Protein Structure Prediction What is PSP Primary sequence





















- Slides: 33

Protein Structure Prediction

What is PSP ?

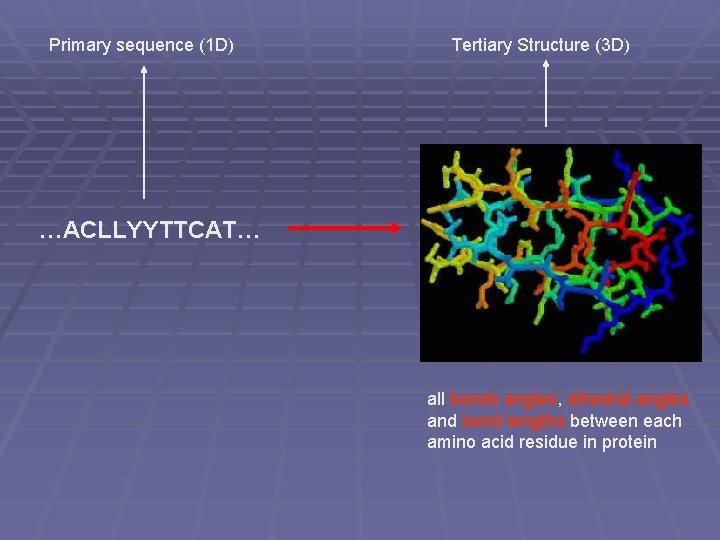
Primary sequence (1 D) Tertiary Structure (3 D) …ACLLYYTTCAT… all bonds angles, dihedral angles and bond lengths between each amino acid residue in protein

“Solving” PSP

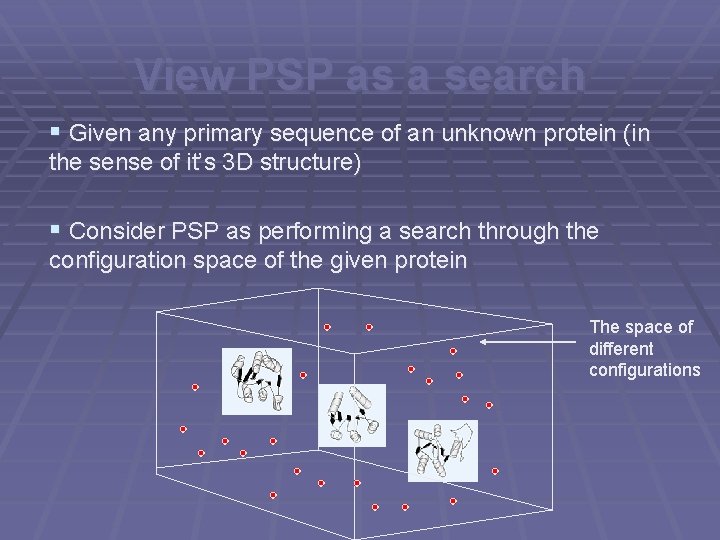
View PSP as a search § Given any primary sequence of an unknown protein (in the sense of it’s 3 D structure) § Consider PSP as performing a search through the configuration space of the given protein The space of different configurations

Steps in solving PSP § Given primary sequence predict the final 3 D structure (1 D 3 D). § 2 Step process (1 D 2 D, 2 D 3 D) § 1 st find configuration for the secondary structure (SS Prediction) § 2 nd find configuration for the side-chains (side-chain conformation)

Required “components” in solving PSP v All methods require the definition of a protein model • A simplified protein structure model • A potential energy function

Simplified structure model of Protein § By the above we mean that the protein in question has simpler physical properties then an actual protein § This is needed as trying to solve PSP is too complex for real proteins § Good simplified models give a good approximation for the actual shape of the protein § Determining a good model is a research area by itself

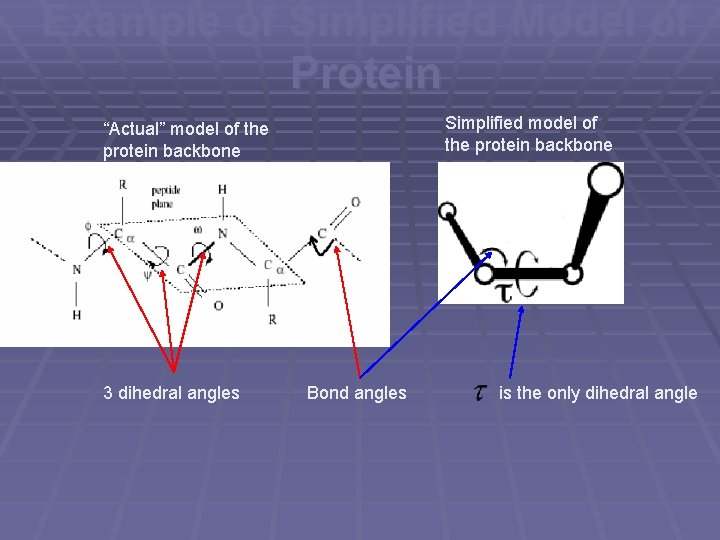
Example of Simplified Model of Protein Simplified model of the protein backbone “Actual” model of the protein backbone 3 dihedral angles Bond angles is the only dihedral angle

Potential Energy Function v How do we know when a predicted structure is the native shape of the protein ? In thermodynamics, A molecule is most stable when it’s free energy is at a minimum native shape is at a free energy minimum • The potential energy function is a simplification of actual forces acting on a real protein molecule and it’s formulation is based on the given simplified structural model

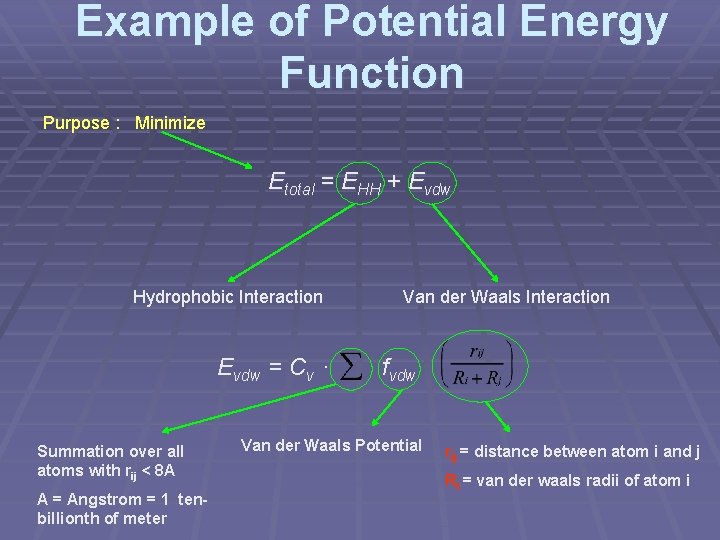
Example of Potential Energy Function Purpose : Minimize Etotal = EHH + Evdw Hydrophobic Interaction Evdw = Cv · Summation over all atoms with rij < 8 A A = Angstrom = 1 tenbillionth of meter Van der Waals Interaction fvdw Van der Waals Potential rij = distance between atom i and j Ri = van der waals radii of atom i

Different approaches to PSP § Ab Initio Methods § Knowledge Based Methods

Ab Initio Methods

What is Ab Initio ? • Ab Initio means from 1 st principles • Use thermodynamic laws to figure out the configuration of the fold of the given protein folding problem • Global/semi-global minimization of the function • 1 D 2 D = secondary structure problem • 2 D 3 D = side-chain conformation

Some Ab Initio Methods § Molecular Dynamic Simulation § Using complex energy functions simulate folding of the primary sequence until it reaches it’s native state (1 D->3 D) § Genetic Algorithm § Used in refining a given potential function so that it can best predict the native state of a protein § Simulated Annealing § Branch and Bound Methods (usually used in side-chain conformation) § Approximation algorithms §Comparative/Homologue Modeling § Threading § Docking

Knowledge Based Methods

Knowledge Based Methods § Using knowledge of currently known protein folds, predict the shape of the target protein § Assumption is the native fold of the target protein is similar to a currently known one i. e in the same family § Unable to predict any novel folds, i. e new fold family

Some Knowledge-Based Methods § Comparative/Homologue Modeling § Threading § Docking

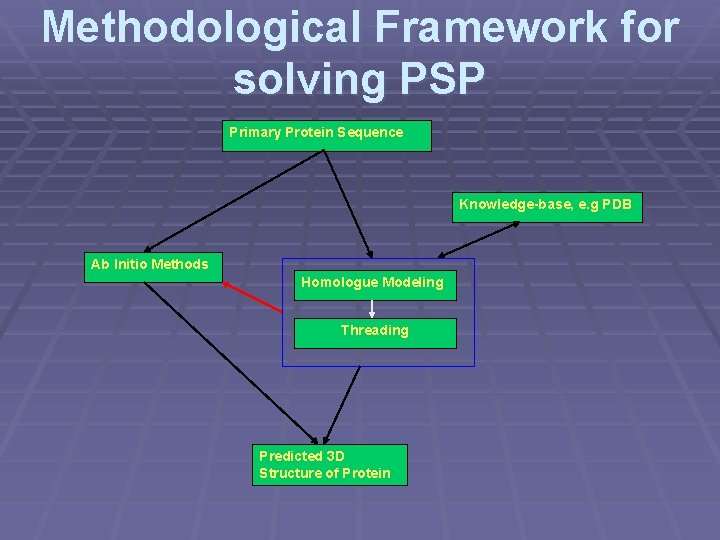
Methodological Framework for solving PSP Primary Protein Sequence Knowledge-base, e. g PDB Ab Initio Methods Homologue Modeling Threading Predicted 3 D Structure of Protein

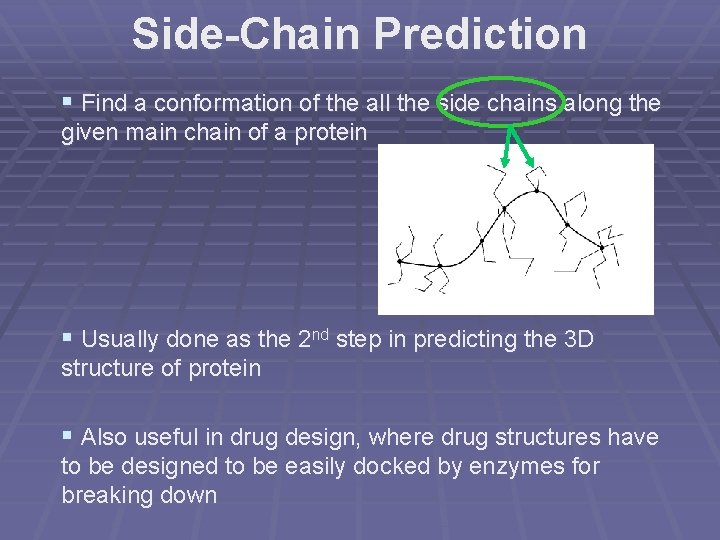
Side-Chain Prediction § Find a conformation of the all the side chains along the given main chain of a protein § Usually done as the 2 nd step in predicting the 3 D structure of protein § Also useful in drug design, where drug structures have to be designed to be easily docked by enzymes for breaking down

Side-Chain Prediction § The main chain fold has been computed and given as input § choose positions of all side chains so as to minimize some potential energy function § Problem if solved Ab Initio is proven to be NP-Complete (reduce Clique to it)

Central Dogma § The more tightly packed Side-chains are, the more stable they will be. § Ponder & Richards have shown that there a fixed set of rotations (rotamers) side-chains can take. § Most methods now make use of this library of rotamers (abt 67 different rotations) § Main concern is the search strategy to find the best conformation

Methods in Side-chain prediction § Simulated Annealing § A* algorithm § Monte Carlo Minimization § Molecular Dynamics Simulation § Dead End Elimination § Genetic Algorithm

Dead End Elimination § Deterministic method to determine the global minimum energy conformation (GMEC) of set of side-chains. § Continuously eliminate rotamers from consideration in the GMEC, until only 1 rotamer is left in each side-chain position (thus giving final conformation). § DEE can be viewed as a mathematical criteria that a rotamer must fulfill in order not to be eliminated

Dead End Elimination § Potential function is described in terms of pair-wise interactions of all rotamers at all positions. § Therefore energy function to minimized can be formulated as Sum of pairwise interaction energy between rotamer r at position j and rotamer u at positions j energy of the given backbone fold Sum of energy of rotamer r at side chain i

Dead End Elimination § Assuming p side-chains and n rotamers for each sidechain § Time complexity of finding the configuration that minimizes the energy function takes O(p*np) § Not feasible to use the original formulation

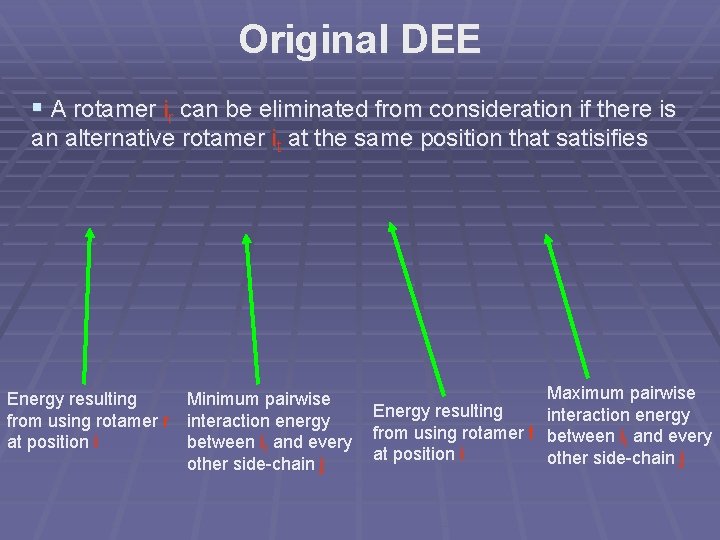
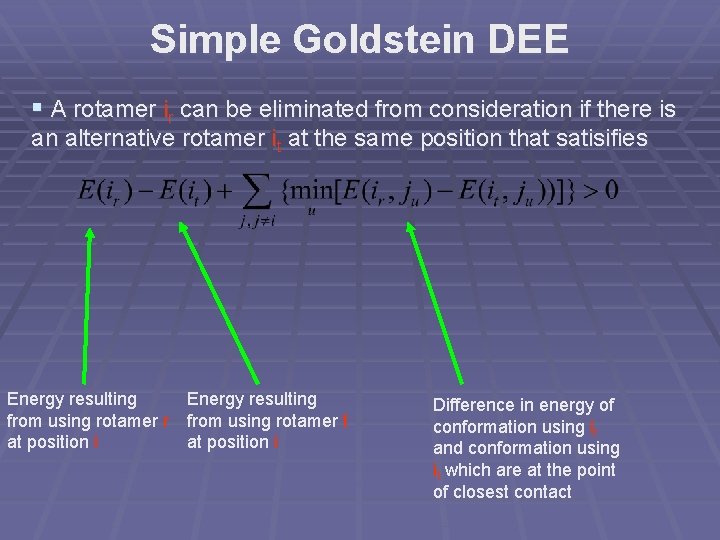
Original DEE § A rotamer ir can be eliminated from consideration if there is an alternative rotamer it at the same position that satisifies Energy resulting Minimum pairwise from using rotamer r interaction energy at position i between ir and every other side-chain j Maximum pairwise Energy resulting interaction energy from using rotamer t between it and every at position i other side-chain j

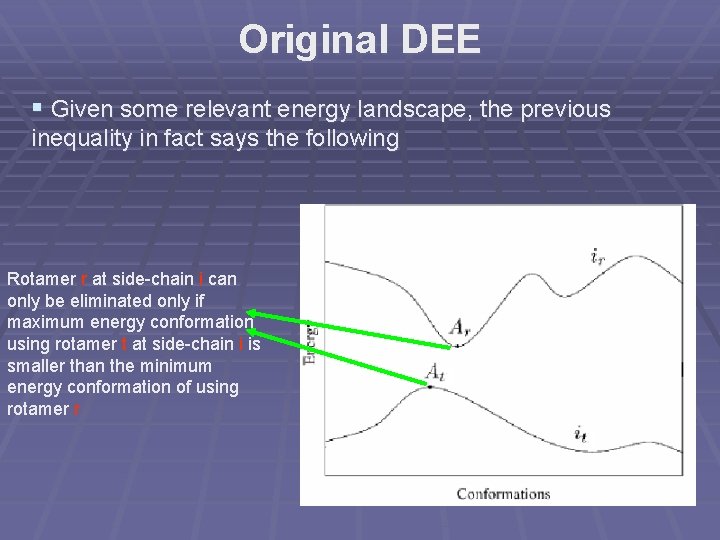
Original DEE § Given some relevant energy landscape, the previous inequality in fact says the following Rotamer r at side-chain i can only be eliminated only if maximum energy conformation using rotamer t at side-chain i is smaller than the minimum energy conformation of using rotamer r

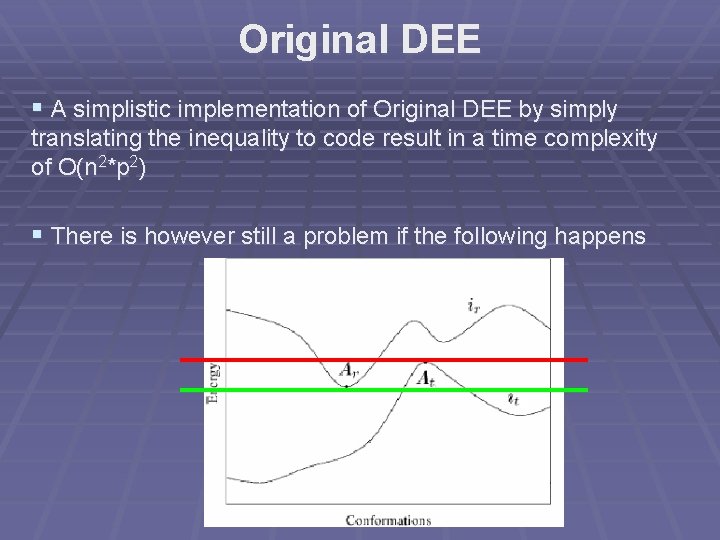
Original DEE § A simplistic implementation of Original DEE by simply translating the inequality to code result in a time complexity of O(n 2*p 2) § There is however still a problem if the following happens

Simple Goldstein DEE § A rotamer ir can be eliminated from consideration if there is an alternative rotamer it at the same position that satisifies Energy resulting from using rotamer r from using rotamer t at position i Difference in energy of conformation using ir and conformation using it which are at the point of closest contact

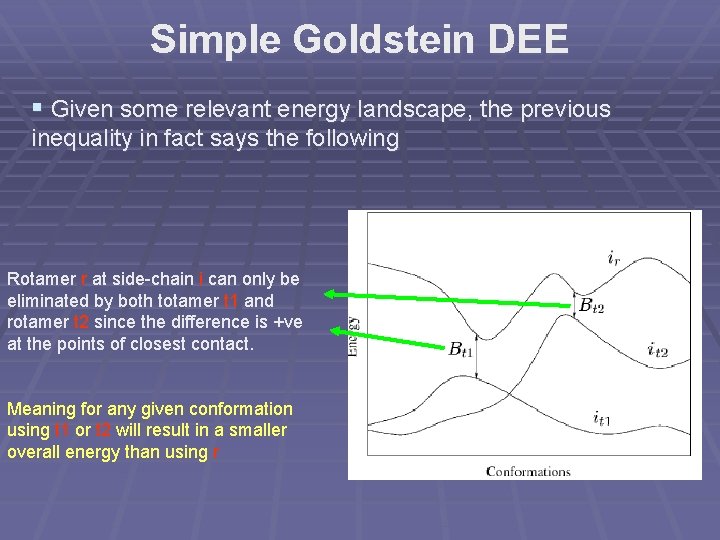
Simple Goldstein DEE § Given some relevant energy landscape, the previous inequality in fact says the following Rotamer r at side-chain i can only be eliminated by both totamer t 1 and rotamer t 2 since the difference is +ve at the points of closest contact. Meaning for any given conformation using t 1 or t 2 will result in a smaller overall energy than using r

Simple Goldstein DEE § A simplistic implementation of Simple Goldstein DEE by simply translating the inequality to code result in a time complexity of O(n 3*p 2) § There is however still a problem if energy profiles of rotamer r and every other rotamer intersect. § More powerful criteria will have to be used § General Gold. Stein DEE, Simple Split DEE and general Split DEE § The more powerful the criterion the higher it’s time complexity

Conclusion § Myriad of methods to attempt to solve the protein prediction problem §Knowledge-based methods have gained a edge over Ab initio methods § However not much improvement in the prediction power of modern heuristics, since the 1 st experiment by Anfisen 3 decades ago § Either problem is too hard / More discovery awaits the adventurous researcher