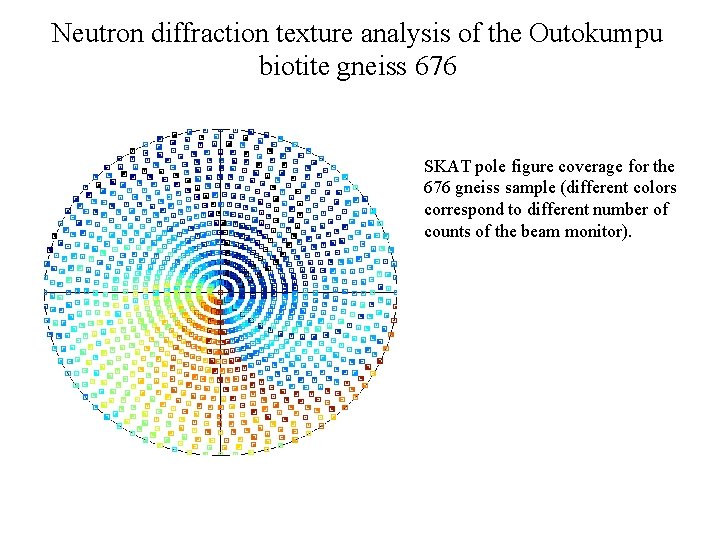
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 SKAT pole figure coverage for the 676 gneiss sample (different colors correspond to different number of counts of the beam monitor).

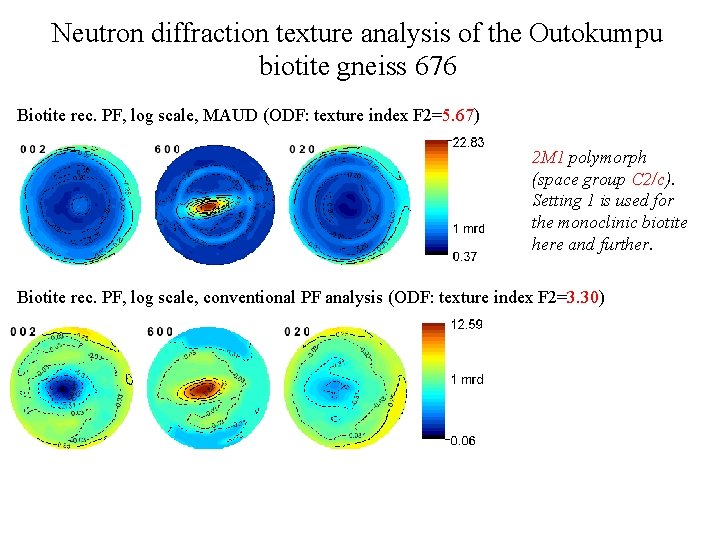
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Biotite rec. PF, log scale, MAUD (ODF: texture index F 2=5. 67) 2 M 1 polymorph (space group C 2/c). Setting 1 is used for the monoclinic biotite here and further. Biotite rec. PF, log scale, conventional PF analysis (ODF: texture index F 2=3. 30)

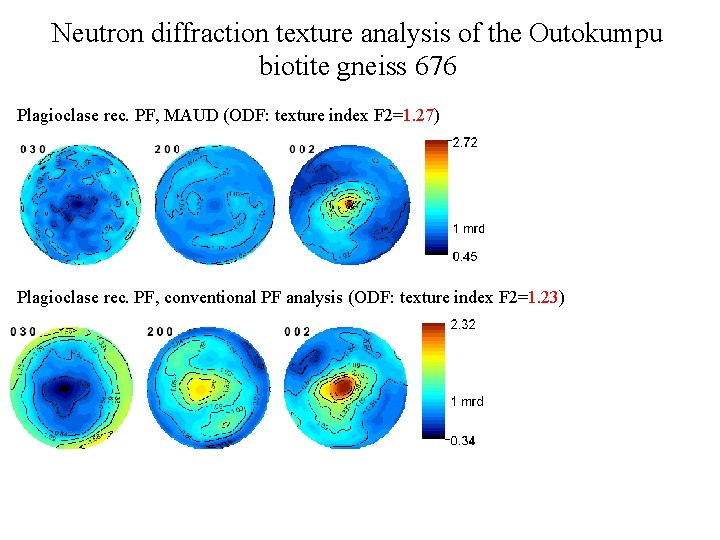
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Plagioclase rec. PF, MAUD (ODF: texture index F 2=1. 27) Plagioclase rec. PF, conventional PF analysis (ODF: texture index F 2=1. 23)

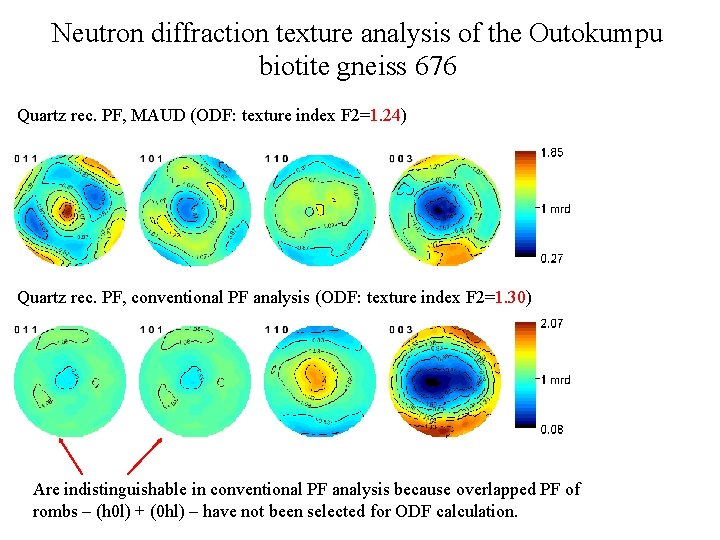
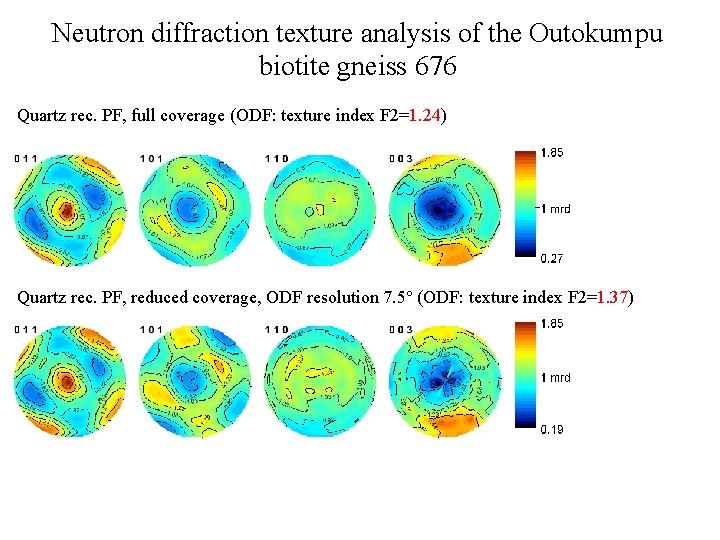
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Quartz rec. PF, MAUD (ODF: texture index F 2=1. 24) Quartz rec. PF, conventional PF analysis (ODF: texture index F 2=1. 30) Are indistinguishable in conventional PF analysis because overlapped PF of rombs – (h 0 l) + (0 hl) – have not been selected for ODF calculation.

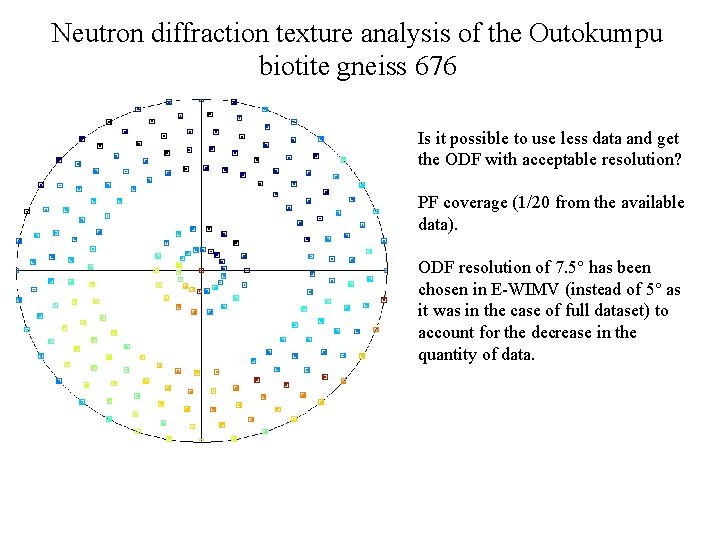
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Is it possible to use less data and get the ODF with acceptable resolution? PF coverage (1/20 from the available data). ODF resolution of 7. 5° has been chosen in E-WIMV (instead of 5° as it was in the case of full dataset) to account for the decrease in the quantity of data.

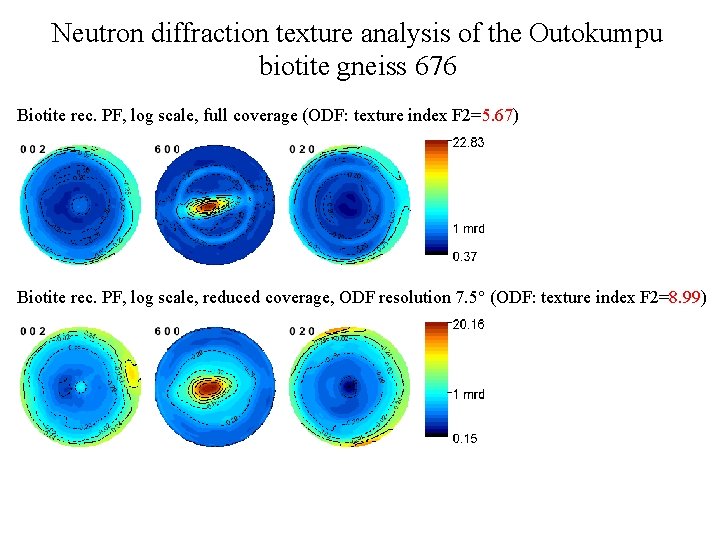
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Biotite rec. PF, log scale, full coverage (ODF: texture index F 2=5. 67) Biotite rec. PF, log scale, reduced coverage, ODF resolution 7. 5° (ODF: texture index F 2=8. 99)

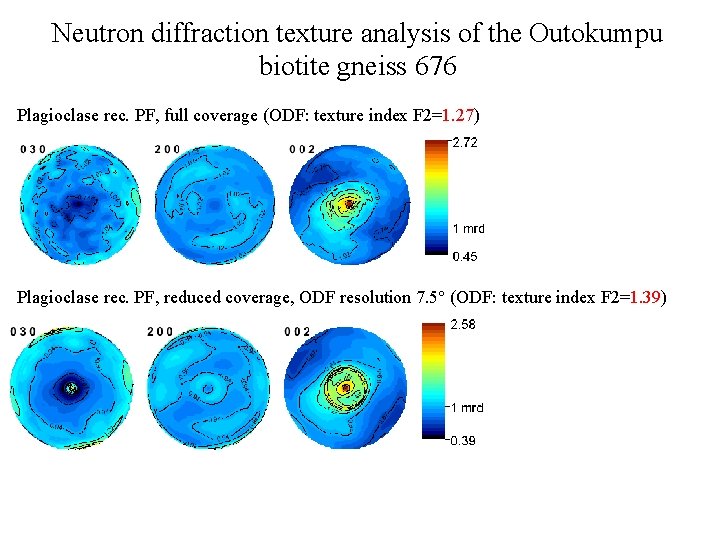
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Plagioclase rec. PF, full coverage (ODF: texture index F 2=1. 27) Plagioclase rec. PF, reduced coverage, ODF resolution 7. 5° (ODF: texture index F 2=1. 39)

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 676 Quartz rec. PF, full coverage (ODF: texture index F 2=1. 24) Quartz rec. PF, reduced coverage, ODF resolution 7. 5° (ODF: texture index F 2=1. 37)

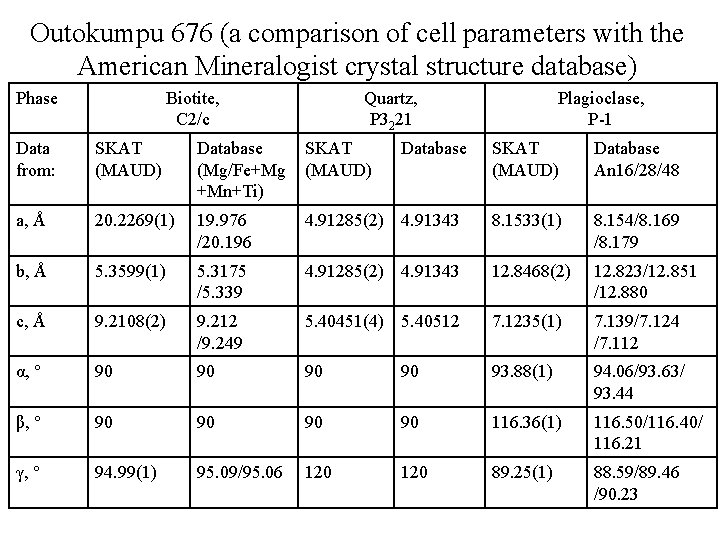
Outokumpu 676 (a comparison of cell parameters with the American Mineralogist crystal structure database) Phase Biotite, C 2/c Quartz, P 3221 Data from: SKAT (MAUD) Database (Mg/Fe+Mg +Mn+Ti) SKAT (MAUD) a, Å 20. 2269(1) 19. 976 /20. 196 b, Å 5. 3599(1) c, Å Database Plagioclase, P-1 SKAT (MAUD) Database An 16/28/48 4. 91285(2) 4. 91343 8. 1533(1) 8. 154/8. 169 /8. 179 5. 3175 /5. 339 4. 91285(2) 4. 91343 12. 8468(2) 12. 823/12. 851 /12. 880 9. 2108(2) 9. 212 /9. 249 5. 40451(4) 5. 40512 7. 1235(1) 7. 139/7. 124 /7. 112 α, ° 90 90 93. 88(1) 94. 06/93. 63/ 93. 44 β, ° 90 90 116. 36(1) 116. 50/116. 40/ 116. 21 γ, ° 94. 99(1) 95. 09/95. 06 120 89. 25(1) 88. 59/89. 46 /90. 23

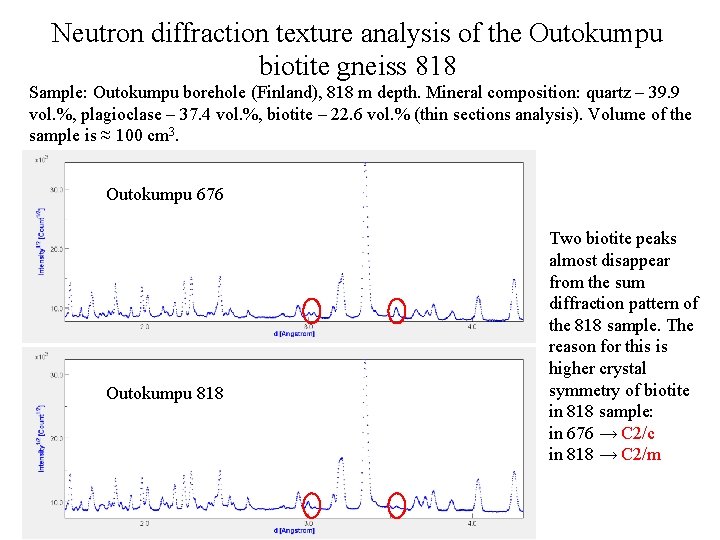
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 Sample: Outokumpu borehole (Finland), 818 m depth. Mineral composition: quartz – 39. 9 vol. %, plagioclase – 37. 4 vol. %, biotite – 22. 6 vol. % (thin sections analysis). Volume of the sample is ≈ 100 cm 3. Outokumpu 676 Outokumpu 818 Two biotite peaks almost disappear from the sum diffraction pattern of the 818 sample. The reason for this is higher crystal symmetry of biotite in 818 sample: in 676 → C 2/c in 818 → C 2/m

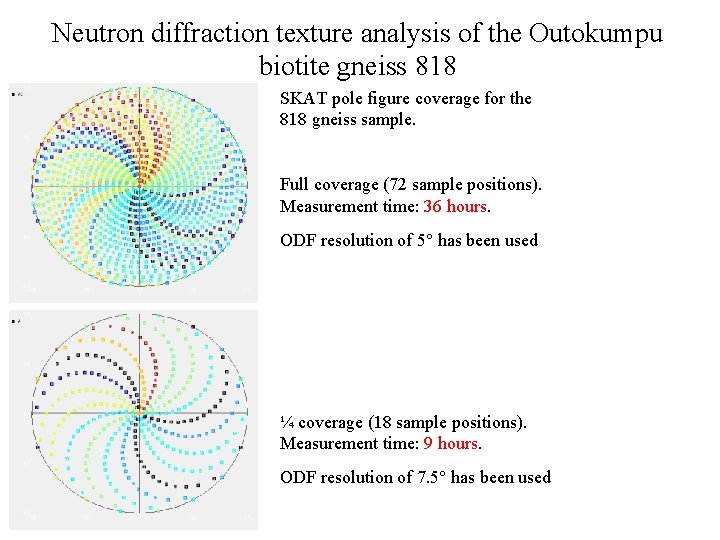
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 SKAT pole figure coverage for the 818 gneiss sample. Full coverage (72 sample positions). Measurement time: 36 hours. ODF resolution of 5° has been used ¼ coverage (18 sample positions). Measurement time: 9 hours. ODF resolution of 7. 5° has been used

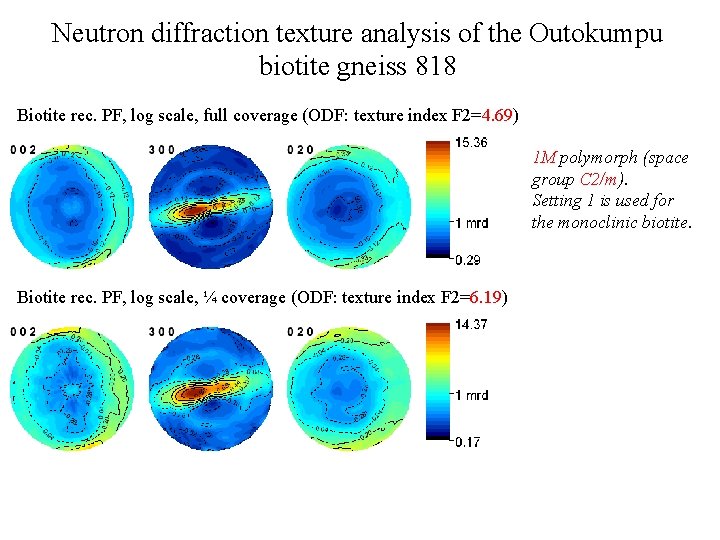
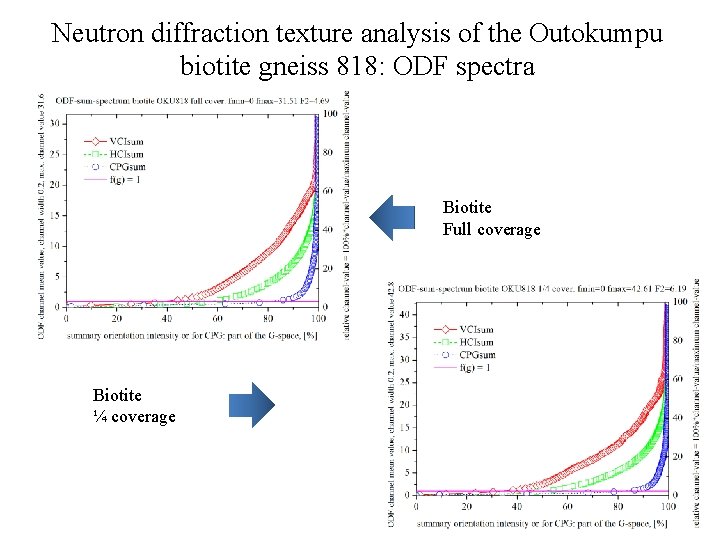
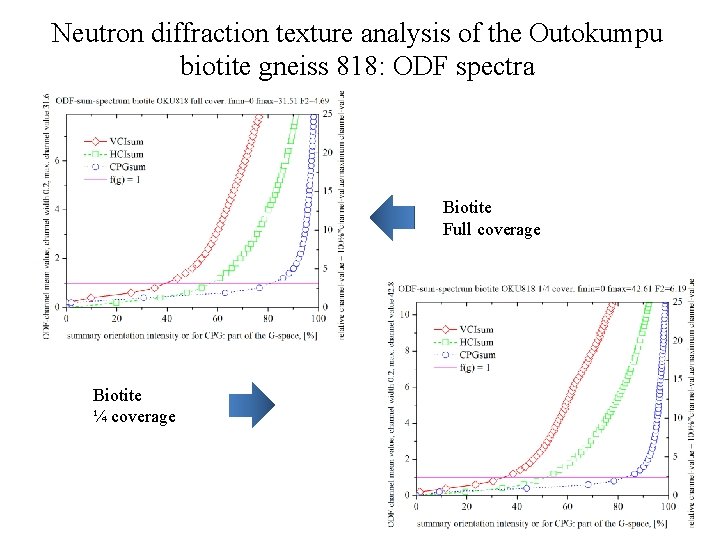
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 Biotite rec. PF, log scale, full coverage (ODF: texture index F 2=4. 69) 1 M polymorph (space group C 2/m). Setting 1 is used for the monoclinic biotite. Biotite rec. PF, log scale, ¼ coverage (ODF: texture index F 2=6. 19)

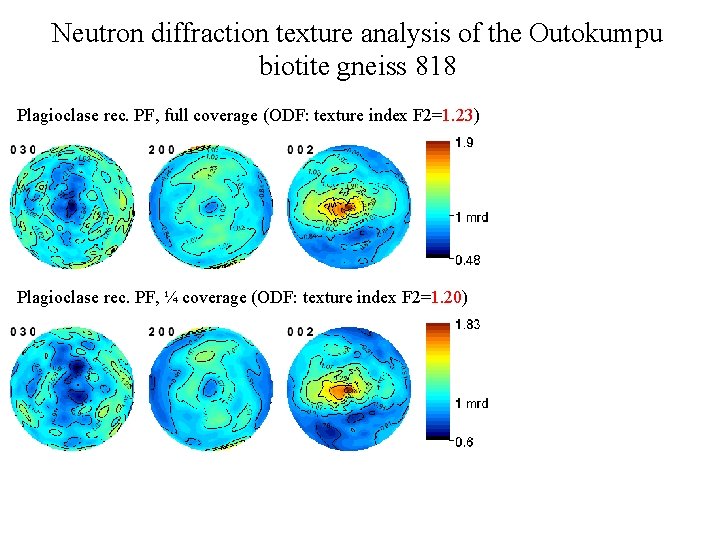
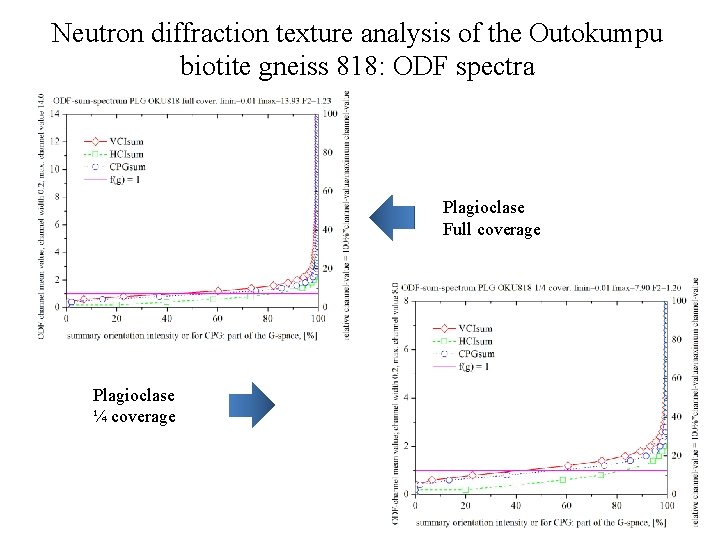
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 Plagioclase rec. PF, full coverage (ODF: texture index F 2=1. 23) Plagioclase rec. PF, ¼ coverage (ODF: texture index F 2=1. 20)

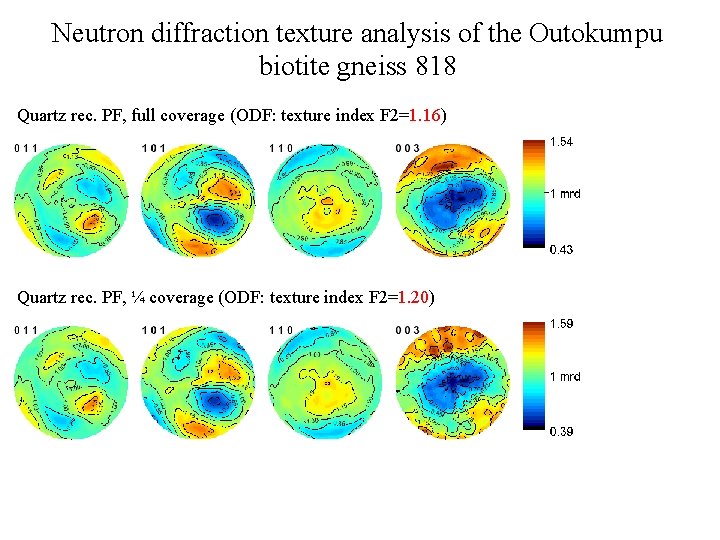
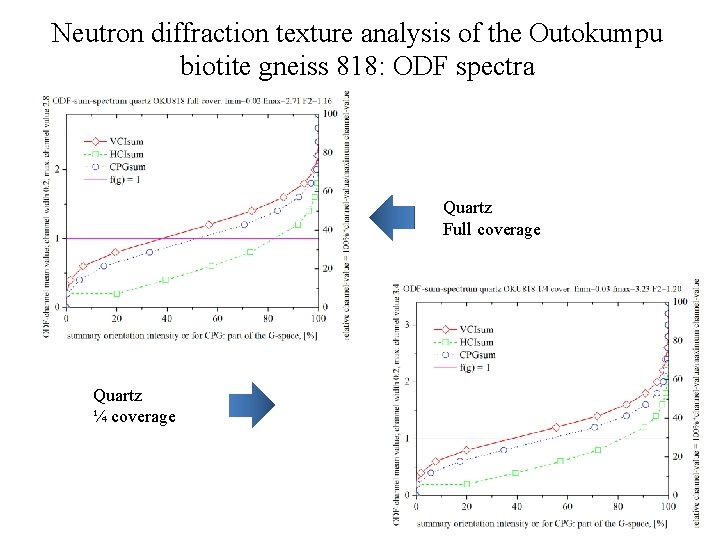
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 Quartz rec. PF, full coverage (ODF: texture index F 2=1. 16) Quartz rec. PF, ¼ coverage (ODF: texture index F 2=1. 20)

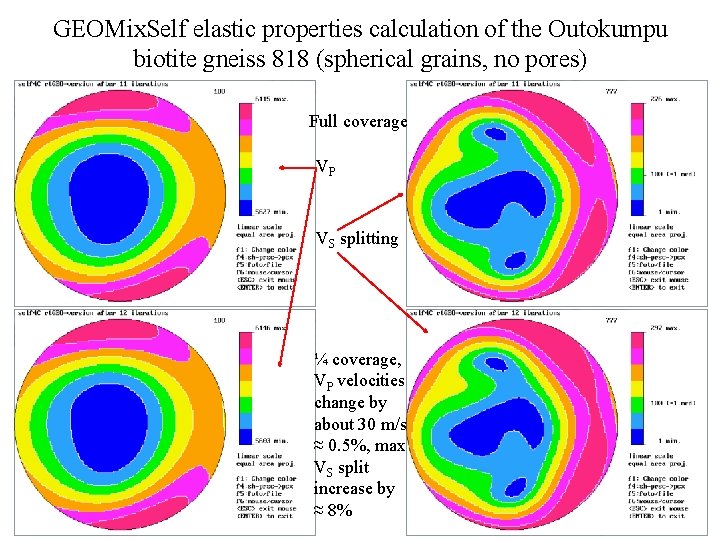
GEOMix. Self elastic properties calculation of the Outokumpu biotite gneiss 818 (spherical grains, no pores) Full coverage VP VS splitting ¼ coverage, VP velocities change by about 30 m/s ≈ 0. 5%, max VS split increase by ≈ 8%

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818: ODF spectra Biotite Full coverage Biotite ¼ coverage

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818: ODF spectra Biotite Full coverage Biotite ¼ coverage

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818: ODF spectra Plagioclase Full coverage Plagioclase ¼ coverage

Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818: ODF spectra Quartz Full coverage Quartz ¼ coverage

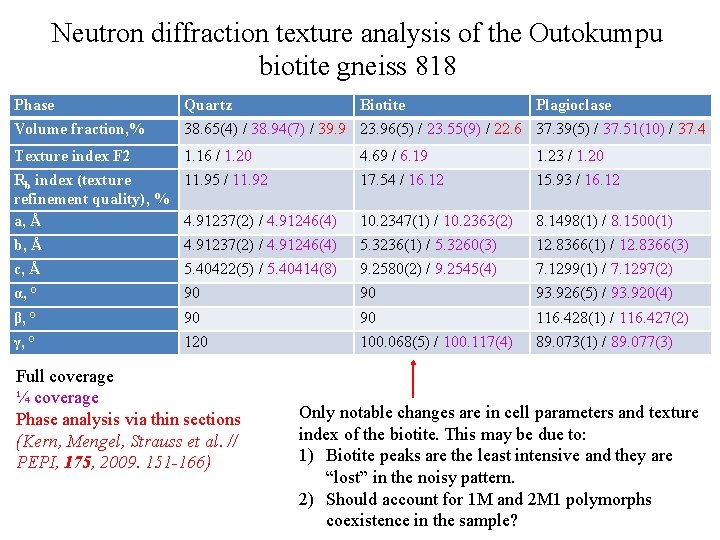
Neutron diffraction texture analysis of the Outokumpu biotite gneiss 818 Phase Quartz Biotite Plagioclase Volume fraction, % 38. 65(4) / 38. 94(7) / 39. 9 23. 96(5) / 23. 55(9) / 22. 6 37. 39(5) / 37. 51(10) / 37. 4 Texture index F 2 1. 16 / 1. 20 4. 69 / 6. 19 1. 23 / 1. 20 Rb index (texture 11. 95 / 11. 92 refinement quality), % a, Å 4. 91237(2) / 4. 91246(4) 17. 54 / 16. 12 15. 93 / 16. 12 10. 2347(1) / 10. 2363(2) 8. 1498(1) / 8. 1500(1) b, Å 4. 91237(2) / 4. 91246(4) 5. 3236(1) / 5. 3260(3) 12. 8366(1) / 12. 8366(3) c, Å 5. 40422(5) / 5. 40414(8) 9. 2580(2) / 9. 2545(4) 7. 1299(1) / 7. 1297(2) α, ° 90 90 93. 926(5) / 93. 920(4) β, ° 90 90 116. 428(1) / 116. 427(2) γ, ° 120 100. 068(5) / 100. 117(4) 89. 073(1) / 89. 077(3) Full coverage ¼ coverage Phase analysis via thin sections (Kern, Mengel, Strauss et al. // PEPI, 175, 2009. 151 -166) Only notable changes are in cell parameters and texture index of the biotite. This may be due to: 1) Biotite peaks are the least intensive and they are “lost” in the noisy pattern. 2) Should account for 1 M and 2 M 1 polymorphs coexistence in the sample?

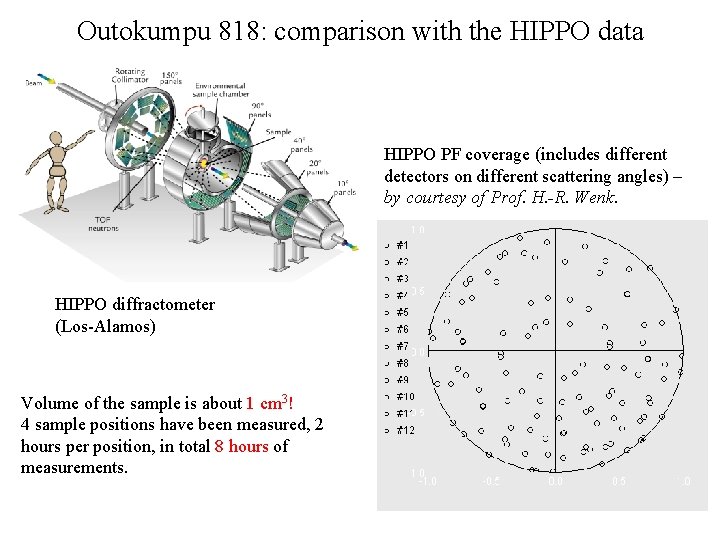
Outokumpu 818: comparison with the HIPPO data HIPPO PF coverage (includes different detectors on different scattering angles) – by courtesy of Prof. H. -R. Wenk. HIPPO diffractometer (Los-Alamos) Volume of the sample is about 1 cm 3! 4 sample positions have been measured, 2 hours per position, in total 8 hours of measurements.

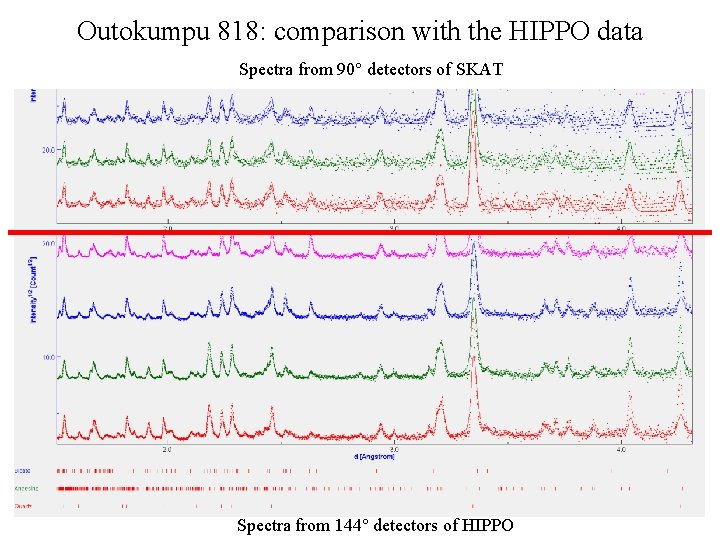
Outokumpu 818: comparison with the HIPPO data Spectra from 90° detectors of SKAT Spectra from 144° detectors of HIPPO

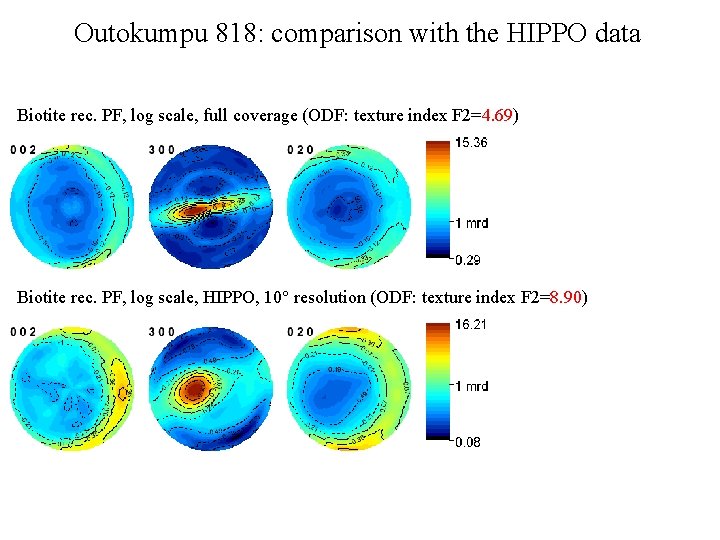
Outokumpu 818: comparison with the HIPPO data Biotite rec. PF, log scale, full coverage (ODF: texture index F 2=4. 69) Biotite rec. PF, log scale, HIPPO, 10° resolution (ODF: texture index F 2=8. 90)

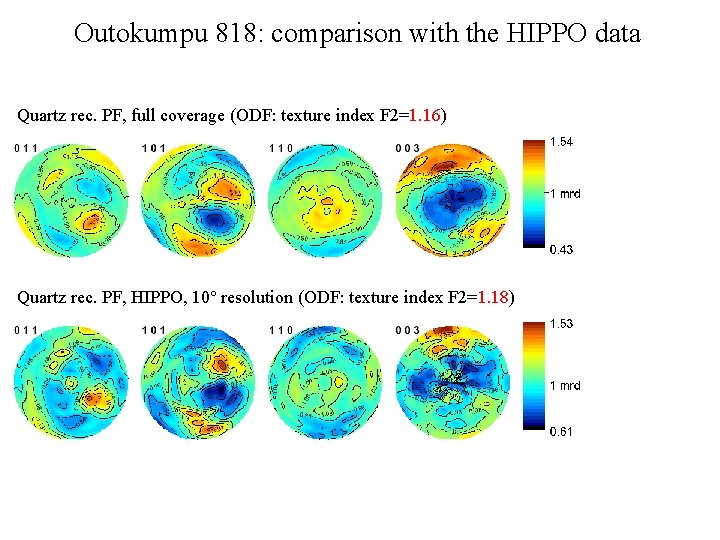
Outokumpu 818: comparison with the HIPPO data Quartz rec. PF, full coverage (ODF: texture index F 2=1. 16) Quartz rec. PF, HIPPO, 10° resolution (ODF: texture index F 2=1. 18)

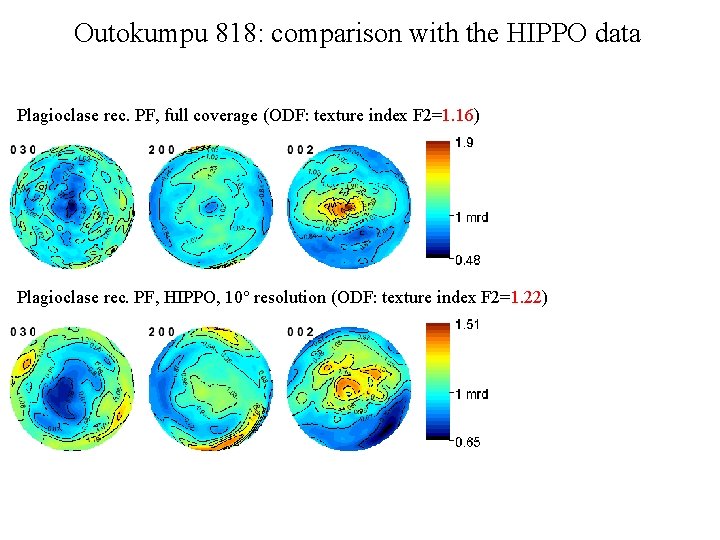
Outokumpu 818: comparison with the HIPPO data Plagioclase rec. PF, full coverage (ODF: texture index F 2=1. 16) Plagioclase rec. PF, HIPPO, 10° resolution (ODF: texture index F 2=1. 22)

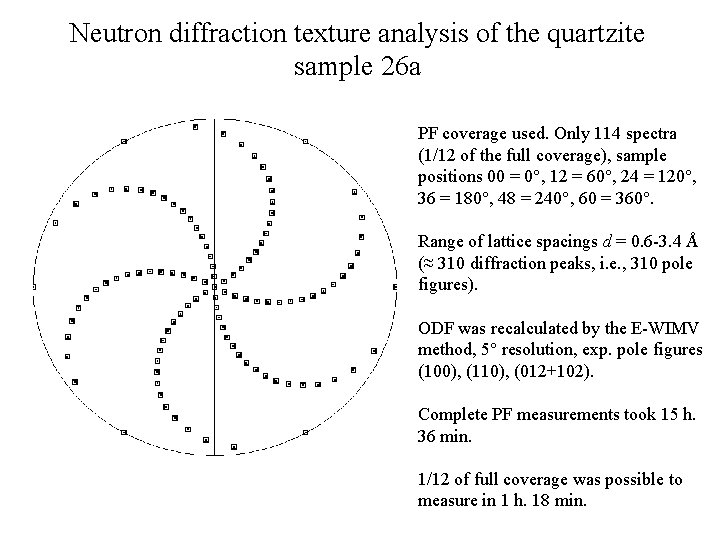
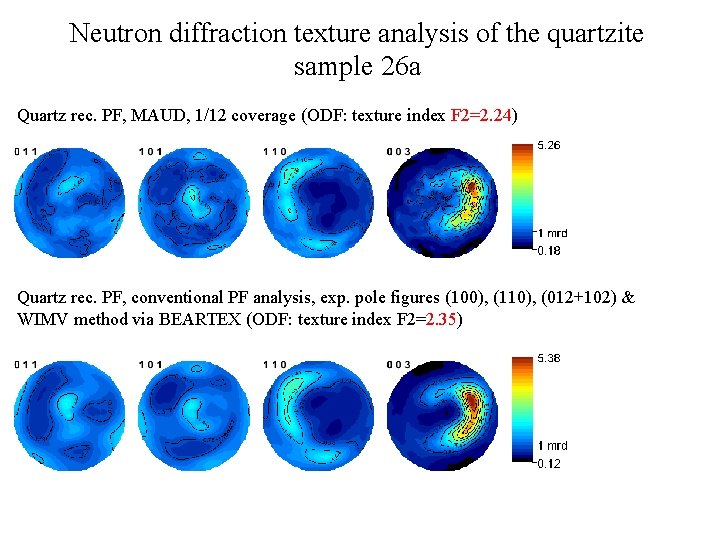
Neutron diffraction texture analysis of the quartzite sample 26 а PF coverage used. Only 114 spectra (1/12 of the full coverage), sample positions 00 = 0°, 12 = 60°, 24 = 120°, 36 = 180°, 48 = 240°, 60 = 360°. Range of lattice spacings d = 0. 6 -3. 4 Å (≈ 310 diffraction peaks, i. e. , 310 pole figures). ODF was recalculated by the E-WIMV method, 5° resolution, exp. pole figures (100), (110), (012+102). Complete PF measurements took 15 h. 36 min. 1/12 of full coverage was possible to measure in 1 h. 18 min.

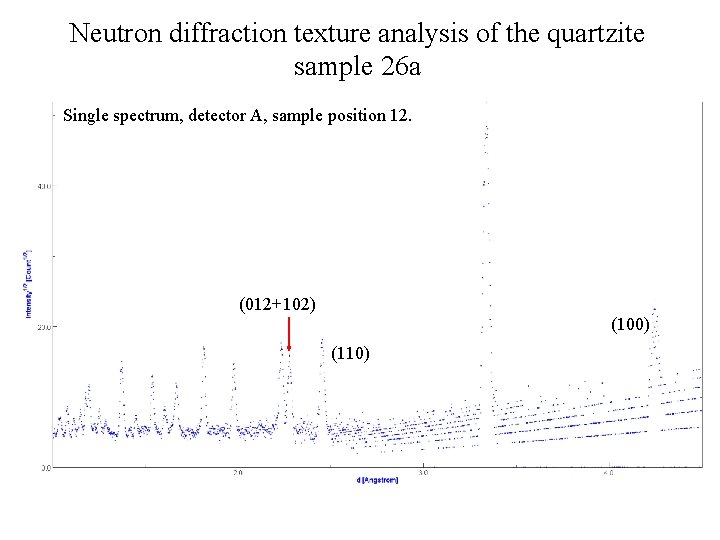
Neutron diffraction texture analysis of the quartzite sample 26 а Single spectrum, detector A, sample position 12. (012+102) (100) (110)

Neutron diffraction texture analysis of the quartzite sample 26 а Quartz rec. PF, MAUD, 1/12 coverage (ODF: texture index F 2=2. 24) Quartz rec. PF, conventional PF analysis, exp. pole figures (100), (110), (012+102) & WIMV method via BEARTEX (ODF: texture index F 2=2. 35)

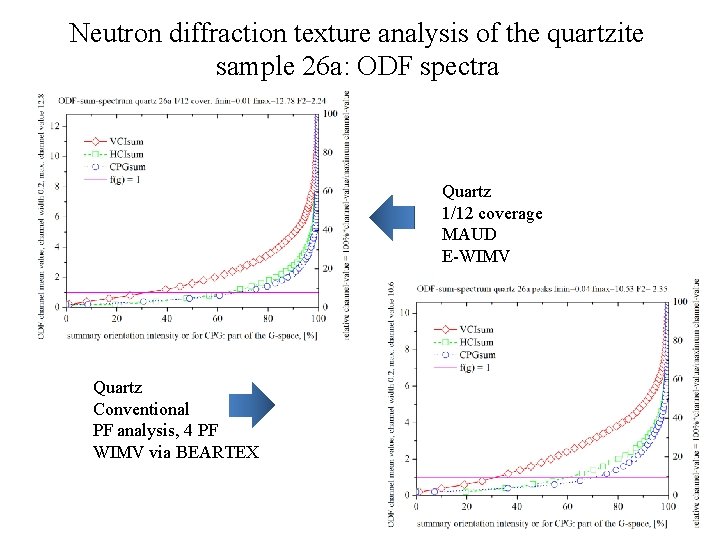
Neutron diffraction texture analysis of the quartzite sample 26 а: ODF spectra Quartz 1/12 coverage MAUD E-WIMV Quartz Conventional PF analysis, 4 PF WIMV via BEARTEX

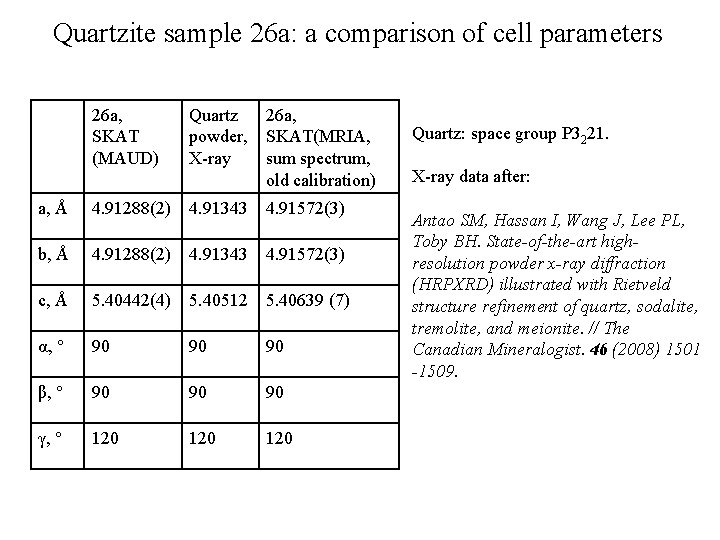
Quartzite sample 26 а: a comparison of cell parameters 26 a, SKAT (MAUD) Quartz powder, X-ray 26 a, SKAT(MRIA, sum spectrum, old calibration) a, Å 4. 91288(2) 4. 91343 4. 91572(3) b, Å 4. 91288(2) 4. 91343 4. 91572(3) c, Å 5. 40442(4) 5. 40512 5. 40639 (7) α, ° 90 90 90 β, ° 90 90 90 γ, ° 120 120 Quartz: space group P 3221. X-ray data after: Antao SM, Hassan I, Wang J, Lee PL, Toby BH. State-of-the-art highresolution powder x-ray diffraction (HRPXRD) illustrated with Rietveld structure refinement of quartz, sodalite, tremolite, and meionite. // The Canadian Mineralogist. 46 (2008) 1501 -1509.

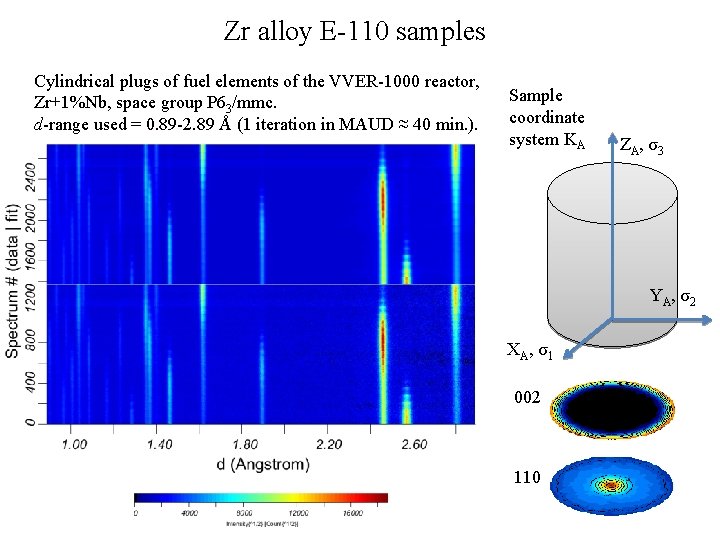
Zr alloy E-110 samples Cylindrical plugs of fuel elements of the VVER-1000 reactor, Zr+1%Nb, space group P 63/mmc. d-range used = 0. 89 -2. 89 Å (1 iteration in MAUD ≈ 40 min. ). Sample coordinate system KA Z A , σ3 Y A , σ2 X A , σ1 002 110

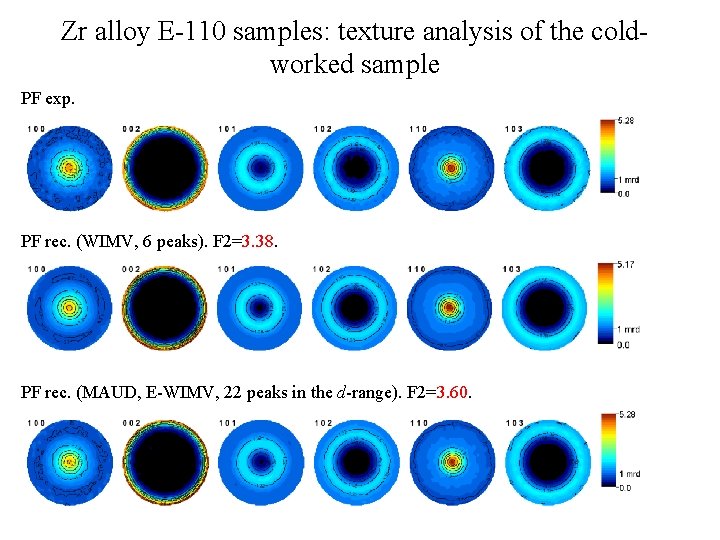
Zr alloy E-110 samples: texture analysis of the coldworked sample PF exp. PF rec. (WIMV, 6 peaks). F 2=3. 38. PF rec. (MAUD, E-WIMV, 22 peaks in the d-range). F 2=3. 60.

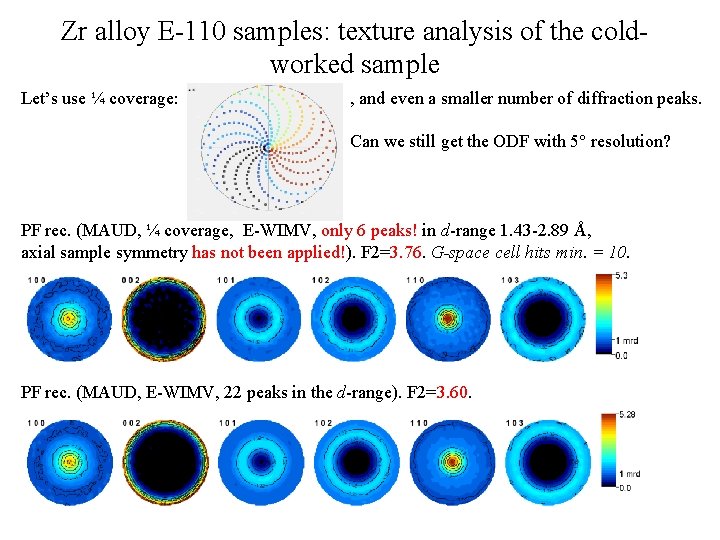
Zr alloy E-110 samples: texture analysis of the coldworked sample Let’s use ¼ coverage: , and even a smaller number of diffraction peaks. Can we still get the ODF with 5° resolution? PF rec. (MAUD, ¼ coverage, E-WIMV, only 6 peaks! in d-range 1. 43 -2. 89 Å, axial sample symmetry has not been applied!). F 2=3. 76. G-space cell hits min. = 10. PF rec. (MAUD, E-WIMV, 22 peaks in the d-range). F 2=3. 60.

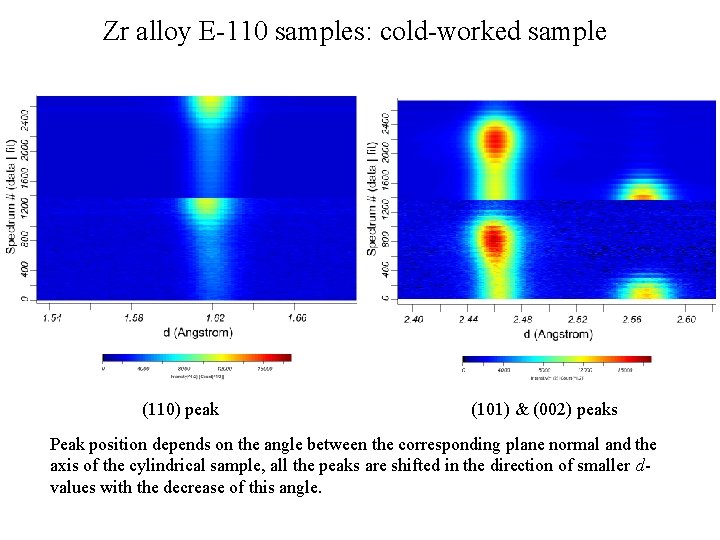
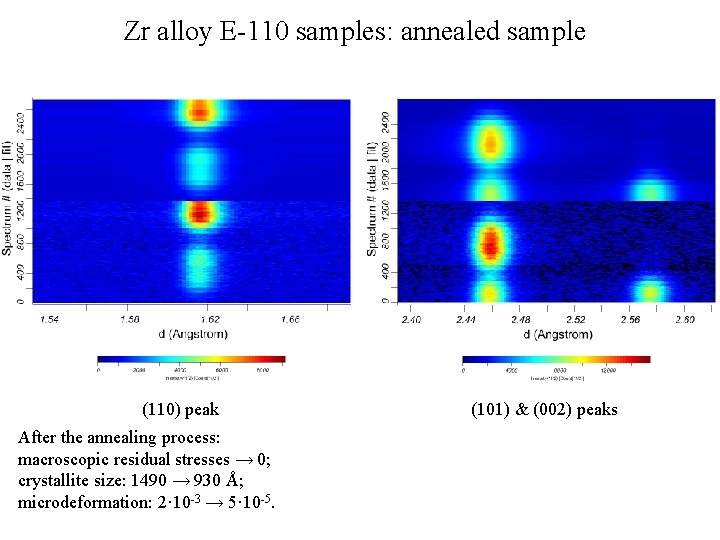
Zr alloy E-110 samples: cold-worked sample (110) peak (101) & (002) peaks Peak position depends on the angle between the corresponding plane normal and the axis of the cylindrical sample, all the peaks are shifted in the direction of smaller dvalues with the decrease of this angle.

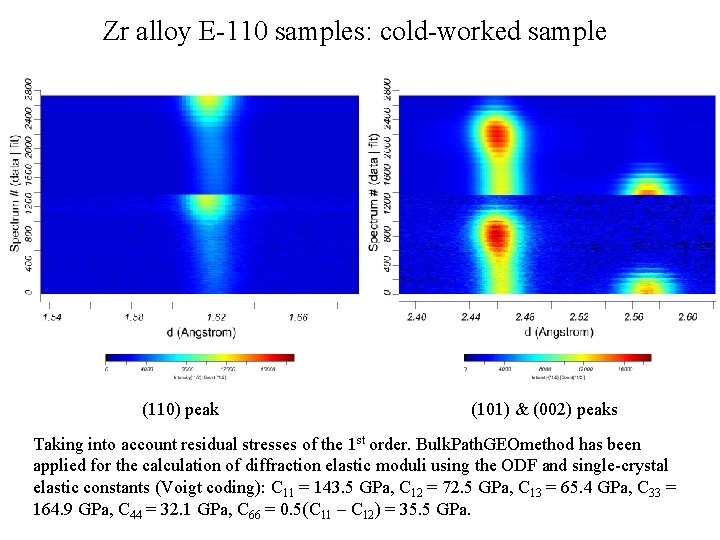
Zr alloy E-110 samples: cold-worked sample (110) peak (101) & (002) peaks Taking into account residual stresses of the 1 st order. Bulk. Path. GEOmethod has been applied for the calculation of diffraction elastic moduli using the ODF and single-crystal elastic constants (Voigt coding): C 11 = 143. 5 GPa, С 12 = 72. 5 GPa, С 13 = 65. 4 GPa, С 33 = 164. 9 GPa, С 44 = 32. 1 GPa, С 66 = 0. 5(C 11 – С 12) = 35. 5 GPa.

Zr alloy E-110 samples: annealed sample (110) peak After the annealing process: macroscopic residual stresses → 0; crystallite size: 1490 → 930 Å; microdeformation: 2· 10 -3 → 5· 10 -5. (101) & (002) peaks

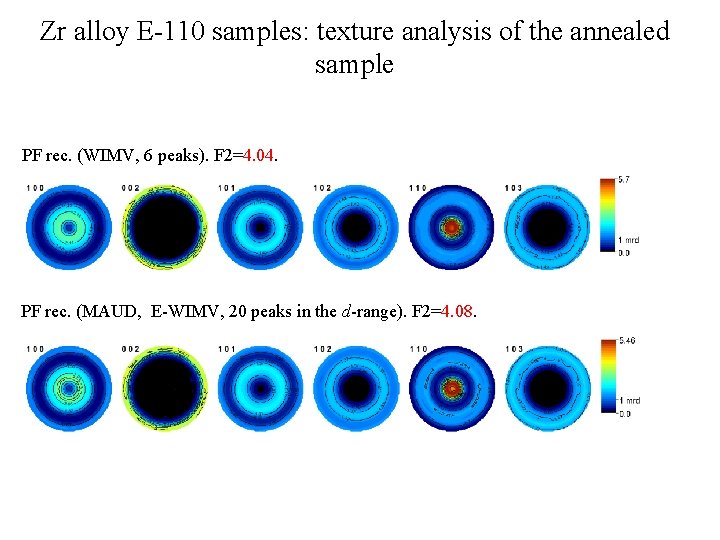
Zr alloy E-110 samples: texture analysis of the annealed sample PF rec. (WIMV, 6 peaks). F 2=4. 04. PF rec. (MAUD, E-WIMV, 20 peaks in the d-range). F 2=4. 08.

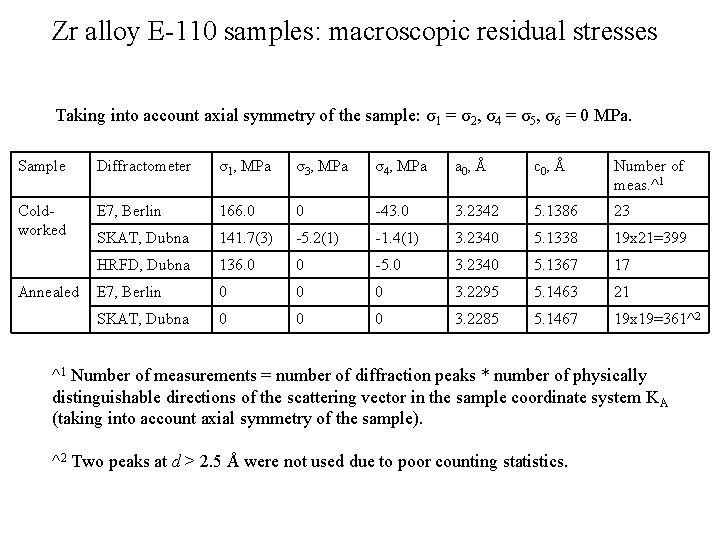
Zr alloy E-110 samples: macroscopic residual stresses Taking into account axial symmetry of the sample: σ1 = σ2, σ4 = σ5, σ6 = 0 MPa. Sample Diffractometer σ1, MPa σ3, MPa σ4, MPa a 0, Å c 0, Å Number of meas. ^1 Coldworked E 7, Berlin 166. 0 0 -43. 0 3. 2342 5. 1386 23 SKAT, Dubna 141. 7(3) -5. 2(1) -1. 4(1) 3. 2340 5. 1338 19 x 21=399 HRFD, Dubna 136. 0 0 -5. 0 3. 2340 5. 1367 17 E 7, Berlin 0 0 0 3. 2295 5. 1463 21 SKAT, Dubna 0 0 0 3. 2285 5. 1467 19 x 19=361^2 Annealed ^1 Number of measurements = number of diffraction peaks * number of physically distinguishable directions of the scattering vector in the sample coordinate system K A (taking into account axial symmetry of the sample). ^2 Two peaks at d > 2. 5 Å were not used due to poor counting statistics.

Current results • The analysis of the SKAT diffractometer data using the Rietveld method (MAUD software) is possible, and now it is easily available. All 1368 spectra could be analyzed in wide d-range even in case of very complex samples (e. g. , > 1000 spectra and > 7∙ 105 diffraction peaks in total…). A number of very different samples measured during different reactor cycles from February 1998 till December 2006 have been processed with MAUD with no problems. • Only 2 programs are needed to get the ODF out of diffraction patterns (SKAT 2 MAUD → MAUD) instead of 4 (e. g. , Geo. TOF → Auto. Index → Berkeley → Beartex). • The d-range and the resolution of SKAT are enough to get necessary information for the ODF reconstruction in case of rocks containing several low-symmetry and/or large-cell minerals, e. g. monoclinic 2 M 1 biotite polymorph (20. 2 x 5. 4 x 9. 2 Å) + triclinic feldspar. But counting statistics should be increased (especially at d > 3. 5 Å) due to modernized reactor, new neutron guide, new (cold) moderator or … • by using less sample positions (e. g. , 1/6 of full coverage), but increasing the measurement time per position (e. g. , 6 times). In some cases utilization of partial experimental PF coverage may drastically (~ 10 times) reduce total measurement time while the ODF reconstruction with 5° resolution will still be possible. • From the SKAT data now its possible to easily get not only ODF but also cell parameters, phase volume fractions, 1 st order residual stresses, etc.

Some things to do and suggestions • Still there is a lot of space for the method testing/development… • Add a few options to SKAT 2 MAUD. • In future, it will be great to measure some standard sample with diffraction peaks in wide d-range – e. g. , Al 2 O 3 – during each reactor cycle (and also the vanadium standard). • Make a few special test experiments to verify MAUD-processed SKAT data on texture, cell parameters, stresses, crystallite sizes, etc. • Some more experiments, tests and discussions concerning the possibility to measure incomplete PF & reduce measurement time per sample.

Acknowledgements I would like to thank Creators of SKAT and MAUD for the great instrument and the great software Dr. Tatiana Ivankina (JINR) for the data on ODF of biotite, plagioclase and quartz for the 676 Outokumpu sample (results of the conventional PF analysis of the SKAT data) Yuri Kovalev (JINR) for the reminder about the Get. Last. Error() function Prof. Siegfried Matthies (JINR) for the software he made to convert MAUD format ODF into conventional STD format Dr. Vyacheslav Sumin (JINR) for HRFD and E 7 data on E-110 zirconium alloy samples and discussion of the results on these samples Prof. Hans-Rudolf Wenk (Berkeley university) for the data on the 818 Outokumpu sample measured on HIPPO diffractometer, and his help with the analysis of incomplete PF of 676 Outokumpu sample measured on SKAT
Thank you!